Funneliformis mosseae Inoculation Enhances Cucurbita pepo L. Plant Growth and Fruit Yield by Reshaping Rhizosphere Microbial Community Structure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials and Growth Conditions
2.2. Plant Growth, Root Morphology, and Root Colonization Analysis
2.3. Microbial Community Analysis
2.4. Statistical Analysis
3. Results
3.1. Effects of F. mosseae Inoculation on C. pepo Growth and Yield
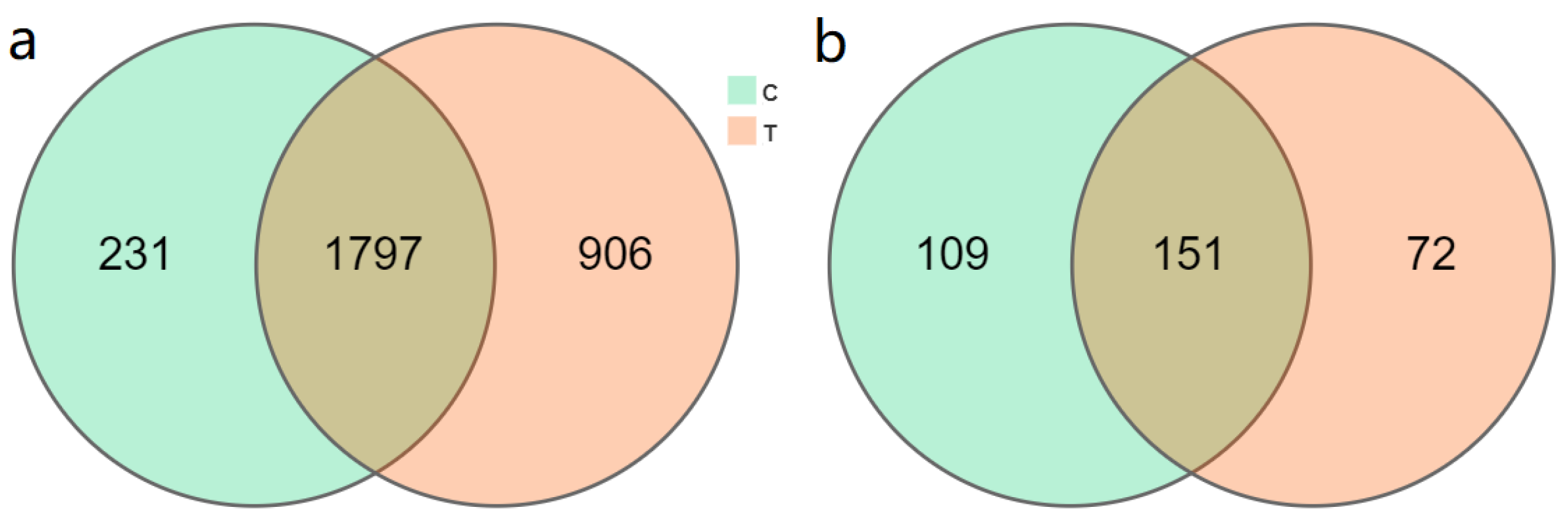
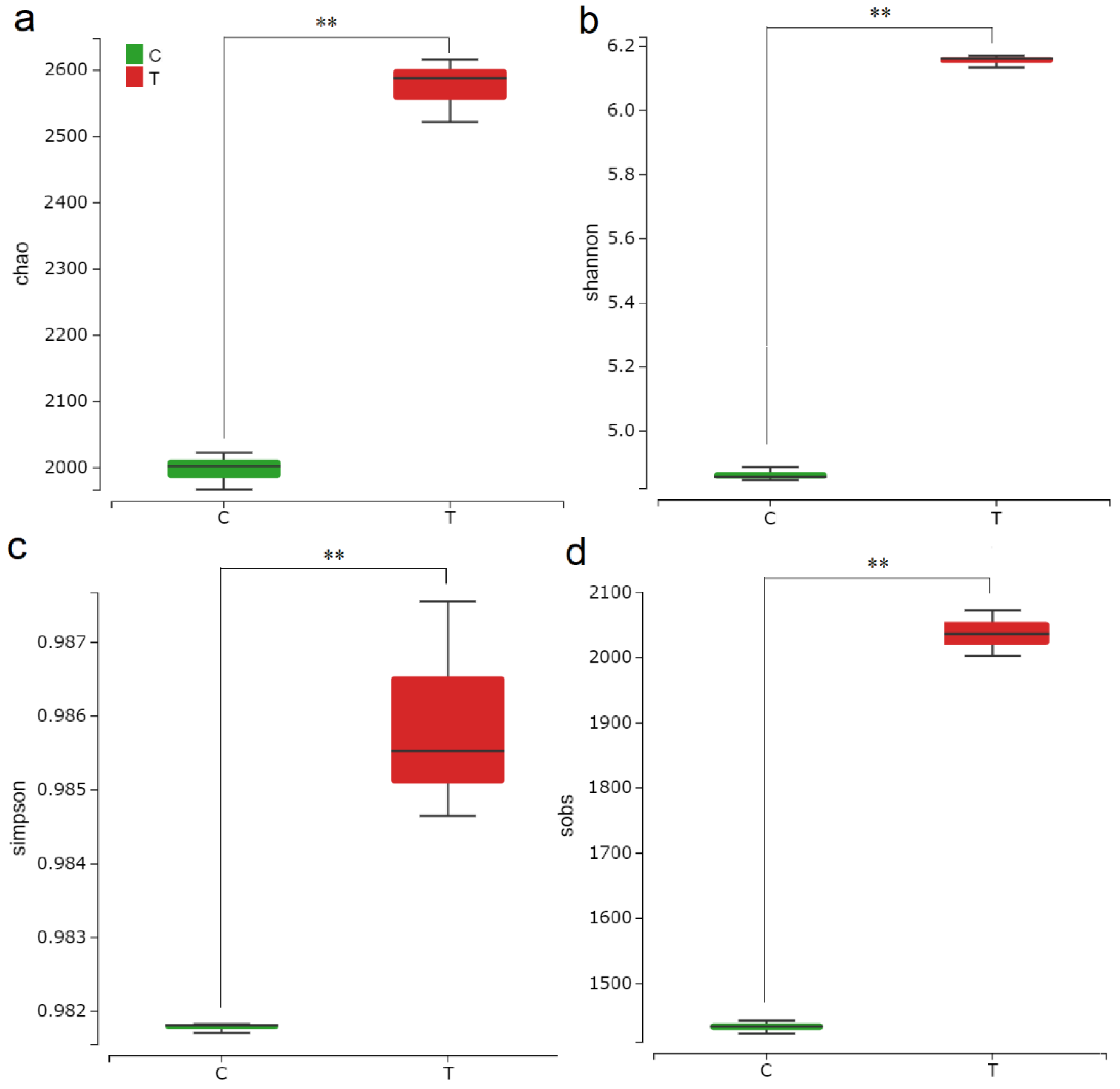
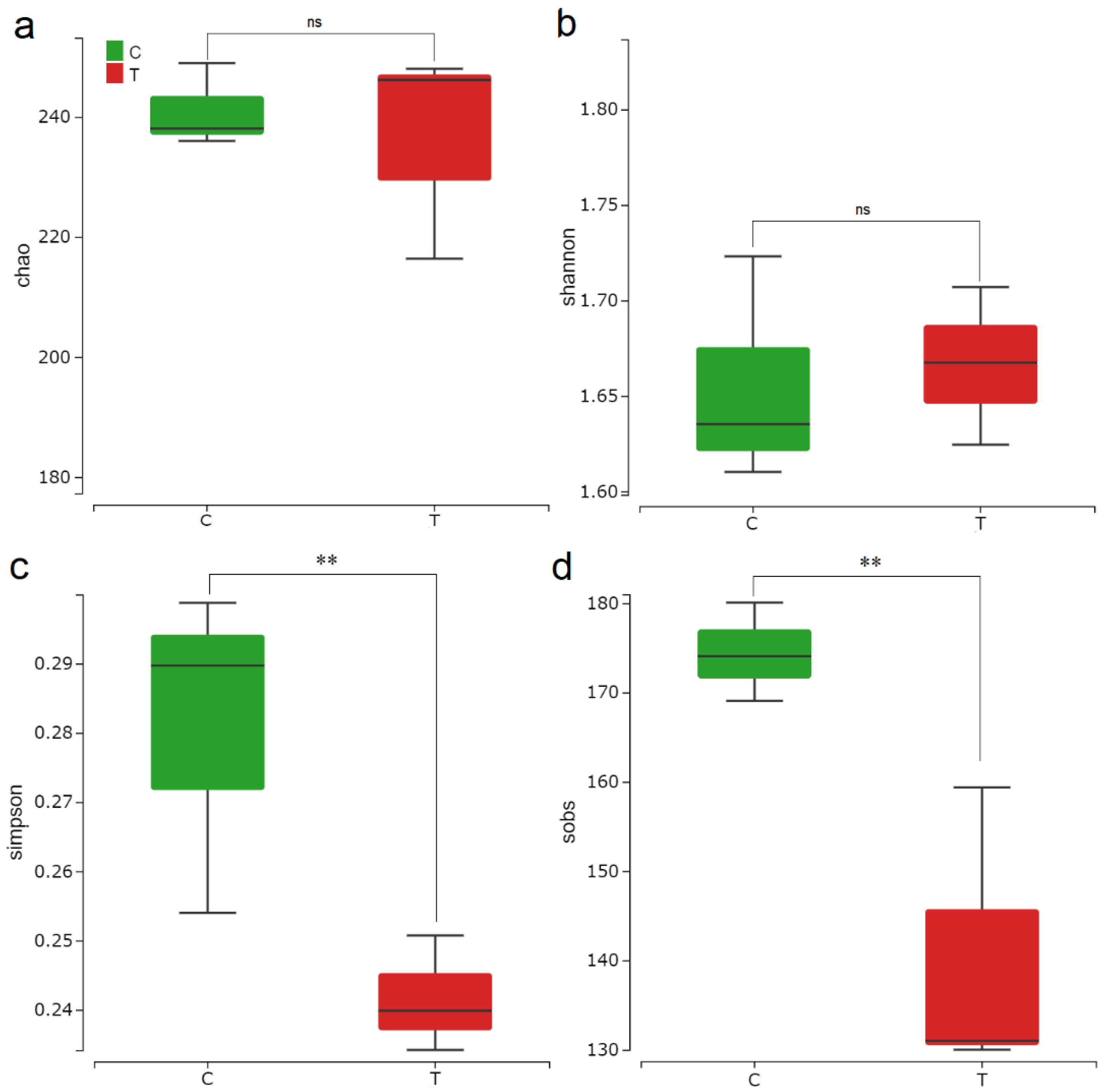
3.2. F. mosseae-Induced Changes in Rhizosphere Microbial Community Diversity
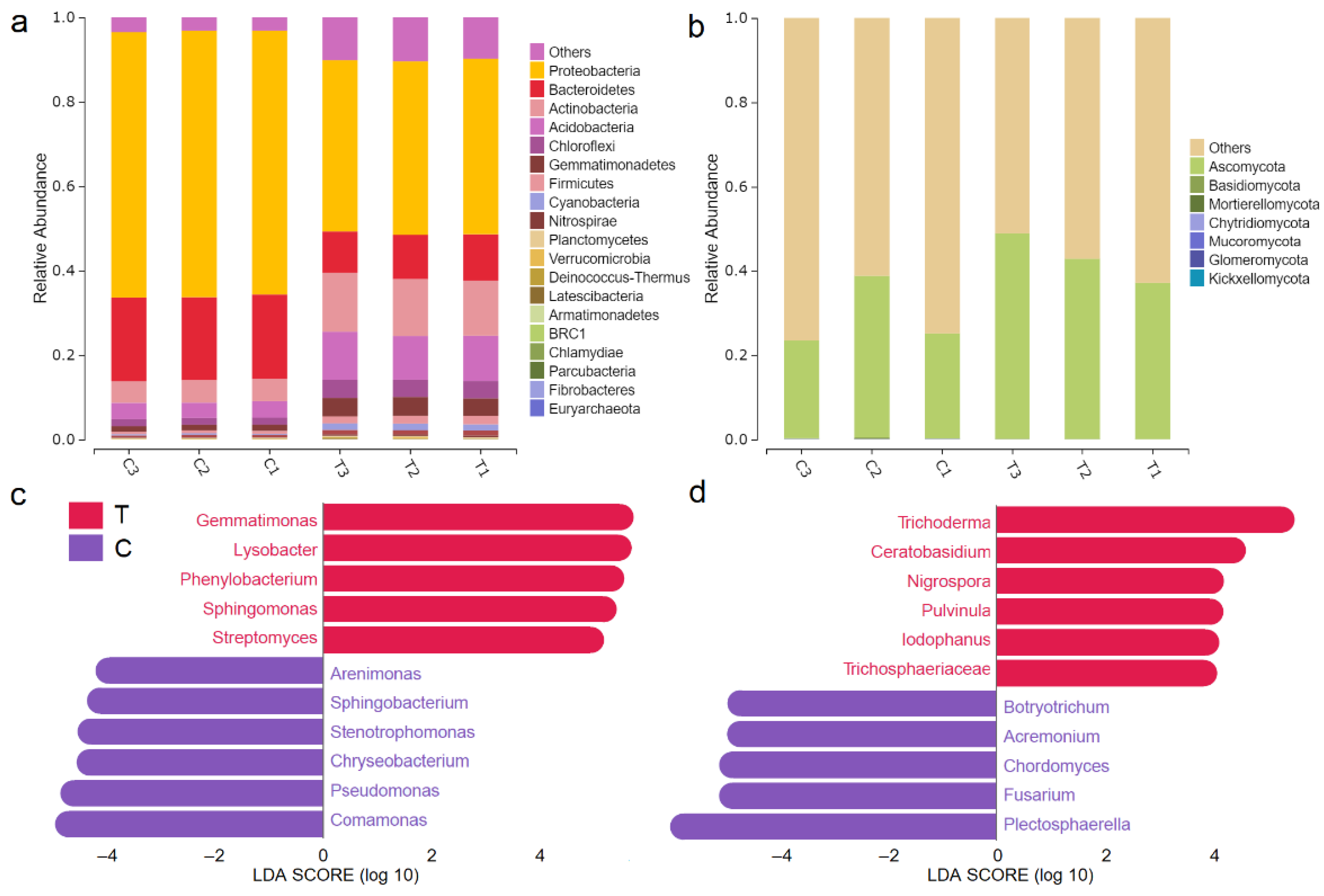
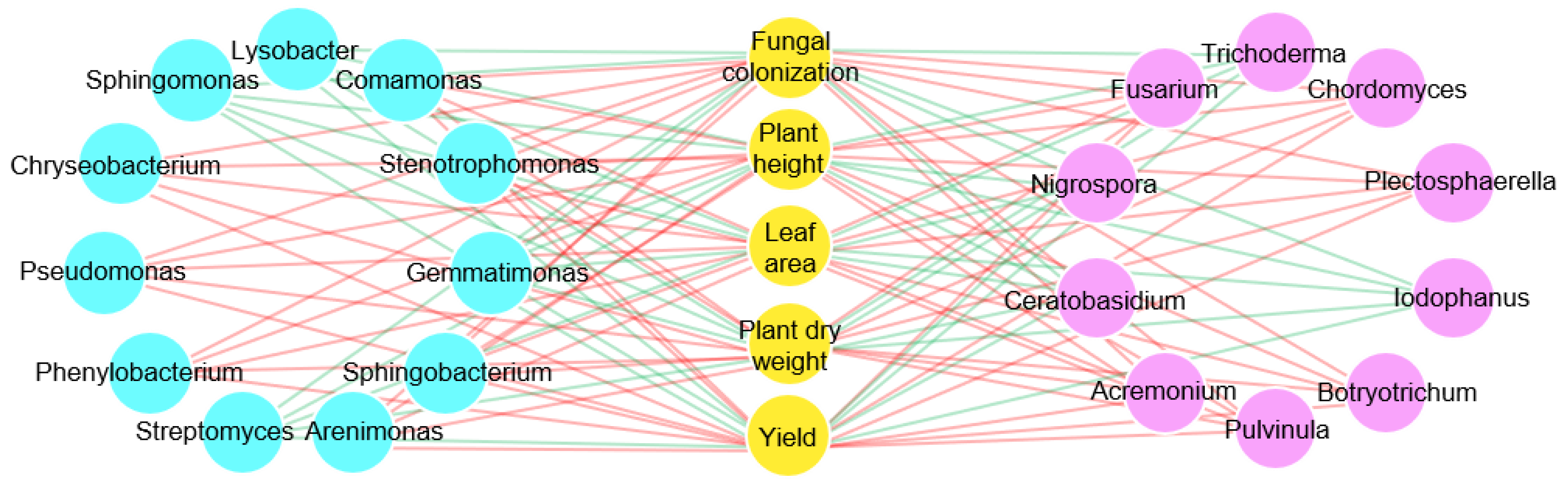
3.3. F. mosseae-Mediated Regulation of Microbial Community Structure in the Rhizosphere
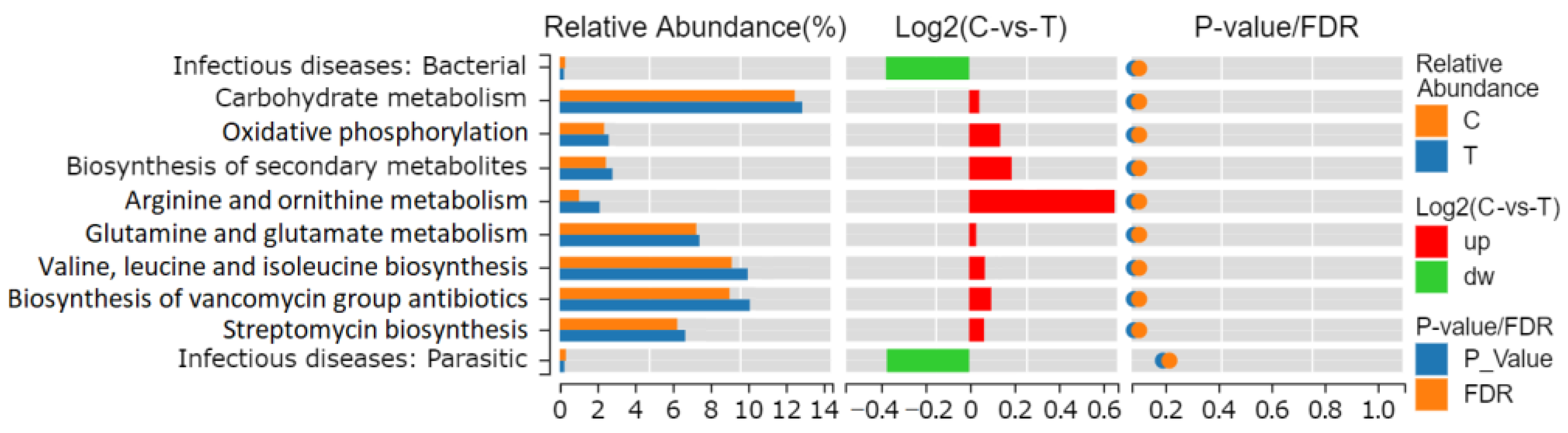
3.4. Microbial Function Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santosh, K.B.; Prianka, H. Melatonin plays multifunctional role in horticultural crops against environmental stresses: A review. Environ. Exp. Bot. 2020, 176, 104063. [Google Scholar]
- Fu, C.; Shi, H.; Li, Q. A review on pharmacological activities and utilization technologies of pumpkin. Plant Food Hum. Nutr. 2006, 61, 70–77. [Google Scholar]
- Younis, Y.M.; Ghirmay, S.; Al-Shihry, S.S. African Cucurbita pepo L.: Properties of seed and variability in fatty acid composition of seed oil. Phytochemistry 2000, 54, 71–75. [Google Scholar] [CrossRef]
- Doran, J.W.; Zeiss, M.R. Soil health and sustainability: Managing the biotic component of soil quality. Appl. Soil. Ecol. 2000, 15, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.L.; Cui, H.L.; Su, J.Q. Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 2019, 24, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, K.; Gerlach, N.; Sacristán, S. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wei, Z.; Yang, K. Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotechnol. 2019, 37, 1513–1520. [Google Scholar] [CrossRef]
- Zhang, C.L.; Zhang, Y.M.; Ding, Z.J. Contribution of microbial inter-kingdom balance to plant health. Mol. Plant 2019, 12, 148–149. [Google Scholar] [CrossRef] [Green Version]
- Smith, F.A.; Smith, S.E. What is the significance of the arbuscular mycorrhizal colonisation of many economically important crop plants? Plant Soil 2011, 348, 63–79. [Google Scholar] [CrossRef]
- Hodge, A.; Fitter, A.H. Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proc. Natl. Acad. Sci. USA 2010, 107, 13754–13759. [Google Scholar] [CrossRef] [Green Version]
- Hélène, J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar]
- Akhtar, M.S.; Siddiqui, Z.A. Biocontrol of a chickpea root-rot disease complex with Glomus intraradices, Pseudomonas putida and Paenibacillus polymyxa. Australas Plant Path. 2007, 36, 175–180. [Google Scholar] [CrossRef]
- Aroca, R.; Porcel, R. How does arbuscular mycorrhizal symbiosis regulate root hydraulic properties and plasma membrane aquaporins in Phaseolus vulgaris under drought, cold or salinity stresses? New Phytol. 2007, 173, 808–816. [Google Scholar] [CrossRef]
- Johnson, N.C.; Graham, J.H.; Smith, F.A. Functioning of mycorrhizal associations along the mutualism parasitism continuum. New Phytol. 1997, 135, 575–586. [Google Scholar] [CrossRef]
- Ahmad, A.; Imran, G.M.; Ding, H.Y.; Iqbal, M.; Cheng, Z.H.; Cai, Z.C. Arbuscular mycorrhizal inoculum coupled with organic substrate induces synergistic effects for soil quality changes, and rhizosphere microbiome structure in long-term monocropped cucumber planted soil. Rhizosphere 2021, 20, 100428. [Google Scholar]
- Linderman, R.G. Mycorrhizae and Plant Health; Pfleger, F.L., Linderman, R.G., Eds.; American Physics Society Press: St. Paul, MN, USA, 1994; pp. 1–25. [Google Scholar]
- Martin, B.N.; Joan, L.; Damase, P.K. Mycorrhizae and rhizobacteria on Precambrian rocky gold mine tailings: I. mine-adapted symbionts promote white spruce health and growth. Front. Plant Sci. 2018, 9, 1267. [Google Scholar]
- Bizos, G.; Efimia, M.; Papatheodorou, T.C.; Ntalli, N.; Vassilis, G.; Aschonitis, N.K. The role of microbial inoculants on plant protection, growth stimulation, and crop productivity of the olive tree (Olea europea L.). Plants 2020, 9, 743. [Google Scholar] [CrossRef]
- Chatzistathis, T.; Orfanoudakis, M.; Alifragis, D.; Therios, I. Colonization of Greek olive cultivars’ root system by arbuscular mycorrhiza fungus: Root morphology, growth, and mineral nutrition of olive plants. Sci. Agric. 2013, 70, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Abd-Alhamid, N.; Hassan, H.S.A.; Haggag, L.F.; Hassan, A.M. Effect of mineral and bio-fertilization on vegetative growth, leaf mineral contents and flowering of manzanillo olive trees. Int. J. ChemTech. Res. 2015, 8, 51–61. [Google Scholar]
- Meng, L.L.; Srivastava, A.K.; Kuca, K.; Wu, Q.S. Earthworm (Pheretima guillelmi)-mycorrhizal fungi (Funneliformis mosseae) association mediates rhizosphere responses in white clover. Appl. Soil. Ecol. 2022, 172, 104371. [Google Scholar] [CrossRef]
- Seifi, E.; Teymoor, Y.S.; Alizadeh, M.; Fereydooni, H. Olive mycorrhization: Influences of genotype, mycorrhiza, and growing periods. Sci. Hortic. Amst. 2014, 180, 214–219. [Google Scholar] [CrossRef]
- Kapulnik, Y.; Tsror, L.; Zipori, I.; Hazanovsky, M.; Wininger, S.; Dag, A. Effect of AMF application on growth, productivity and susceptibility to Verticillium wilt of olives grown under desert conditions. Symbiosis 2010, 52, 103–111. [Google Scholar] [CrossRef]
- Giovannini, C.; Garcia-Mina, J.M.; Ciavatta, C.; Marzadori, C. Ureic nitrogen transformation in multi-layer soil columns treated with urease and nitrification inhibitors. J. Agric. Food Chem. 2009, 57, 4883–4887. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, Y.; Zeeshan, M.; Qin, S.; Ma, J.; Liu, L.; Yang, J.; Zhou, X.; Huang, J. Arbuscular mycorrhizal fungi reverse selenium stress in Zea mays seedlings by improving plant and soil characteristics. Ecotoxicol. Environ. Saf. 2021, 228, 113000. [Google Scholar] [CrossRef] [PubMed]
- Puschel, D.; Rydlova, J.; Sudova, R.; Gryndler, M.; Vosatka, M. The potential of mycorrhizal inoculation and organic amendment to increase yields of Galega orientalis and Helianthus tuberosus in a spoil-bank substrate. J. Plant Nutr. Soil. Sci. 2011, 174, 664–672. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, S.; Huang, H.; Luo, L.; Wen, B. Arsenic accumulation and speciation in maize as affected by inoculation with arbuscular mycorrhizal fungus Glomus mosseae. J. Agric. Food Chem. 2009, 57, 3695–3701. [Google Scholar] [CrossRef]
- Escrig, P.V.; Iglesias, D.J.; Corma, A.; Primo, J.; Primo-Millo, E.; Cabedo, N. Euphorbia characias as bioenergy crop: A study of variations in energy value components according to phenology and water status. J. Agric. Food Chem. 2013, 61, 10096–10109. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, Y.; Hernandez-Viezcas, J.A.; Servin, A.D.; Hong, J.; Niu, G.; Peralta-Videa, J.R.; Duarte-Gardea, M.; Gardea-Torresdey, J.L. Influence of CeO2 and ZnO nanoparticles on cucumber physiological markers and bioaccumulation of Ce and Zn: A life cycle study. J. Agric. Food Chem. 2013, 61, 11945–11951. [Google Scholar] [CrossRef]
- Zafari, M.; Ebadi, A.; Jahanbakhsh, S.; Sedghi, M. Safflower (Carthamus tinctorius) biochemical properties, yield, and oil content affected by 24-epibrassinosteroid and genotype under drought stress. J. Agric. Food Chem. 2020, 68, 6040–6047. [Google Scholar] [CrossRef]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Lear, G.; Dickie, I.; Banks, J.; Boyer, S.; Buckley, H.L.; Buckley, T.R.; Cruickshank, R.; Dopheide, A.; Handley, K.M.; Hermans, S.; et al. Methods for the extraction, storage, amplification and sequencing of DNA from environmental samples. N. Zldn. J. Ecol. 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.; He, X.; Baer, M.; Beirinckx, S.; Tian, T.; Moya, Y.; Zhang, X.; Deichmann, M.; Frey, F.P.; Bresgen, V.; et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 2021, 7, 481–499. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Hu, X.; Li, M.; Miao, J.; Du, J.; Wu, R. Composition and metabolic activities of the bacterial community in shrimp sauce at the flavor-forming stage of fermentation as revealed by metatranscriptome and 16S rRNA gene sequencings. J. Agric. Food Chem. 2016, 64, 2591–2603. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Huttenhower, C. Toward an efficient method of identifying core genes for evolutionary and functional microbial phylogenies. PLoS ONE 2011, 6, e24704. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; VegaThurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wu, S.C.; Cao, Z.H.; Li, Z.G.; Cheung, K.C.; Wong, M.H. Effects of biofertilizer containing N-fixer, P and K solubilizers and AM fungi on maize growth: A greenhouse trial. Geoderma 2004, 125, 155–166. [Google Scholar] [CrossRef]
- Al-Karaki, G.N. Growth of mycorrhizal tomato and mineral acquisition under salt stress. Mycorrhiza 2000, 10, 51–54. [Google Scholar] [CrossRef]
- Chen, M.L.; Yang, G.; Sheng, Y.; Li, P.Y.; Qiu, H.Y.; Zhou, X.T.; Huang, L.Q.; Chao, Z. Glomus mosseae inoculation improves the root system architecture, photosynthetic efficiency and flavonoids accumulation of liquor ice under nutrient stress. Front. Plant Sci. 2017, 8, 931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.S.; Xia, R.X. Arbuscular mycorrhizal fungi influence growth, osmotic adjustment and photosynthesis of citrus under well-watered and water stress conditions. J. Plant Physiol. 2006, 163, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Al-Karaki, G.N.; Hammad, R.; Rusan, M. Response of two tomato cultivars differing in salt tolerance to inoculation with mycorrhizal fungi under salt stress. Mycorrhiza 2001, 11, 43–47. [Google Scholar] [CrossRef]
- Jog, R.; Pandya, M.; Nareshkumar, G.; Rajkumar, S. Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiology 2014, 160 Pt 4, 778–788. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Liu, Y.; Li, Y.; Sun, Y.; Yang, B.; Lai, H.; Xue, Q. Effects and mechanism of two Streptomyces strains on promoting plant growth and increasing grain yield of maize. Chin. J. Appl. Ecol. 2017, 28, 315–326. [Google Scholar]
- Adikari, T.B.; Joseph, C.M.; Yang, G.P. Evaluation of bacteria isolated from rice for plant growth promotion and biological control of seedling disease of rice. Can. J. Microbiol. 2001, 47, 916–924. [Google Scholar] [CrossRef]
- Trivedi, P.; Duan, Y.P.; Wang, N. Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl. Environ. Microbiol. 2010, 76, 3427–3436. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Yap, M.; Behringer, G. Volatile organic compounds emitted by Trichoderma species mediate plant growth. Fungal Biol. Biotechnol. 2016, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Marcela, F.C.; Angelica, Q.; Christian, D.; Christian, S.; Maria, X. Evaluation of actinomycete strains for key traits related with plant growth promotion and mycorrhiza helping activities. Appl. Soil. Ecol. 2010, 45, 209–217. [Google Scholar]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef] [PubMed]
- El-Tarabily, K.A. Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase-producing streptomycete actinomycetes. Plant Soil. 2008, 308, 161–174. [Google Scholar] [CrossRef]
- Vilaene, T.; Langendries, S.; Beirinckx, S.; Maes, M.; Goormactig, S. Streptomyces as a plant’s best friend? FEMS Microbiol. Ecol. 2016, 92, fiw119. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.W.; Gan, Y.T.; Xu, B.L. Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 2019, 19, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.G.; Xu, X.X.; Huo, Y.Q. Trichoderma-inoculation and mowing synergistically altered soil available nutrients, rhizosphere chemical compounds and soil microbial community, potentially driving alfalfa growth. Front. Microbiol. 2018, 9, 3241. [Google Scholar] [CrossRef]
- Miransari, M. Interactions between arbuscular mycorrhizal fungi and soil bacteria. Appl. Microbiol. 2011, 89, 917–930. [Google Scholar] [CrossRef]
- Sharmah, D.; Jha, D.K. Diversity of arbuscular mycorrhizal fungi in undisturbed forest, slash-and-burn field, and monoculture forest of Indo-Burma megadiverse region. Braz. J. Bot. 2014, 37, 339–351. [Google Scholar] [CrossRef]
- Postma, J.; Stevens, L.H.; Wiegers, G.L. Biological control of Pythium aphanidermatum in cucumber with a combined application of Lysobacter enzymogenes strain 3.1T8 and chitosan. Biol. Control 2009, 48, 301–309. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, J.; Zhang, L. Lysobacter ximonensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 786–789. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.F.; Hou, X.Y.; Jiang, C.Y.; Zhai, T.T.; Miao, R.; Deng, J.J.; Yao, Z.H.; Zhang, R.S. A native Trichoderma harzianum strain Th62 displays antagonistic activities against phytopathogenic fungi and promotes the growth of Celosia cristata. Horticult. Environ. Biotechnol. 2021, 62, 169–179. [Google Scholar]
- Pierson, L.S.; Thomashow, L.S. Cloning and hetero logous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30–84. Mol. Plant Microbe Interact. 1992, 5, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Liu, Y.; Li, W.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.H.; Zhang, N.; Miao, Y.Z.; Shen, Q.R.; et al. Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Root Colonization Rate (%) | Plant Height (cm) | Stem Diameter (cm) | Leaf Area (cm2) | Plant Fresh Weight (g plant−1) | Plant Dry Weight (g plant−1) | Relative Water Content (%) | Fruit Yield (kg plant−1) |
|---|---|---|---|---|---|---|---|---|
| Uninoculated control | – | 60.2 | 2.06 | 577 | 327 | 80.7 | 75.2 | 4.31 |
| Inoculation | 23.4 | 65.1 ** | 2.01 | 625 ** | 388 ** | 89.2 ** | 82.3 ** | 4.62 ** |
| Coefficient of variation (%) | 10.4 | 16.2 | 2.6 | 20.4 | 15.5 | 14.4 | 9.2 | 12.8 |
| Treatment | Root Length (m) | Root Volume (cm3) | Root Surface Area (cm2) | Root Diameter (mm) |
|---|---|---|---|---|
| Uninoculated control | 9.4 | 6.2 | 26.8 | 0.8 |
| Inoculation | 12.3 ** | 7.7 ** | 33.2 ** | 1.0 ** |
| Coefficient of variation (%) | 14.2 | 13.9 | 20.1 | 12.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Fu, W.; Sun, C.; Cai, S.; Tang, C. Funneliformis mosseae Inoculation Enhances Cucurbita pepo L. Plant Growth and Fruit Yield by Reshaping Rhizosphere Microbial Community Structure. Diversity 2022, 14, 932. https://doi.org/10.3390/d14110932
Wang J, Fu W, Sun C, Cai S, Tang C. Funneliformis mosseae Inoculation Enhances Cucurbita pepo L. Plant Growth and Fruit Yield by Reshaping Rhizosphere Microbial Community Structure. Diversity. 2022; 14(11):932. https://doi.org/10.3390/d14110932
Chicago/Turabian StyleWang, Junsong, Wenjiang Fu, Chenyu Sun, Shuai Cai, and Cheng Tang. 2022. "Funneliformis mosseae Inoculation Enhances Cucurbita pepo L. Plant Growth and Fruit Yield by Reshaping Rhizosphere Microbial Community Structure" Diversity 14, no. 11: 932. https://doi.org/10.3390/d14110932
APA StyleWang, J., Fu, W., Sun, C., Cai, S., & Tang, C. (2022). Funneliformis mosseae Inoculation Enhances Cucurbita pepo L. Plant Growth and Fruit Yield by Reshaping Rhizosphere Microbial Community Structure. Diversity, 14(11), 932. https://doi.org/10.3390/d14110932





