Strong Genetic Differentiation between Generalist Populations of Venturia inaequalis and Populations from Partially Resistant Apple Cultivars Carrying Rvi3 or Rvi5
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Resistance of Indicator Cultivars
2.2. Pathogenicity Testing by Detached Leaf Assay
2.3. Culturing Monosporic Isolates of V. inaequalis
2.4. DNA Extraction and SSR Marker Analyses
2.5. Statistical Analyses of the Genetic Data
3. Results
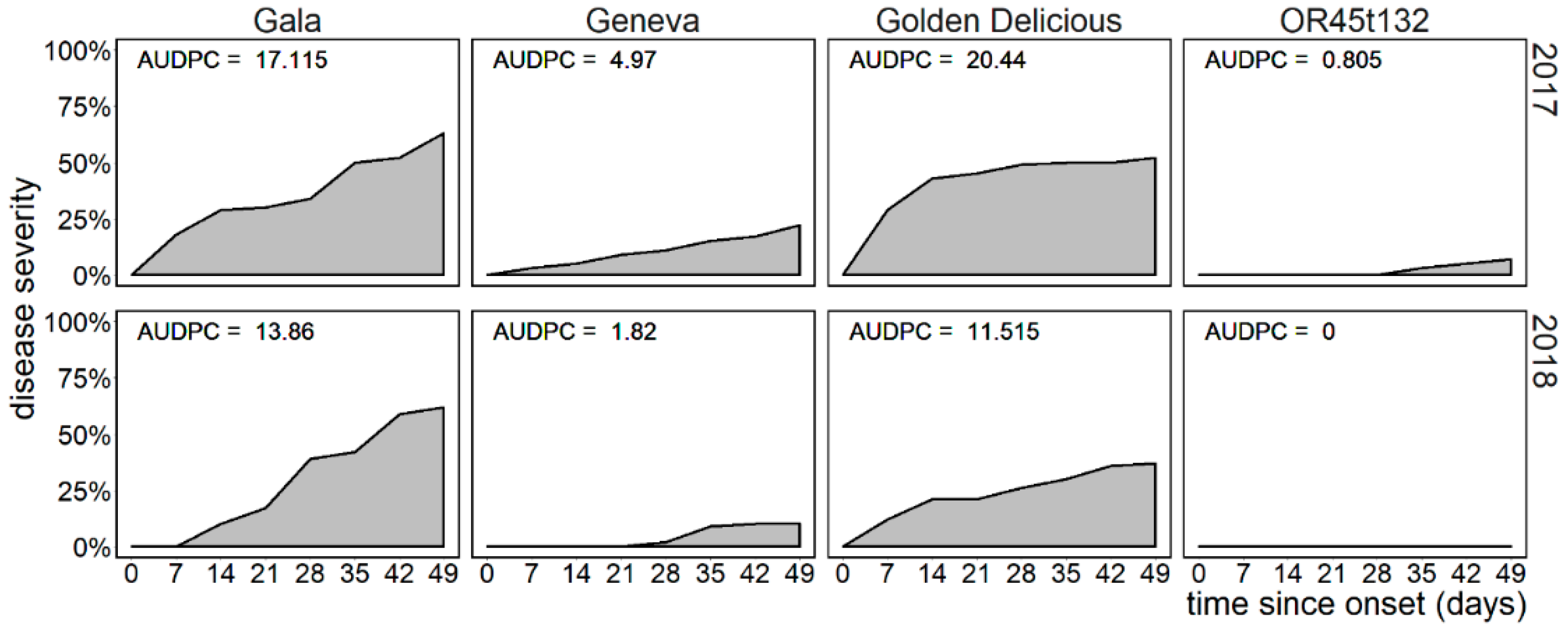
3.1. Field Scab Severity
3.2. Detached Leaf Assay
3.3. Polymorphism of SSR Markers
3.4. Population Genetic Diversity
3.5. Population Structure
3.6. AMOVA and PCoA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacHardy, W.E. Apple Scab: Biology, Epidemiology, and Management; American Phytopathological Society: St. Paul, MN, USA, 1996. [Google Scholar]
- Höfer, M.; Flachowsky, H.; Schröpfer, S.; Peil, A. Evaluation of Scab and Mildew Resistance in the Gene Bank Collection of Apples in Dresden-Pillnitz. Plants 2021, 10, 1227. [Google Scholar] [CrossRef] [PubMed]
- Holb, I.J.; Heijne, B.; Jeger, M.J. Summer epidemics of apple scab: The relationship between measurements and their implications for the development of predictive models and threshold levels under different disease control regimes. J. Phytopathol. 2003, 151, 335–343. [Google Scholar] [CrossRef]
- Patocchi, A.; Wehrli, A.; Dubuis, P.H.; Auwerkerken, A.; Leida, C.; Cipriani, G.; Passey, T.; Staples, M.; Didelot, F.; Philion, V.; et al. Ten years of VINQUEST: First insight for breeding new apple cultivars with durable apple scab resistance. Plant Dis. 2020, 104, 2074–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holb, I.J.; Heijne, B.; Withagen, J.C.M.; Gáll, J.M.; Jeger, M.J. Analysis of summer epidemic progress of apple scab at different apple production systems in the Netherlands and Hungary. Phytopathology 2005, 95, 1001–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosby, J.A.; Janick, J.; Pecknold, P.C.; Korban, S.S.; O’Connor, P.A.; Ries, S.M.; Goffreda, J.; Voordeckers, A. Breeding apples for scab resistance: 1945–1990. Acta Hortic. 1992, 317, 43–70. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.K.; Tellier, A. Plant-parasite coevolution: Bridging the gap between genetics and ecology. Annu. Rev. Phytopathol. 2011, 49, 345–367. [Google Scholar] [CrossRef] [Green Version]
- Bus, V.G.; Rikkerink, E.H.; Caffier, V.; Durel, C.E.; Plummer, K.M. Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus. Annu. Rev. Phytopathol. 2011, 49, 391–413. [Google Scholar] [CrossRef] [Green Version]
- Jänsch, M.; Paris, R.; Amoako-Andoh, F.; Keulemans, W.; Davey, M.W.; Pagliarani, G.; Tartarini, S.; Patocchi, A. A phenotypic, molecular and biochemical characterization of the first cisgenic scab-resistant apple variety ‘Gala’. Plant Mol. Biol. Rep. 2014, 32, 679–690. [Google Scholar] [CrossRef]
- Joshi, S.G.; Schaart, J.G.; Groenwold, R.; Jacobsen, E.; Schouten, H.J.; Krens, F.A. Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol. Biol. 2011, 75, 579–591. [Google Scholar] [CrossRef]
- Schouten, H.J.; Brinkhuis, J.; van der Burgh, A.; Schaart, J.G.; Groenwold, R.; Broggini, G.A.; Gessler, C. Cloning and functional characterization of the Rvi15 (Vr2) gene for apple scab resistance. Tree Genet. Genomes 2014, 10, 251–260. [Google Scholar] [CrossRef]
- Yousaf, Y.; Baldi, P.; Piazza, S.; Patocchi, A.; Malnoy, M. Functional characterization of resistance gene Rvi 12 (Vb) against apple scab. In Proceedings of the 7th International Horticulture Research Conference, Online, 1–30 July 2020. [Google Scholar]
- Bus, V.G.; Laurens, F.N.; Van De Weg, W.E.; Rusholme, R.L.; Rikkerink, E.H.; Gardiner, S.E.; Bassett, H.C.M.; Kodde, L.P.; Plummer, K.M. The Vh8 locus of a new gene-for-gene interaction between Venturia inaequalis and the wild apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila R12740-7A. New Phytol. 2005, 166, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.S.; Mohammed, I.; Sofi, T.A.; Ahanger, F.A.; Shah, M.D.; Ahmad, M.; Hamid, A.; Mir, A.A.; Nabi, A.; Padder, B.A. Distribution of apple scab race flora and identification of resistant sources against Venturia inaequalis in Kashmir. Plant Pathol. J. 2015, 14, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Parisi, L.; Fouillet, V.; Schouten, H.J.; Groenwold, R.; Laurens, F.; Didelot, F.; Evans, K.; Fischer, C.; Gennari, F.; Kemp, H.; et al. Variability of the pathogenicity of Venturia inaequalis in Europe. Acta Hortic. 2004, 663, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Parisi, L.; Lespinasse, Y.; Guillaumes, J.; Krüger, J. A new race of Venturia inaequalis virulent to apples with resistance due to the Vf gene. Phytopathology 1993, 83, 533–537. [Google Scholar] [CrossRef]
- Papp, D.; Gao, L.; Thapa, R.; Olmstead, D.; Khan, A. Field apple scab susceptibility of a diverse Malus germplasm collection identifies potential sources of resistance for apple breeding. CABI Agric. Biosci. 2020, 1, 16. [Google Scholar] [CrossRef]
- Baumgartner, I.O.; Patocchi, A.; Frey, J.E.; Peil, A.; Kellerhals, M. Breeding elite lines of apple carrying pyramided homozygous resistance genes against apple scab and resistance against powdery mildew and fire blight. Plant Mol. Biol. Rep. 2015, 33, 1573–1583. [Google Scholar] [CrossRef]
- Lasserre-Zuber, P.; Caffier, V.; Stievenard, R.; Lemarquand, A.; Le Cam, B. Pyramiding Quantitative Resistance with a Major Resistance Gene in Apple: From Ephemeral to Enduring Effectiveness in Controlling Scab. Plant Dis. 2018, 102, 2220–2223. [Google Scholar] [CrossRef] [Green Version]
- Zelmene, K.; Kārkliņa, K.; Ikase, L.; Lācis, G. Inheritance of Apple (Malus× domestica (L.) Borkh) Resistance against Apple Scab (Venturia inaequalis (Cooke) Wint.) in Hybrid Breeding Material Obtained by Gene Pyramiding. Horticulturae 2022, 8, 772. [Google Scholar] [CrossRef]
- Peil, A.; Patocchi, A.; Hanke, M.V.; Bus, V.G. Apple cultivar Regia possessing both Rvi2 and Rvi4 resistance genes is the source of a new race of Venturia inaequalis. Eur. J. Plant Pathol. 2018, 151, 533–539. [Google Scholar] [CrossRef]
- Bastiaanse, H.; Bassett, H.C.; Kirk, C.; Gardiner, S.E.; Deng, C.; Groenworld, R.; Chagné, D.; Bus, V.G. Scab resistance in ‘Geneva’ apple is conditioned by a resistance gene cluster with complex genetic control. Mol. Plant Pathol. 2016, 17, 159–172. [Google Scholar] [CrossRef]
- Mundt, C.C. Pyramiding for resistance durability: Theory and practice. Phytopathology 2018, 108, 792–802. [Google Scholar] [CrossRef] [Green Version]
- Rex Consortium. Combining selective pressures to enhance the durability of disease resistance genes. Front. Plant Sci. 2016, 7, 1916. [Google Scholar] [CrossRef] [Green Version]
- Stam, R.; McDonald, B.A. When resistance gene pyramids are not durable—The role of pathogen diversity. Mol. Plant Pathol. 2018, 19, 521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madden, L.V.; Hughes, G.; Van Den Bosch, F. The Study of Plant Disease Epidemics; American Phytopathological Society: St. Paul, MN, USA, 2007. [Google Scholar]
- Mills, W. Effect of temperature on the incubation period of apple scab. N.Y. State Coll. Agric. Wkly. News Lett. Insect Pests Plant Dis. 1946, 8, 24–25. [Google Scholar]
- Nicholson, R.L.; Van Scoyoc, S.; Kuc, J.; Williams, E.B. Response of Detached Apple Leaves to Venturia inaequalis. Phytopathology 1972, 63, 649–650. [Google Scholar] [CrossRef]
- Conolly, J.J. Fluorescence Phenotyping of Venturia inaequalis Resistance in Apple. Master’s Thesis, University of Auckland, Auckland, New Zealand, 2022. [Google Scholar]
- Papp, D.; Singh, J.; Gadoury, D.; Khan, A. New North American isolates of Venturia inaequalis can overcome apple scab resistance of Malus floribunda 821. Plant Dis. 2020, 104, 649–655. [Google Scholar] [CrossRef]
- Tenzer, I.; degli Ivanissevich, S.; Morgante, M.; Gessler, C. Identification of microsatellite markers and their application to population genetics of Venturia inaequalis. Phytopathology 1999, 89, 748–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guérin, F.; Franck, P.; Loiseau, A.; Devaux, M.; Le Cam, B. Isolation of 21 new polymorphic microsatellite loci in the phytopathogenic fungus Venturia inaequalis. Mol. Ecol. Notes 2004, 4, 268–270. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.P.; Royo, L.J.; Álvarez, I.; Goyache, F. MolKin v2. 0: A computer program for genetic analysis of populations using molecular coancestry information. J. Hered. 2005, 96, 718–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Liu, J.X. StructureSelector: A web based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Laine, A.L.; Tellier, A. Heterogeneous selection promotes maintenance of polymorphism in host-parasite interactions. Oikos 2008, 117, 1281–1288. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puechmaille, S.J. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations. In Variability within and among Natural Populations; The University of Chicago Press: Chicago, IL, USA, 1978; Volume 4. [Google Scholar]
- Gladieux, P.; Zhang, X.G.; Róland-Ruiz, I.; Caffier, V.; Leroy, T.; Devaux, M.; van Glabeke, A.; Coart, E.; Le Cam, B. Evolution of the population structure of Venturia inaequalis, the apple scab fungus, associated with the domestication of its host. Mol. Ecol. 2010, 19, 658–674. [Google Scholar] [CrossRef]
- Ebrahimi, L.; Fotuhifar, K.B.; Nikkhah, M.J.; Naghavi, M.R.; Baisakh, N. Population genetic structure of apple scab (Venturia inaequalis (Cooke) G. Winter) in Iran. PLoS ONE 2016, 11, e0160737. [Google Scholar] [CrossRef] [Green Version]
- Dar, M.S.; Padder, B.A.; Ahmad, M.; Sofi, T.A.; Mir, A.A.; Nabi, A.; Shah, M.D. Population structure of Venturia inaequalis, a hemibiotrophic fungus, under different host resistance specificities in the Kashmir valley. Arch. Microbiol. 2020, 202, 2245–2253. [Google Scholar] [CrossRef]
- Kaymak, S.; Boyraz, N.; Daniels, J. Molecular markers to evaluate genetic diversity among Venturia inaequalis isolates obtained from apple plantations in Isparta Province. Turk. J. Agric. For. 2016, 40, 489–498. [Google Scholar] [CrossRef]
- Li, X.; Tao, F.; Fan, S.; Li, H.; Yang, J.; Gao, L. Genetic diversity of Venturia inaequalis isolates (Apple scab) in China and U.K. determined by SSR markers. PLoS ONE 2021, 16, e0252865. [Google Scholar] [CrossRef] [PubMed]
- Michalecka, M.; Masny, S.; Leroy, T.; Puławska, J. Population structure of Venturia inaequalis, a causal agent of apple scab, in response to heterogeneous apple tree cultivation. BMC Evol. Biol. 2018, 18, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sitther, V.; Garrido, P.A.; Molineros, J.E.; Garzon, C.D.; Jiménez-Gasco, M.M. Genetic diversity of apple- and crabapple-infecting isolates of Venturia inaequalis in Pennsylvania, the United States, determined by microsatellite markers. For. Pathol. 2018, 48, e12405. [Google Scholar] [CrossRef]
- Sokolova, O.; Moročko-Bičevska, I.; Lācis, G. Genetic Diversity of Venturia inaequalis in Latvia Revealed by Microsatellite Markers. Pathogens 2022, 11, 1165. [Google Scholar] [CrossRef]
- Boehm, E.W.; Freeman, S.; Shabi, E.; Michailides, T.J. Microsatellite primers indicate the presence of asexual populations of Venturia inaequalis in coastal Israeli apple orchards. Phytoparasitica 2003, 31, 236–251. [Google Scholar] [CrossRef]
- Koopman, T.A.; Meitz-Hopkins, J.C.; der Merwe, A.E.B.-V.; Tobutt, K.R.; Bester, C.; Lennox, C.L. Genetic diversity and gene flow of four South African Venturia inaequalis (apple scab) populations. Phytopathology 2017, 107, 455–462. [Google Scholar] [CrossRef] [Green Version]
- Barbara, D.J.; Roberts, A.L.; Xu, X.M. Virulence characteristics of apple scab (Venturia inaequalis) isolates from monoculture and mixed orchards. Plant Pathol. 2008, 57, 552–561. [Google Scholar] [CrossRef]
- Passey, T.A.J.; Shaw, M.W.; Xu, X.M. Differentiation in populations of the apple scab fungus Venturia inaequalis on cultivars in a mixed orchard remain over time. Plant Pathol. 2016, 65, 1133–1141. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Harvey, N.; Roberts, A.; Barbara, D. Population variation of apple scab (Venturia inaequalis) within mixed orchards in the UK. Eur. J. Plant Pathol. 2013, 135, 97–104. [Google Scholar] [CrossRef]
- Lemaire, C.; De Gracia, M.; Leroy, T.; Michalecka, M.; Lindhard-Pedersen, H.; Guerin, F.; Gladieux, P.; Le Cam, B. Emergence of new virulent populations of apple scab from nonagricultural disease reservoirs. New Phytol. 2016, 209, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Leroy, T.; Lemaire, C.; Dunemann, F.; Le Cam, B. The genetic structure of a Venturia inaequalis population in a heterogeneous host population composed of different Malus species. BMC Evol. Biol. 2013, 13, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastiaanse, H.; Muhovski, Y.; Mingeot, D.; Lateur, M. Candidate defense genes as predictors of partial resistance in ‘Président Roulin’against apple scab caused by Venturia inaequalis. Tree Genet. Genomes 2015, 11, 125. [Google Scholar] [CrossRef]
- Durel, C.E.; Parisi, L.; Laurens, F. Does the Vf gene maintain a residual resistance to apple scab despite its breakdown by Venturia inaequalis race 6 strains? In Proceedings of the Eucarpia symposium on Fruit Breeding and Genetics, Dresden, Germany, 6–10 September 1999. [Google Scholar]
- Mushtaq, R.; Pandit, A.; Ali, M.T.; Raja, R.H.S.; Sharma, M.K.; Nazir, N.; Khalil, A. Phenological features of four exotic apple cultivars on M9T337 rootstock under high density plantation in North Himalayan region of India. Curr. J. Appl. Sci. Technol. 2018, 29, 43317. [Google Scholar] [CrossRef]
- Mansoor, S.; Ahmed, N.; Sharma, V.; Jan, S.; Nabi, S.U.; Mir, J.I.; Mir, M.A.; Masoodi, K.Z. Elucidating genetic variability and population structure in Venturia inaequalis associated with apple scab disease using SSR markers. PLoS ONE 2019, 14, e0224300. [Google Scholar] [CrossRef] [Green Version]
- Bock, C.H.; Charlton, N.D.; Shiller, J.; Randall, J.J.; Young, C.A. Population genetic diversity and structure of the pecan scab pathogen, Venturia effusa, on cv. Desirable and native seedlings, and the impact of marker number. Plant Pathol. 2022, 71, 1103–1119. [Google Scholar] [CrossRef]
- Xhaard, C.; Barre’s, B.; Andrieux, A.; Bousset, L.; Halkett, F.; Frey, P. Disentangling the genetic origins of a plant pathogen during disease spread using an original molecular epidemiology approach. Mol. Ecol. 2012, 21, 2383–2398. [Google Scholar] [CrossRef]
- Wu, X.X.; Xu, X.F.; Ma, D.X.; Chen, R.Z.; Li, T.Y.; Cao, Y.Y. Virulence structure and its genetic diversity analyses of Blumeria graminis f. sp. tritici isolates in China. BMC Evol. Biol. 2019, 19, 183. [Google Scholar] [CrossRef]
- Cieplak, M.; Nucia, A.; Ociepa, T.; Okoń, S. Virulence Structure and Genetic Diversity of Blumeria graminis f. sp. avenae from Different Regions of Europe. Plants 2022, 11, 1358. [Google Scholar] [CrossRef]
- Ekroth, A.K.; Rafaluk-Mohr, C.; King, K.C. Host genetic diversity limits parasite success beyond agricultural systems: A meta-analysis. In Proceedings of the Royal Society B, Online, 18 November 2019. [Google Scholar]



| Marker | Na | Ne | I | h |
|---|---|---|---|---|
| 1tc1a | 14 | 6.55 | 2.20 | 0.84 |
| 1tc1b | 8 | 2.12 | 1.18 | 0.52 |
| 1tc1g | 22 | 14.97 | 2.87 | 0.93 |
| 1aac3b | 2 | 1.09 | 0.18 | 0.08 |
| Vitg11/70 | 7 | 3.69 | 1.48 | 0.72 |
| Vicacg8/42 | 10 | 4.51 | 1.74 | 0.77 |
| Vica10/154 | 14 | 3.79 | 1.81 | 0.73 |
| Population | Host Cultivar | N | Nh | Na | Ne | I | h | PIC |
|---|---|---|---|---|---|---|---|---|
| Vi-Gala | Gala | 23 | 22 | 7.42 | 4.79 | 1.61 | 0.70 | 8.89 |
| Vi-Geneva | Geneva | 14 | 12 | 3.00 | 2.16 | 0.78 | 0.44 | 8.53 |
| Vi-GD | Golden Delicious | 10 | 10 | 5.00 | 4.00 | 1.34 | 0.64 | 8.86 |
| Vi-OR | OR45T132 | 19 | 12 | 2.28 | 1.72 | 0.48 | 0.27 | 8.23 |
| Vi-Gala | Vi-Geneva | Vi-GD | Vi-OR | |
|---|---|---|---|---|
| - | 0.001 | 0.152 * | 0.001 | Vi-Gala |
| 0.104 | - | 0.001 | 0.001 | Vi-Geneva |
| 0.020 | 0.162 | - | 0.001 | Vi-GD |
| 0.286 | 0.410 | 0.347 | - | Vi-OR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papp, D.; Gangadharappa Harigondra, S.; Paredes, C.; Karacs-Végh, A.; Penksza, K.; T.-Járdi, I.; Papp, V. Strong Genetic Differentiation between Generalist Populations of Venturia inaequalis and Populations from Partially Resistant Apple Cultivars Carrying Rvi3 or Rvi5. Diversity 2022, 14, 1050. https://doi.org/10.3390/d14121050
Papp D, Gangadharappa Harigondra S, Paredes C, Karacs-Végh A, Penksza K, T.-Járdi I, Papp V. Strong Genetic Differentiation between Generalist Populations of Venturia inaequalis and Populations from Partially Resistant Apple Cultivars Carrying Rvi3 or Rvi5. Diversity. 2022; 14(12):1050. https://doi.org/10.3390/d14121050
Chicago/Turabian StylePapp, David, Shambhulinga Gangadharappa Harigondra, Cristina Paredes, Anita Karacs-Végh, Károly Penksza, Ildikó T.-Járdi, and Viktor Papp. 2022. "Strong Genetic Differentiation between Generalist Populations of Venturia inaequalis and Populations from Partially Resistant Apple Cultivars Carrying Rvi3 or Rvi5" Diversity 14, no. 12: 1050. https://doi.org/10.3390/d14121050
APA StylePapp, D., Gangadharappa Harigondra, S., Paredes, C., Karacs-Végh, A., Penksza, K., T.-Járdi, I., & Papp, V. (2022). Strong Genetic Differentiation between Generalist Populations of Venturia inaequalis and Populations from Partially Resistant Apple Cultivars Carrying Rvi3 or Rvi5. Diversity, 14(12), 1050. https://doi.org/10.3390/d14121050







