The Introduction of Two New Species of Aquatic Fungi from Anzali Lagoon, Northern Iran
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Cultivation
2.2. DNA Extraction, PCR and Sequencing
2.3. Phylogenetic Analyses
3. Results
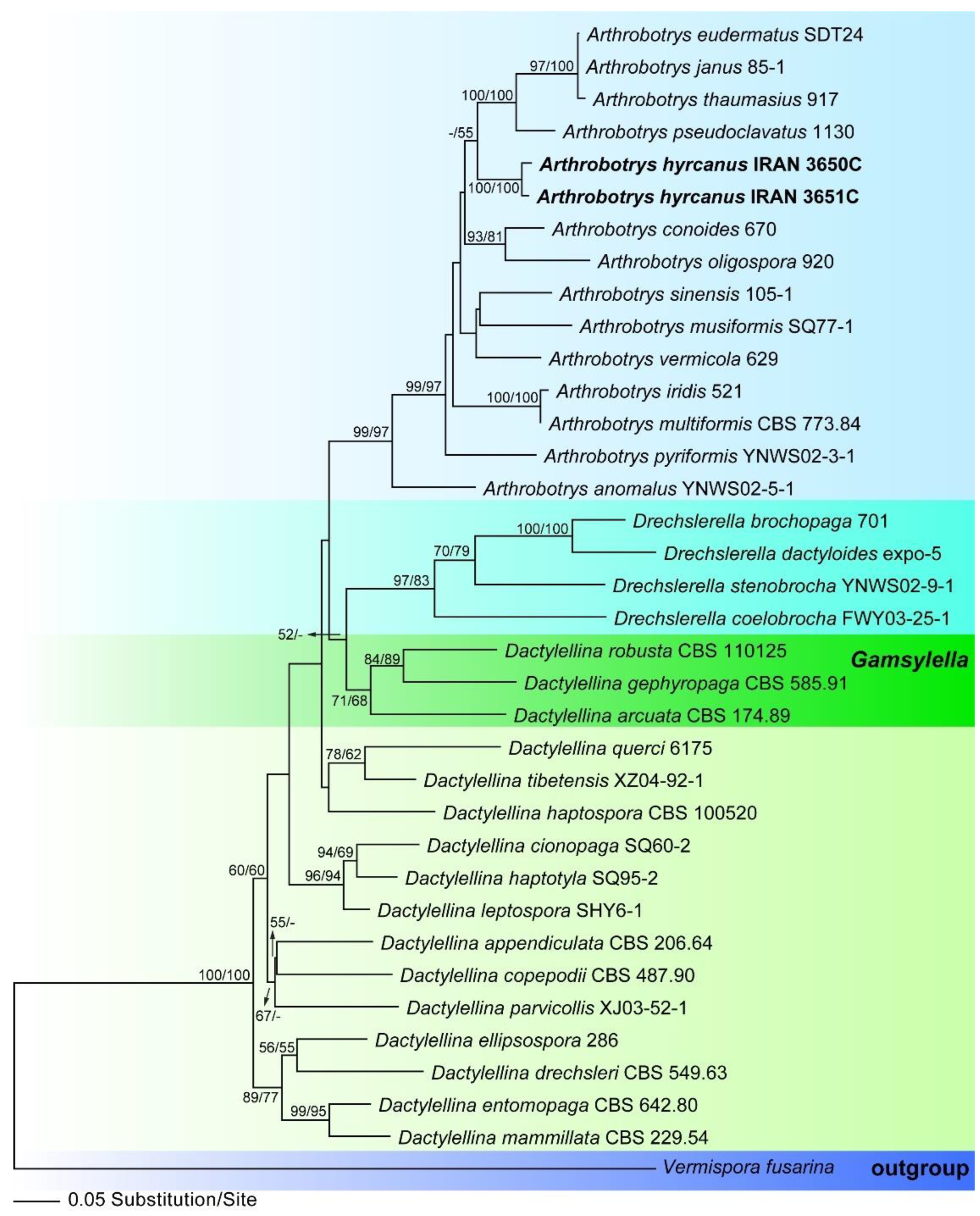
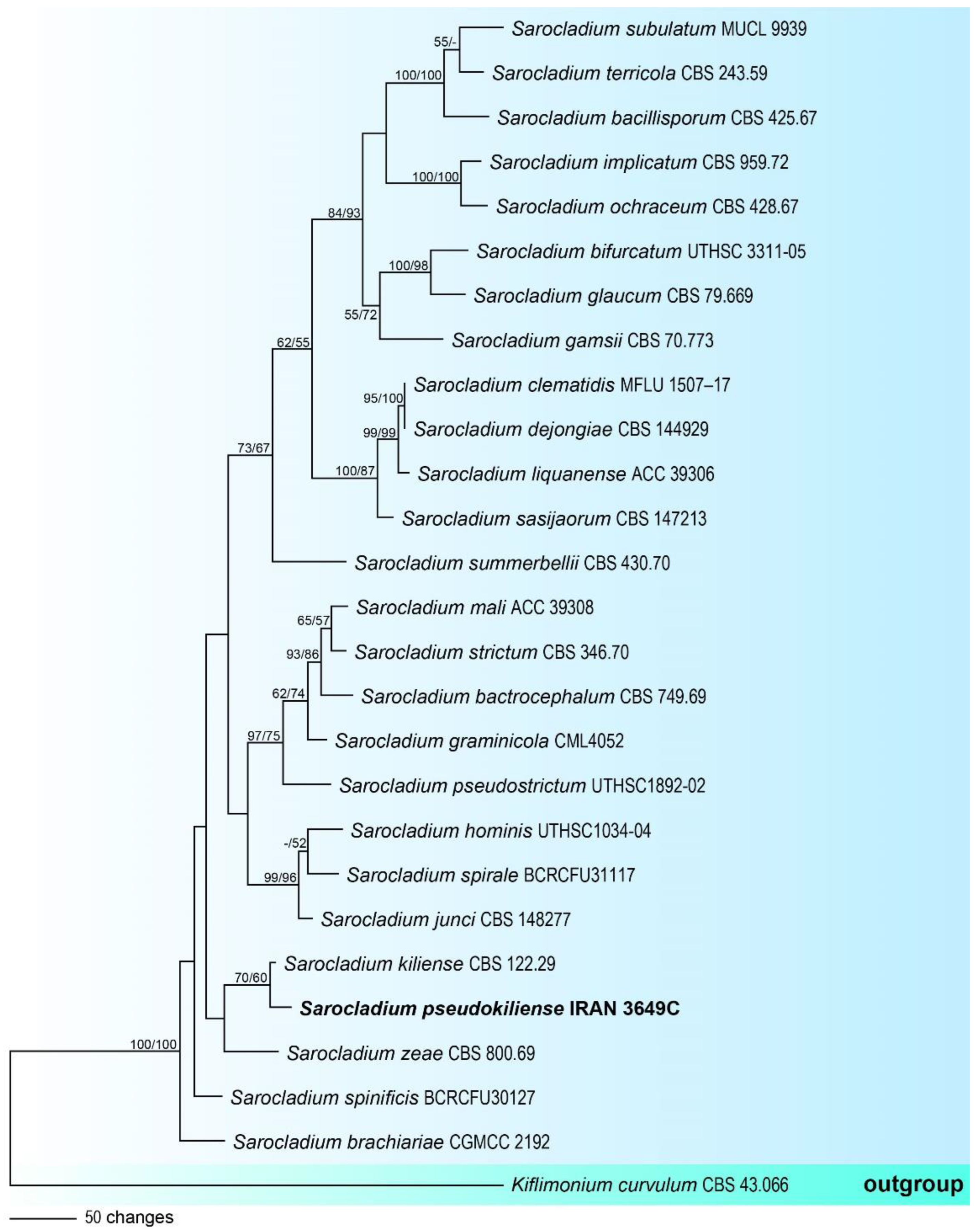
3.1. Molecular Phylogeny
3.2. Taxonomy
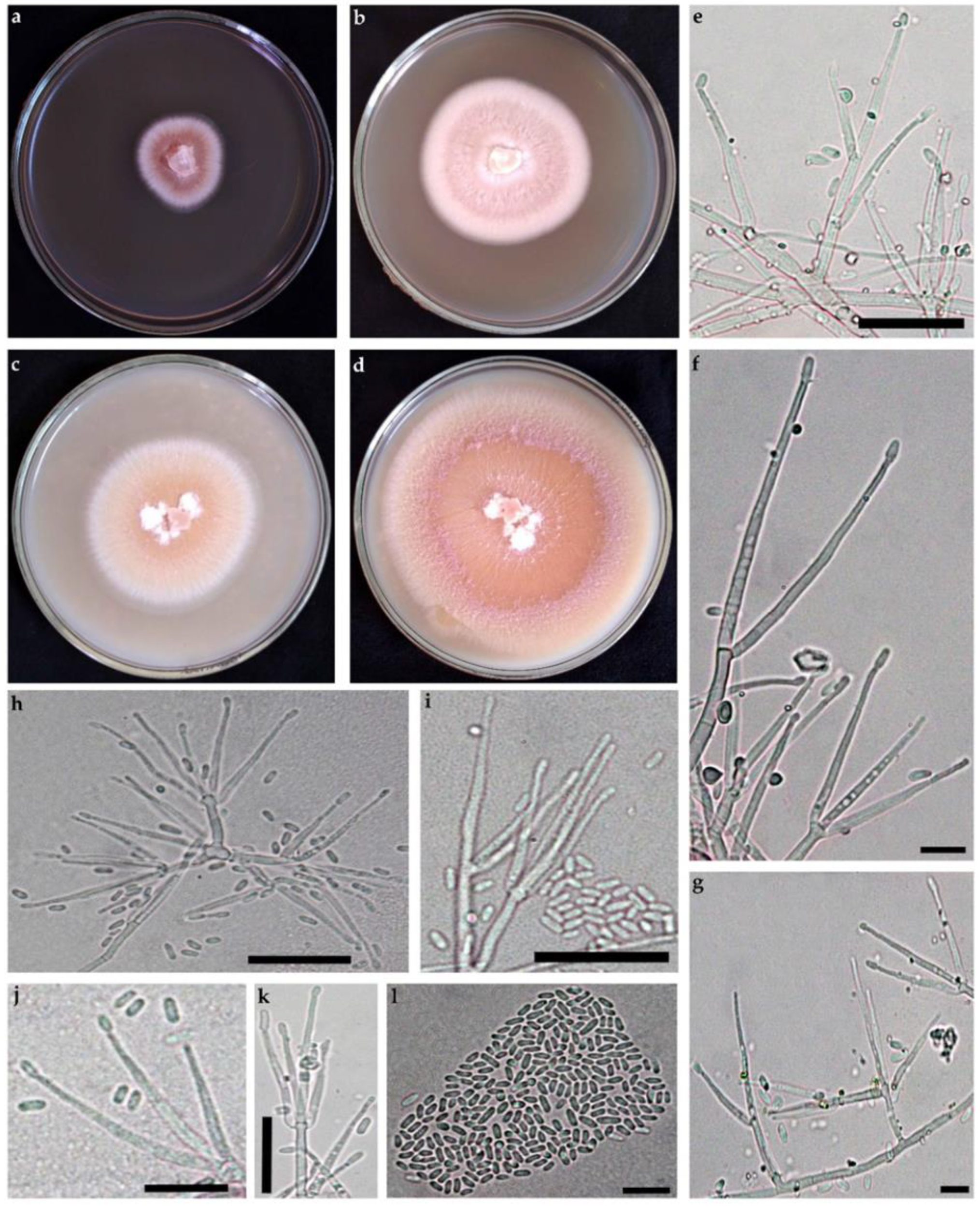
3.2.1. Arthrobotrys hyrcanus, sp. nov., Masigol, Rezakhani, Pourmoghaddam, Khodaparast (Figure 3)
- MycoBank No: 845353
- Etymology: hyrcanus derived from “Hyrcania”, an ancient biogeographical region located in the south of the Caspian Sea where the specimens were collected.
- Holotype: Iran, Guilan Province, Anzali County, Anzali Lagoon, 37°28′16″ N, 49°27′44″ E, on rotten leaves, 11 August 2017, F. Rezakhani, (GUM 1904, ex-holotype culture IRAN 3650C); ITS, LSU, and TEF1 sequences GenBank MH367058, MH367076, and OP351540, respectively.
- Mycelium hyaline, scanty, hyphae septate, branched, 1.5–3 μm wide. Conidiophores growing from mycelium on the substratum, single, erect, 2–5-septate, 70–312 μm long, 5–6 μm wide, bearing a single conidium at the apex. Conidia hyaline, clavate or spindle-shaped, narrowing at the basal, 2–9-septate, rarely 9-septate, 44.2–135.2 × 10–14.4 μm. The proportion of conidia with 2, 3, 4, 5, 6, 7, and 8 septa is 7, 10, 18, 22, 12, 19, and 11%, respectively. Some conidia had small tubercles at the one or both ends and could germinate from these tubercles. Conidia could produce secondary conidiophores and secondary conidia. The secondary conidia are clavate, 20.8–33.8 × 2.6–4.8 μm and 1-septate. Chlamydospores present in cultures after 3 wk.
- Culture characteristics: Colonies on CMA whitish, rapidly growing and extending to a diameter of 9 cm at 25 °C within 7 days.
- Other specimen examined: Iran, Guilan Province, Anzali County, Anzali Lagoon, 37°28′16″ N, 49°27′44″ E, on rotten leaves, 11 Aug 2017, F. Rezakhani (IRAN 3651C); ITS, LSU and TEF1 sequences GenBank MH367063, MH367081, and OP351541, respectively.
- Notes: This species is similar to A. dianchiensis, but it can be distinguished from the latter by the larger primary [44.2–135.2 × 10–14.4 vs. 37.5–100 (70) × 10–17.5 (14.3) µm] and secondary (20.8–33.8 × 2.6–4.8 vs. 23.9 × 5 µm) conidia, septation of primary conidia, and the presence of chlamydospores. Table 3 compares morphological characters of some species that may be confused with A. hyrcanus.

3.2.2. Sarocladium pseudokiliense, sp. nov., Rezakhani, Khodaparast, Masigol, and Grossart (Figure 4)
- MycoBank No: 845356
- Etymology: pseudokiliense, referring to the morphological similarity to Sarocladium kiliense.
- Holotype: Iran, Guilan Province, Anzali County, Anzali Lagoon, 37°28′16″N, 49°27′44″E, on rotten leaves, 11 September 2017, F. Rezakhani (GUM 1905, ex-holotype culture IRAN 3649C); ITS, LSU, TEF1 and TUB2 sequences GenBank MH367070, MH367052, OP351542, and OP351543, respectively.
- Mycelium consisting of hyaline, smooth-walled, branched, septate, 1.5–2.5 µm wide. Conidiophores erect, arising directly from vegetative hyphae or ropes of hyphae, straight, simple, hyaline, smooth-walled, up to 21 µm long. Phialides subcylindrical to acicular, 15–45 µm long, 1.5–2 µm wide at the base, thin- and smooth-walled, hyaline with inconspicuous periclinal thickening; adelophialides and schizophialides not observed. Conidia solitary, cylindrical, 3–6 × 1–1.5 µm, hyaline, thin- and smooth-walled, arranged in slimy heads. Chlamydospores or sexual morph not observed.
- Culture characteristics: Colonies on OA at 25 °C attaining 80–85 mm in 14 d, at first orange with white margin, becoming pink to purple. On PDA at 25 °C attaining 40–45 mm in 14 d, at first orange with white margin, becoming white to cream.
- Notes: Sarocladium is an acremonium-like genus that contains several important plant and human pathogens [29,56]. The description of this species is based on a single specimen, which shows phylogenetically close to S. kiliense, but it can be distinguished from the latter by the shape of conidia (in S. kiliense conidia are ellipsoidal to cylindrical), the absence of adelophialides and chlamydospores.
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grossart, H.-P.; Rojas-Jimenez, K. Aquatic fungi: Targeting the forgotten in microbial ecology. Curr. Opin. Microbiol. 2016, 31, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Ittner, L.D.; Junghans, M.; Werner, I. Aquatic Fungi: A Disregarded Trophic Level in Ecological Risk Assessment of Organic Fungicides. Front. Environ. Sci. 2018, 6, 105. [Google Scholar] [CrossRef]
- Masigol, H.; Khodaparast, S.A.; Woodhouse, J.N.; Rojas-Jimenez, K.; Fonvielle, J.; Rezakhani, F.; Mostowfizadeh-Ghalamfarsa, R.; Neubauer, D.; Goldhammer, T.; Grossart, H.P. The contrasting roles of aquatic fungi and oomycetes in the degradation and transformation of polymeric organic matter. Limnol. Oceanogr. 2019, 64, 2662–2678. [Google Scholar] [CrossRef] [Green Version]
- Masigol, H.; Woodhouse, J.N.; van West, P.; Mostowfizadeh-Ghalamfarsa, R.; Rojas-Jimenez, K.; Goldhammer, T.; Khodaparast, S.A.; Grossart, H.-P. Phylogenetic and Functional Diversity of Saprolegniales and Fungi Isolated from Temperate Lakes in Northeast Germany. J. Fungi 2021, 7, 968. [Google Scholar] [CrossRef] [PubMed]
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in aquatic food webs. Front. Microbiol. 2014, 5, 166. [Google Scholar] [CrossRef]
- Klawonn, I.; Van den Wyngaert, S.; Parada, A.E.; Arandia-Gorostidi, N.; Whitehouse, M.J.; Grossart, H.-P.; Dekas, A.E. Characterizing the “fungal shunt”: Parasitic fungi on diatoms affect carbon flow and bacterial communities in aquatic microbial food webs. Proc. Natl. Acad. Sci. USA 2021, 118, e2102225118. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Stielow, J.B.; Lévesque, C.A.; Seifert, K.A.; Meyer, W.; Irinyi, L.; Smits, D.; Renfurm, R.; Verkley, G.J.M.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia—Mol. Phylogeny Evol. Fungi 2015, 35, 242–263. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.-W.; Cao, Y.; Wu, S.-H.; Vlasák, J.; Li, D.-W.; Li, M.-J.; Dai, Y.-C. Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry 2015, 114, 7–15. [Google Scholar] [CrossRef]
- Shearer, C.A.; Descals, E.; Kohlmeyer, B.; Kohlmeyer, J.; Marvanová, L.; Padgett, D.; Porter, D.; Raja, H.A.; Schmit, J.P.; Thorton, H.A.; et al. Fungal biodiversity in aquatic habitats. Biodivers. Conserv. 2007, 16, 49–67. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Wurzbacher, C.; James, T.Y.; Kagami, M. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol. 2016, 19, 28–38. [Google Scholar] [CrossRef] [Green Version]
- Wijayawardene, N.N.; Bahram, M.; Sánchez-Castro, I.; Dai, D.-Q.; Ariyawansa, K.G.; Jayalal, U.; Suwannarach, N.; Tedersoo, L. Current Insight into Culture-Dependent and Culture-Independent Methods in Discovering Ascomycetous Taxa. J. Fungi 2021, 7, 703. [Google Scholar] [CrossRef] [PubMed]
- Swe, A.; Jeewon, R.; Pointing, S.B.; Hyde, K.D. Diversity and abundance of nematode-trapping fungi from decaying litter in terrestrial, freshwater and mangrove habitats. Biodivers. Conserv. 2009, 18, 1695–1714. [Google Scholar] [CrossRef]
- Fuentes, M.; Quinones, R.A. Carbon utilization profile of the filamentous fungal species Fusarium fujikuroi, Penicillium decumbens, and Sarocladium strictum isolated from marine coastal environments. Mycologia 2016, 108, 1069–1081. [Google Scholar] [CrossRef]
- Rezakhani, F.; Khodaparast, S.A.; Masigol, H.; Roja-Jimenez, K.; Grossart, H.-P.; Bakhshi, M. A preliminary report of aquatic hyphomycetes isolated from Anzali lagoon (Gilan province, North of Iran). Rostaniha 2019, 20, 123–143. [Google Scholar] [CrossRef]
- Zhang, F.; Boonmee, S.; Bhat, J.D.; Xiao, W.; Yang, X.-Y. New Arthrobotrys Nematode-Trapping Species (Orbiliaceae) from Terrestrial Soils and Freshwater Sediments in China. J. Fungi 2022, 8, 671. [Google Scholar] [CrossRef]
- Jaffee, B.A. Do Organic Amendments Enhance the Nematode-Trapping Fungi Dactylellina haptotyla and Arthrobotrys oligospora? J. Nematol. 2004, 36, 267. [Google Scholar]
- Zhang, K.Q.; Li, T.F.; Liu, X.Z. Biology of Nematophagous Fungi; China Science & Technology Press: Beijing, China, 2001. [Google Scholar]
- Zhang, K.Q.; Hyde, K.D. Nematode-Trapping Fungi; Springer Science & Business: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Yu, Z.; Qiao, M.; Zhang, Y.; Baral, H.O.; Zhang, K. Orbilia vermiformis sp. nov. and its anamorph. Mycotaxon 2007, 99, 271–278. [Google Scholar]
- Zhang, K.Q.; Mo, M.H. Flora Fungorum Sinicorum: Arthrobotrys et Gengra Cetera Cognata; Science Press: Beijing, China, 2006; Volume 33. [Google Scholar]
- Martín-Rodríguez, A.J.; Reyes, F.; Martín, J.; Pérez-Yépez, J.; León-Barrios, M.; Couttolenc, A.; Espinoza, C.; Trigos, Á.; Martín, V.S.; Norte, M.; et al. Inhibition of Bacterial Quorum Sensing by Extracts from Aquatic Fungi: First Report from Marine Endophytes. Mar. Drugs 2014, 12, 5503–5526. [Google Scholar] [CrossRef] [Green Version]
- Descal, E. Techniques for Handling Ingoldian Fungi. In Book Methods to Study Litter Decomposition; Graça, M.A., Bärlocher, F., Gessner, M.O., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 129–142. [Google Scholar]
- Galloway, L.D.; Burgess, R. Applied Mycology and Bacteriology; Leonard Hill, Ltd.: London, UK, 1952. [Google Scholar]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium Species: An Illustrated Manual for Identification; Pennsylvania State University Press: University Park, PA, USA, 1983. [Google Scholar]
- Downes, F.P.; Ito, K. Compendium of Methods for the Microbiological Examination of Foods, 4th ed.; APHA: Washington, DC, USA, 2001. [Google Scholar]
- Murray, P.R.; Baron, E.J.; Jorgensen, J.H.; Pfaller, M.A.; Yolken, R.H. Manual of Clinical Microbiology; ASM: Washington, DC, USA, 2003. [Google Scholar]
- Yu, Z.; Mo, M.; Zhang, Y.; Zhang, K.-Q. Taxonomy of Nematode-Trapping Fungi from Orbiliaceae, Ascomycota; Nematode-Trapping Fungi. Fungal Diversity Research, Series; Zhang, K.Q., Hyde, K., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 23. [Google Scholar] [CrossRef]
- Giraldo, A.; Gené, J.; Sutton, D.; Madrid, H.; de Hoog, G.; Cano, J.; Decock, C.; Crous, P.; Guarro, J. Phylogeny of Sarocladium (Hypocreales). Persoonia 2015, 34, 10–24. [Google Scholar] [CrossRef] [Green Version]
- Montero-Pau, J.; Gomez, A.; Munoz, J. Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnol. Oceanogr. Methods 2008, 6, 218–222. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; PCR Protocols: A Guide to Methods and, Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1999; pp. 315–322. [Google Scholar]
- Borneman, J.; Hartin, R.J. PCR Primers That Amplify Fungal rRNA Genes from Environmental Samples. Appl. Environ. Microbiol. 2000, 66, 4356–4360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woudenberg, J.H.C.; Aveskamp, M.M.; De Gruyter, J.; Spiers, A.G.; Crous, P.W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Per.—Mol. Phylogeny Evol. Fungi 2009, 22, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [Green Version]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP* 4.0b10: Phylogenetic Analysis Using Parsimony (*and Other Methods); Sinauer Associates: Sunderland, UK, 2002. [Google Scholar]
- Yang, Y.E.; Yang, Z.; Liu, X. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. USA 2007, 104, 8379–8384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, X.Z. A new generic approach to the taxonomy of predatory anamorphic Orbiliaceae (Ascomycotina). Mycotaxon 2006, 97, 153–161. [Google Scholar]
- Hagedorn, G.; Scholler, M. A reevaluation of predatory orbiliaceous fungi. I. Phylogenetic analysis using rDNA sequence data. Sydowia 1999, 51, 27–48. [Google Scholar]
- Liou, G.Y.; Tzean, S.S. Phylogeny of the genus Arthrobotrys and allied nematode-trapping fungi based on rDNA sequences. Mycologia 1997, 89, 876–884. [Google Scholar] [CrossRef]
- Li, Y.; Hyde, K.D.; Jeewon, R.; Lei, C.; Vijaykrishna, D.; Zhang, K. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia 2005, 97, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jeewon, R.; Hyde, K.D.; Mo, M.-H.; Zhang, K.-Q. Two new species of nematode-trapping fungi: Relationships inferred from morphology, rDNA and protein gene sequence analyses. Mycol. Res. 2006, 110, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, A.; Gené, J.; Cano, J.; de Hoog, S.; Guarro, J. Two new species of Acremonium from Spanish soils. Mycologia 2012, 104, 1456–1465. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Gueidan, C.; Schroers, H.-J.; de Hoog, G.S.; Starink, M.; Rosete, Y.A.; Guarro, J.; Scott, J.A. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011, 68, 139–162. [Google Scholar] [CrossRef]
- Liu, X.; Guo, Z.; Huang, G. Sarocladium brachiariae sp. nov., an endophytic fungus isolated from Brachiaria brizantha. Mycosphere 2017, 8, 827–834. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Jones, E.B.G.; Bhat, D.J.; Stadler, M.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Crous, P.W.; Luangsa-Ard, J.J.; Wingfield, M.; Carnegie, A.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.; Baseia, I.; Cano-Lira, J.; et al. Fungal Planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, H.; Sutton, D.A.; García, D.; Fothergill, A.W.; Cano, J.; Gené, J.; Summerbell, R.C.; Rinaldi, M.G.; Guarro, J. Spectrum of Clinically Relevant Acremonium Species in the United States. J. Clin. Microbiol. 2011, 49, 243–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anjos, R.M.; Moreira, S.I.; Costa, S.S.; Abreu, L.M.; Alves, E.; Cardoso, P.G. Sarocladium graminicola, a new endophytic species from tropical grasses. Mycol. Prog. 2020, 19, 605–614. [Google Scholar] [CrossRef]
- Crous, P.W.; Osieck, E.R.; Jurjević, Ž.; Boers, J.; Van Iperen, A.; Starink-Willemse, M.; Dima, B.; Balashov, S.; Bulgakov, T.; Johnston, P.; et al. Fungal Planet description sheets: 1284–1382. Persoonia 2021, 47, 178–374. [Google Scholar] [CrossRef]
- Hou, Y.M.; Zhang, X.; Zhang, N.N.; Naklumpa, W.; Zhao, W.Y.; Liang, X.F.; Zhang, R.; Sun, G.Y.; Gleason, M.L. Genera Acremonium and Sarocladium Cause Brown Spot on Bagged Apple Fruit in China. Plant Dis. 2019, 103, 1889–1901. [Google Scholar] [CrossRef]
- Crous, P.W.; Hernández-Restrepo, M.; Schumacher, R.; Cowan, D.; Maggs-Kölling, G.; Marais, E.; Wingfield, M.; Yilmaz, N.; Adan, O.; Akulov, A.; et al. New and Interesting Fungi. 4. Fungal Syst. Evol. 2021, 7, 255–343. [Google Scholar] [CrossRef]
- Yeh, Y.-H.; Kirschner, R. Sarocladium spinificis, a new endophytic species from the coastal grass Spinifex littoreus in Taiwan. Bot. Stud. 2014, 55, 25. [Google Scholar] [CrossRef] [Green Version]
- Ou, J.-H.; Lin, G.-C.; Chen, C.-Y. Sarocladium species associated with rice in Taiwan. Mycol. Prog. 2020, 19, 67–80. [Google Scholar] [CrossRef]
- Masigol, H.; Khodaparast, S.A.; Mostowfizadeh-Ghalamfarsa, R.; Rojas-Jimenez, K.; Woodhouse, J.N.; Neubauer, D.; Grossart, H.-P. Taxonomical and functional diversity of Saprolegniales in Anzali lagoon, Iran. Aquat. Ecol. 2020, 54, 323–336. [Google Scholar] [CrossRef] [Green Version]
- Masigol, H.; Mostowfizadeh-Ghalamfarsa, R.; Grossart, H.P. The current status of Saprolegniales in Iran: Calling mycologists for better taxonomic and ecological resolutions. Mycol. Iran. 2021, 8, 85. [Google Scholar]
- Masigol, H.; Khodaparast, S.A.; Mostowfizadeh-Ghalamfarsa, R.; Mousanejad, S.; Rojas-Jimenez, K.; Grossart, H.P. Notes on Dictyuchus species (Stramenopila, Oomycetes) from Anzali lagoon, Iran. Mycol. Iran. 2018, 5, 79–89. [Google Scholar] [CrossRef]
| Species | Strain Code | GenBank Accession Number | Reference | |
|---|---|---|---|---|
| ITS | TEF1 | |||
| Arthrobotrys | ||||
| A. anomalus | YNWS02-5-1 | AY773451 | AY773393 | [42] |
| A. conoides | 670 | AY773455 | AY773397 | [42] |
| A. eudermatus | SDT24 | AY773465 | HE608633 | [42] |
| Arthrobotryshyrcanus | IRAN 3650C | MH367058 | OP351540 | This study |
| Arthrobotryshyrcanus | IRAN 3651C | MH367063 | OP351541 | This study |
| A. iridis | 521 | AY773452 | AY773394 | [42] |
| A. janus | 85-1 | AY773459 | AY773401 | [42] |
| A. multiformis | CBS 773.84 | MH861834 | N/A | [43] |
| A. musiformis | SQ77-1 | AY773469 | AY773411 | [42] |
| A. oligospora | 920 | AY773462 | AY773404 | [42] |
| A.pseudoclavatus | 1130 | AY773446 | AY773388 | [42] |
| A. pyriformis | YNWS02-3-1 | AY773450 | AY773392 | [42] |
| A. sinensis | 105-1 | AY773445 | AY773387 | [42] |
| A. sphaeroides | SDT24 | AY773465 | AY773407 | [42] |
| A. thaumasius | 917 | AY773461 | AY773403 | [42] |
| A. vermicola | 629 | AY773454 | AY773396 | [42] |
| Dactylellina | ||||
| D. appendiculata | CBS 206.64 | AF106531 | DQ358227 | [44] |
| D. arcuata | CBS 174.89 | AF106527 | DQ999852 | [45] |
| D. cionopaga | SQ60-2 | AY773468 | AY773410 | [42] |
| D. copepodii | CBS 487.90 | U51964 | DQ999835 | [42,46] |
| D. drechsleri | CBS 549.63 | DQ999819 | DQ999840 | [42] |
| D. ellipsospora | 286 | AY773449 | AY773391 | [42] |
| D. entomopaga | CBS 642.80 | AY965758 | DQ358228 | [44,47] |
| D. gephyropaga | CBS 585.91 | AY965756 | DQ999846 | [42] |
| D. haptospora | CBS 100520 | DQ999820 | DQ999850 | [42] |
| D. haptotyla | SQ95-2 | AY773470 | AY773412 | [42] |
| D. leptospora | SHY6-1 | AY773466 | AY773408 | [42] |
| D. mammillata | CBS 229.54 | AY902794 | DQ999843 | [42,48] |
| D. parvicollis | XJ03-52-1 | AY773472 | AY773414 | [42] |
| D. querci | 6175 | AY773453 | AY773395 | [42] |
| D. robusta | CBS 110125 | DQ999821 | DQ999851 | [42] |
| D. tibetensis | XZ04-92-1 | DQ999833 | DQ999848 | [42] |
| Drechslerella brochopaga | 701 | AY773456 | AY773398 | [42] |
| Drechslerella coelobrocha | FWY03-25-1 | AY773464 | AY773406 | [42] |
| Drechslerella dactyloides | expo-5 | AY773463 | AY773405 | [42] |
| Drechslerella stenobrocha | YNWS02-9-1 | AY773460 | AY773402 | [42] |
| Vermispora fusarina | YXJ13-5 | AY773447 | AY773389 | [42] |
| Species | Strain Code | Status | GenBank Accession Number | Reference | ||
|---|---|---|---|---|---|---|
| ITS | LSU | ACT1 | ||||
| Kiflimonium curvulum | CBS 430.66 | HT | HE608638 | HE608656 | HE608630 | [49] |
| Sarocladium bacillisporum | CBS 425.67 | HT | HE608639 | HE608658 | HE608633 | [49] |
| S. bactrocephalum | CBS 749.69 | HT | HG965006 | HQ231994 | HG964956 | [29,50] |
| S. bifurcatum | UTHSC 05-3311 | HT | HG965009 | HG965057 | HG964959 | [29] |
| S.brachiariae | CGMCC 2192 | HT | EU880834 | KP715271 | N/A | [51] |
| S.clematidis | MFLU 17–1507 | HT | MN629287 | MN629285 | N/A | [52] |
| S.dejongiae | CBS 144929 | HT | MK069419 | MK069415 | N/A | [53] |
| S.gamsii | CBS 707.73 | HT | HG965015 | HG965063 | HG964965 | [29] |
| S.glaucum | CBS 796.69 | HT | FN691454 | HE608657 | HE608631 | [49,54] |
| S.graminicola | CML 4052 | HT | MK017855 | MK017871 | MK017838 | [55] |
| S.hominis | UTHSC04-1034 | HT | HG965012 | HG965060 | HG964962 | [29] |
| S.implicatum | CBS 959.72 | HT | HG965023 | HG965072 | HG964974 | [29] |
| S.junci | CBS 148277 | HT | OK664734 | OK663773 | OK651128 | [56] |
| S.kiliense | CBS 122.29 | HT | FN691446 | HQ232052 | HG964975 | [29,50,54] |
| S.liquanense | ACC 39306 | HT | MF987659 | MF987651 | MF987663 | [57] |
| S.mali | ACC 39308 | HT | MF987662 | MF987653 | MF987665 | [57] |
| S.ochraceum | CBS 428.67 | HG965025 | HQ232070 | HG964977 | [29,50] | |
| S. pseudokiliense | IRAN 3649C | HT | MH367052 | MH367070 | N/A | This study |
| S.pseudostrictum | UTHSC02-1892 | HT | HG965029 | HG965073 | HG964981 | [29] |
| S.sasijaorum | CBS 147213 | HT | MW883448 | MW883839 | MW890032 | [58] |
| S.spinificis | BCRCFU30127 | KF269096 | JQ954463 | N/A | [59] | |
| S.spirale | BCRCFU31117 | HT | LC461491 | LC464181 | LC464350 | [60] |
| S.strictum | CBS 346.70 | HT | FN691453 | HQ232141 | HG964982 | [29,50,54] |
| S.subulatum | MUCL 9939 | HT | HG965031 | HG965075 | HG964984 | [29] |
| S.summerbellii | CBS 430.70 | HT | HG965034 | HG965078 | HG964987 | [29] |
| S.terricola | CBS 243.59 | HT | FN706553 | HE608659 | HE608632 | [29,49,54] |
| S.zeae | CBS 800.69 | HT | FN691451 | HQ232152 | HG965000 | [29,50,54] |
| Species | Size of Primary Conidia | Number of Primary Conidial Septa | Chlamydospores |
|---|---|---|---|
| Arthrobotrys dianchiensis | 37.5–100 (70) × 10–17.5 (14.3) | 1–7 (mainly 2–5) | Not mentioned |
| Arthrobotrys hyrcanus | 44.2–135.2 × 10–14.4 | 2–9 | present |
| Arthrobotrys multiformis | 47–198 × 7–20 | 4–12 | present |
| Arthrobotrys shizishanna | 22.5–73.8 (50.6) × 5–10 (6.6) | 2–9 | present |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masigol, H.; Rezakhani, F.; Pourmoghaddam, M.J.; Khodaparast, S.A.; Grossart, H.-P. The Introduction of Two New Species of Aquatic Fungi from Anzali Lagoon, Northern Iran. Diversity 2022, 14, 889. https://doi.org/10.3390/d14100889
Masigol H, Rezakhani F, Pourmoghaddam MJ, Khodaparast SA, Grossart H-P. The Introduction of Two New Species of Aquatic Fungi from Anzali Lagoon, Northern Iran. Diversity. 2022; 14(10):889. https://doi.org/10.3390/d14100889
Chicago/Turabian StyleMasigol, Hossein, Forough Rezakhani, Mohammad Javad Pourmoghaddam, Seyed Akbar Khodaparast, and Hans-Peter Grossart. 2022. "The Introduction of Two New Species of Aquatic Fungi from Anzali Lagoon, Northern Iran" Diversity 14, no. 10: 889. https://doi.org/10.3390/d14100889
APA StyleMasigol, H., Rezakhani, F., Pourmoghaddam, M. J., Khodaparast, S. A., & Grossart, H. -P. (2022). The Introduction of Two New Species of Aquatic Fungi from Anzali Lagoon, Northern Iran. Diversity, 14(10), 889. https://doi.org/10.3390/d14100889







