Conservation and Phylogeography of Plants: From the Mediterranean to the Rest of the World
Abstract
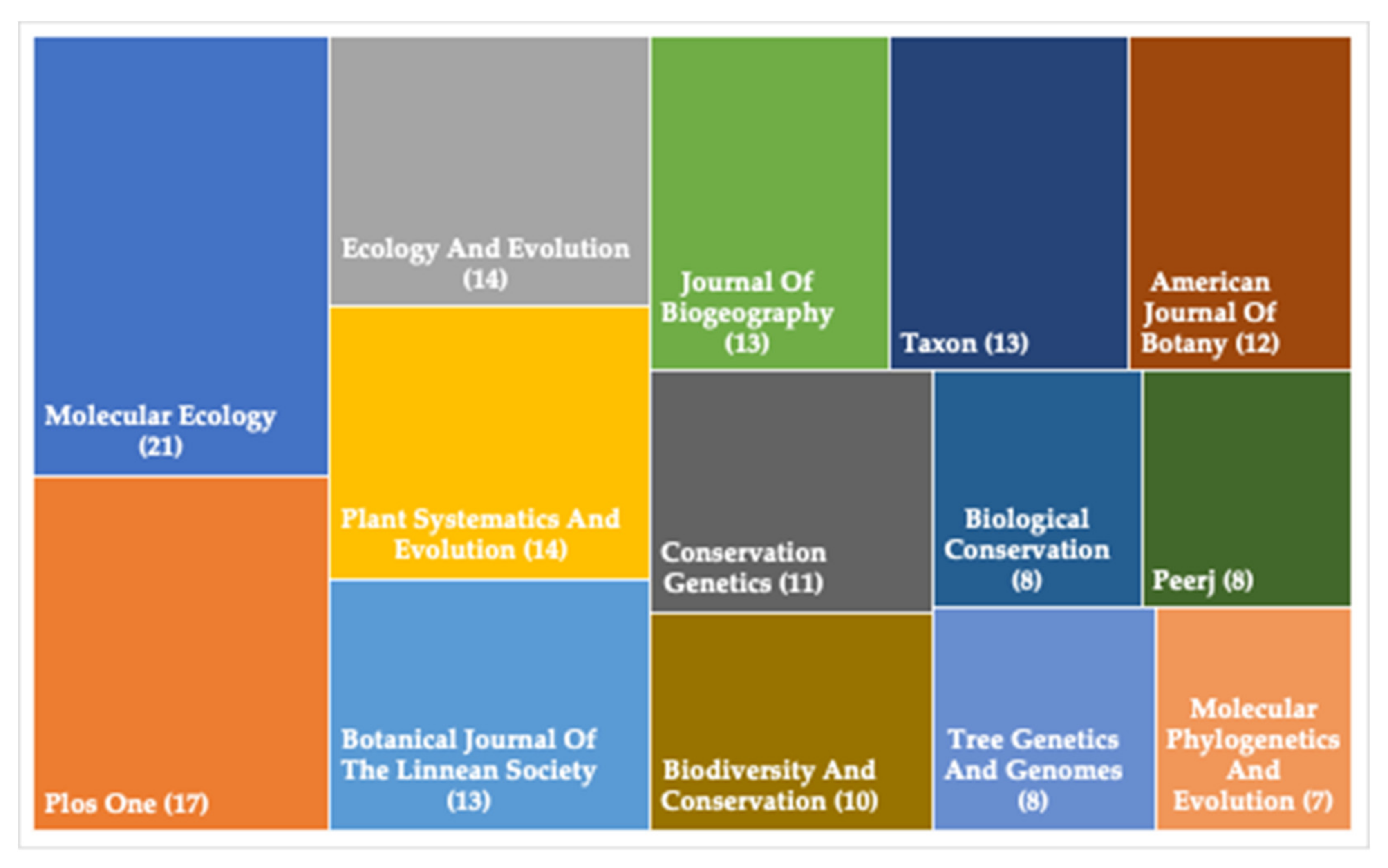
:1. Phylogeographical Studies on Plant Species
2. The Mediterranean Biome, Phylogeography Studies and Plant Conservation
Phylogeography and Conservation in MTEs
3. The Convergence of Phylogeography and Conservation
3.1. Phylogeography and In Situ Conservation
3.2. Phylogeography and Ex Situ Conservation
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A
| Authors | Year | Taxa | Area | Genetic Technique | MTEs |
|---|---|---|---|---|---|
| Prentice et al. | 2003 | Silene hifacensis | Spain | Allozymes and cpDNA | Western Mediterranean |
| Lihová et al. | 2004 | Cardamine amara | Eurasia | AFLP | Mediterranean |
| González-Astorga et al. | 2005 | Dioon angustifolium | Mexico | Allozymes | - |
| Segarra-Moragues et al. | 2005 | Borderea chouardii | Spain | SSRs and RAPDs | Western Mediterranean |
| Grassi et al. | 2006 | Vitis vinifera subsp. silvestris | Mediterranean | SSRs | Mediterranean |
| Pérez-Collazos & Catalán | 2006 | Vella pseudocytisus subsp. paui | Spain | Allozymes and AFLP | Western Mediterranean |
| Agrimonti et al. | 2007 | Myrtus communis | Sardinia and Calabria | AFLP | Central Mediterranean |
| Bucci et al. | 2007 | Pinus pinaster | Western Mediterranean area | SSRs | Western Mediterranean |
| Qiu et al. | 2007 | Dipteronia dyerana | China | ISSRs | - |
| Setsuko et al. | 2007 | Magnolia stellata | Japan | nSSR and cpSSR | - |
| Pérez-Collazos et al. | 2008 | Boleum asperum | Spain | AFLP | Western Mediterranean |
| Chen et al. | 2009 | Rheum tanguticum | China | SSRs | - |
| González-Pérez et al. | 2009 | Anagyris latifolia | Canary Islands | RAPD | Macaronesia |
| Jordán-Pla et al. | 2009 | Leucojum valentinum | Spain | RAPD | Western Mediterranean |
| Rasmussen et al. | 2010 | Abies religiosa subsp. mexicana | Mexico | SSRs | - |
| Segarra-Moragues & Catalán | 2010 | Borderea pyrenaica | Pyrenees | SSRs | Western Mediterranean |
| Douaihy et al. | 2011 | Juniperus excelsa | E Mediterranean | nSSRs | Eastern Mediterranean |
| Ferreira et al. | 2011 | Picconia azorica | Macaronesian islands | RFLPs and SSRs | Macaronesia |
| Migliore et al. | 2011 | Mercurialis corsica | Corsica | AFLP and cpDNA | Central Mediterranean |
| Aguirre-Planter et al. | 2012 | Abies spp. | Mesoamerica | cpDNA | - |
| del Hoyo et al. | 2012 | Glandora oleifolia | Pyrenees | Allozymes and RAPD | Western Mediterranean |
| Juan et al. | 2012 | Juniperus oxycedrus subsp. macrocarpa | Iberian Peninsula | AFLP and cpDNA | Western Mediterranean |
| Nicoletti et al. | 2012 | Campanula sabatia | Italy | AFLP | Central Mediterranean |
| Zhao et al. | 2012 | Leucomeris decora | China | cpDNA | - |
| Balnco-Pastor et al. | 2013 | Linaria glacialis | Spain | cpDNA | Western Mediterranean |
| Dettori et al. | 2013 | Ferula arrigonii | Corsica and Sardinia | AFLP | Central Mediterranean |
| Lesser et el. | 2013 | Pinus ponderosa | EEUU | nSSRs | - |
| Lopez & Barreiro | 2013 | Centaurea borjae | N Spain | AFLP and cpDNA | Western Mediterranean |
| Sánchez-Gómez et al. | 2013 | Tetraclinis articulata | Western Mediterranean | ISSRs | Western Mediterranean |
| Carcía-Castaño et al. | 2014 | Chamaerops humilis | Central and Western Mediterranean | Central and Western Mediterranean | |
| Christe et al. | 2014 | Zelkova spp. | Sicily, Crete and Transcaucasia | cpDNA | Central and Eastern Mediterranean |
| Fernández-Mazuecos et al. | 2014 | Naufraga balearica | Majorca | AFLP and cpDNA | Western Mediterranean |
| Liu et al. | 2014 | Acer mono var. mono | China | nSSRs | - |
| Poudel et al. | 2014 | Taxus spp. | Himalaya | cpDNA and SSRs | - |
| Poudel et al. | 2014 | Taxus contorta | Pakistan | cpDNA and SSRs | - |
| Rumeu et al. | 2014 | Juniperus cedrus | Canary Islands | AFLP and cpDNA | Macaronesia |
| Sánchez et al. | 2014 | Pinus caribaea var. bahamensis | Caribe | cpDNA and nDNA | - |
| Bjedov et al. | 2015 | Vaccinum spp. | E Europe | RAPD | Eastern Mediterranean |
| Cánovas et al. | 2015 | Viola cazorlensis | Spain | ISSRs | Western Mediterranean |
| Coates et al. | 2015 | Banksia brownii | Canada | nSSRs | - |
| Deacon et al. | 2015 | Quercus oleoides | Costa Rica | SSRs | - |
| Gentili et al. | 2015 | Ribes spp. | Sardinia | ISSRs | Central Mediterranean |
| Giovino et al. | 2015 | Pacratium maritimum | Europe | cpDNA | Mediterranean |
| Huerta-Ramos et al. | 2015 | Ipomoea sagittata | Mexico | cpADN and ITS | - |
| Larridon et al. | 2015 | Copiapoa spp. | S America | cpDNA | Chile |
| Miao et al. | 2015 | Taxus yunnanensis | China | SSRs | - |
| Silva et al. | 2015 | Sonchus section Pustulati | W Mediterranean | AFLP, cpDNA and ITS | Western Mediterranean |
| Vitales et al. | 2015 | Cheirolophus uliginosus | Iberian Peninsula | AFLP, cpDNA and ITS | Western Mediterranean |
| Kajtoch et al. | 2016 | 18 taxa | E Europe | - | Eastern Mediterranean |
| Martin et al. | 2016 | Silene nutans | Europe | SNPs and nSSRs | Mediterranean |
| Martín-Hernanz et al. | 2016 | Coronopus navasii | Spain | cpDNA and ITS | Western Mediterranean |
| Peñas et al. | 2016 | Astragalus edulis | W Mediterranean | AFLP and cpDNA | Western Mediterranean |
| Plenk et al. | 2016 | Gentianella bohemica | C Europe | AFLP | Central Mediterranean |
| Rešetnik et al. | 2016 | Salvia officinalis | Eastern Europe | SSRs | Eastern Mediterranean |
| Wen et al. | 2016 | Salsola junatovii | China | cpDNA and ITS | - |
| Bao et al. | 2017 | Prunus mira | Europe | SSRs | Mediterranean |
| Belletti et al. | 2017 | Abies alba | Italy | SSRs | Central Mediterranean |
| Bouchard et al. | 2017 | Dryopteris fragrans | North America | ISSRs | - |
| Duwe et al. | 2017 | Arnica montana | Europe | nSSR | Mediterranean |
| Frey et al. | 2017 | Trapa natans | C Europe | AFLP and ITS | - |
| Guzmán et al. | 2017 | Galvezia leucantha | Galapagos Islands | cpDNA and ITS | - |
| Hrivnák et al. | 2017 | Abies taxa | E Mediterranean | nSSRs | Eastern Mediterranean |
| Hu et al. | 2017 | Juglans hopeiensis | China | cpDNA and nDNA | - |
| Jiménez-Mejías etal. | 2017 | Castrilanthemum debeauxii | Spain | AFLP and cpDNA | Western Mediterranean |
| Kwak et al. | 2017 | Abies koreana | Korea | SSRs | - |
| Lázaro-Nogal et al. | 2017 | Cneorum tricoccon | Western Mediterranean | SSRs | Western Mediterranean |
| Pouget et al. | 2017 | Acis nicaeensis | French–Italian Riviera | cpDNA | Central Mediterranean |
| Sanz et al. | 2017 | Artemisia umbelliformis | W Europe | AFLP | Western Mediterranean |
| Wei et al. | 2017 | Camellia flavida | China | nDNA and cpDNA | - |
| Chung et al. | 2018 | Orchids | Korea | Allozymes | - |
| Chung et al. | 2018 | Lilium cernuum | Nort east Asia | Allozymes | - |
| De Luca et al. | 2018 | Phaseolus vulgaris | Italy | nSSRs | Central Mediterranean |
| Gentili et al. | 2018 | Leucojum aestivum | N Italy | AFLP | Central Mediterranean |
| Gentili et al. | 2018 | Ribes sardoum | Cerdeña | SSRs | Central Mediterranean |
| Gutiérrez-Ortega et al. | 2018 | Dioon sonorense | west coast Mexico | ISSRs | - |
| Mairal et al. | 2018 | Canarina eminii and Canarina canariensis | Afro-Macaronesian forests | nSSR | Western Mediterranean & Macaronesia |
| Martín-Hernanz et al. | 2018 | Helianthemum genus | Spain | SSRs | Western Mediterranean |
| Menezes et al. | 2018 | Campanulaceae | Macaronesian islands | cpDNA and ITS | Macaronesia |
| Pelser et al. | 2018 | Rafflesia speciosa | Panay and Negros Islands | SSRs | - |
| Sekiewicz et al. | 2018 | Cuppresus atlantica, C. dupreziana and C. sempervirens | Mediterranean | nSSRs and cpDNA | Mediterranean |
| Tamaki et al. | 2018 | Pseudotsuga japonica | Japan | SSRs | - |
| Van Rossum et al. | 2018 | Silene nutans | Europe | cpDNA | Mediterranean |
| Wang et al. | 2018 | Bretschneidera sinensis | Asia | cpDNA | - |
| Yan et al. | 2018 | Quercus section Cyclobalanopsis | East Asia | cpDNA | - |
| Bezemer et al. | 2019 | Eucalyptus caesia | south-west Australia | SSRs | Australia |
| Gargiulo et al. | 2019 | Asperula crassifolia | Western Mediterranean | cpDNA and SSRs | Western Mediterranean |
| Grdiša et al. | 2019 | Sideritis scardica | southern Balkan Peninsula | AFLP | Central Mediterranean |
| Kusuma et al. | 2019 | Vatica bantamensis | Indonesia | ISSRs | - |
| Louati et al. | 2019 | Argania spinosa | Tunisia | ISSRs | Southern Mediterranean |
| Ramírez-Rodríguez et al. | 2019 | Delphinium fissum subsp. Sordidum | Spain | cpDNA | Western Mediterranean |
| Romdhane et al. | 2019 | Pennisetum glaucum | Tunisia | nSSRs | Southern Mediterranean |
| Rutherford et al. | 2019 | Eucalyptus tetrapleura | New South Wales | SNPs | - |
| Stefenon et al. | 2019 | Araucaria angustifolia | Brazil | cpDNA | - |
| Walas et al. | 2019 | Aesculus hippocastanum | Greece | nSSRs | Central Mediterranean |
| Xu et al. | 2019 | Paeonia qiui, P. jishanensis, and P. rockii | China | nSSRs and cpDNA | - |
| Zhao et al. | 2019 | Tugarinovia mongolica | Asia | cpDNA | - |
| Amaral Fraga et al. | 2020 | Coleocephalocereus purpureus | Eastern Brazil | SSRs | - |
| Durán et al. | 2020 | Dracaena spp. | Macaronesian islands | cpDNA | Macaronesia |
| Fassou et al. | 2020 | Helleborus odorus subsp. cyclophyllus | Balkan Peninsula | ISSRs | Central Mediterranean |
| Galuszynski & Potts | 2020 | - | southern Cape of South Africa | - | South Africa |
| Kropf et al. | 2020 | Adonis vernalis | Central Europe | AFLP and cpDNA | - |
| Kvesić et al. | 2020 | Acer campestre | Bosnia and Herzegovina | SSRs | Central Mediterranean |
| Liber et al. | 2020 | Degenia velebitica | north-western Dinaric Alps | AFLP | - |
| López-Alvarado et al. | 2020 | Centaurea spp. | Central Mediterranean | Central Mediterranean | |
| Meloni et al. | 2020 | Ruta corsica and R. lamarmorae | Corsica and Sardinia | SSRs | Central Mediterranean |
| Sękiewicz et al. | 2020 | Cupressus atlantica | High Atlas | SSRs | Western Mediterranean |
| Shahzad et al. | 2020 | Dipteronia sinensis | Qinling Mountains | plastid genome and SNPs | - |
| Urquía et al. | 2020 | Psidium galapageium | Galapagos Islands | SSRs | - |
| Alipour et al. | 2021 | Populus caspica | Hyrcanian forests | SSRs | - |
| Bobo-Pinilla et al. | 2021 | Jacobaea auricula | Spain | AFLP and cpDNA | Western Mediterranean |
| Bobo-Pinilla et al. | 2021 | Astragalus edulis | western Europe | AFLP and cpDNA | Western Mediterranean |
| Casazza et al. | 2021 | Lilium pomponium | Maritime and Ligurian Alps | AFLP | - |
| Culshaw et al. | 2021 | Camptoloma genus | Rand flora distribution | cpDNA and ITS | Mediterranean |
| Freitas da Costa et al. | 2021 | Araucaria angustifolia | Southern Brazil | SSRs | - |
| Galuszynski | 2021 | Cyclopia genus | southern Cape of South Africa | cpDNA | South Africa |
| Garcia-Jacas et al. | 2021 | Seseli farrenyi | W Mediterranean basin | SSRs | Western Mediterranean |
| Hellwig et al. | 2021 | Pisum fulvum | Israel and the Palestinian territories | RAD | Eastern Mediterranean |
| Jones et al. | 2021 | Astragalus spp. | Utah, USA | RAD | - |
| Kim et al. | 2021 | Fraxinus chiisanensis | Korea | cpDNA and ITS | - |
| Kougioumoutzis et al. | 2021 | 7043 native plant taxa | Greece | Multiple DNA markers | Central Mediterranean |
| Lin et al. | 2021 | Pinus subsect. strobus | China | cpDNA and mtDNA | - |
| Lin et al. | 2021 | Myripnois dioica | Northern China | RAD | - |
| Medail et al. | 2021 | Acis nicaeensis | French-Italian Riviera | cpDNA | Central Mediterranean |
| Roxo et al. | 2021 | subtribe Daucinae | Macaronesian islands | Genome size | Macaronesia |
| Vaculná et al. | 2021 | Adenophora liliifolia | Central Europe | AFLP and cpDNA | - |
| Yun & Kim | 2021 | Saussurea polylepis | Korea | SSRs | - |
| Žerdoner Čalasan et al. | 2021 | Sisymbrium genus | Old-World | cpDNA and ITS | Mediterranean |
References
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; Neigel, J.E.; Reeb, C.A.; Saunders, N.C. Intraspecific Phylogeography: The Mitochondrial DNA Bridge Between Population Genetics and Systematics. Annu. Rev. Ecol. Syst. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000; ISBN 9780674666382. [Google Scholar]
- Riddle, B.R.; Hafner, D.J. Phylogeography in Historical Biogeography: Investigating the Biogeographic Histories of Populations, Species, and Young Biotas. In Biogeography in a Changing World; Ebach, M., Tangney, R., Eds.; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and Community Ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef] [Green Version]
- Hickerson, M.J.; Carstens, B.C.; Cavender-Bares, J.; Crandall, K.A.; Graham, C.H.; Johnson, J.B.; Rissler, L.; Victoriano, P.F.; Yoder, A.D. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol. Phylogenet. Evol. 2010, 54, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Petit, R.J.; Hampe, A.; Cheddadi, R. Climate changes and tree phylogeography in the Mediterranean. Taxon 2005, 54, 877–885. [Google Scholar] [CrossRef]
- Sanmartín, I. Historical Biogeography: Evolution in Time and Space. Evol. Educ. Outreach 2012, 5, 555–568. [Google Scholar] [CrossRef] [Green Version]
- Buisson, E.; De Almeida, T.; Durbecq, A.; Arruda, A.J.; Vidaller, C.; Alignan, J.; Toma, T.S.P.; Hess, M.C.M.; Pavon, D.; Isselin-Nondedeu, F.; et al. Key issues in Northwestern Mediterranean dry grassland restoration. Restor. Ecol. 2021, 29. [Google Scholar] [CrossRef]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 2015, 1. [Google Scholar] [CrossRef] [Green Version]
- Forest, F.; Grenyer, R.; Rouget, M.; Davies, T.J.; Cowling, R.M.; Faith, D.P.; Balmford, A.; Manning, J.C.; Procheş, Ş.; van der Bank, M.; et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 2007, 445, 757–760. [Google Scholar] [CrossRef]
- Médail, F. Plant Biogeography and Vegetation Patterns of the Mediterranean Islands. Bot. Rev. 2021. [CrossRef]
- Avise, J.C. Phylogeography: Retrospect and prospect. J. Biogeogr. 2009, 36, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Kidd, D.M.; Ritchie, M.G. Phylogeographic information systems: Putting the geography into phylogeography. J. Biogeogr. 2006, 33, 1851–1865. [Google Scholar] [CrossRef]
- Laikre, L.; Hoban, S.; Bruford, M.W.; Segelbacher, G.; Allendorf, F.W.; Gajardo, G.; Rodríguez, A.G.; Hedrick, P.W.; Heuertz, M.; Hohenlohe, P.A.; et al. Post-2020 goals overlook genetic diversity. Science 2020, 367, 1083–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. A Primer of Conservation Genetics; Cambridge University Press: Cambridge, UK, 2004; ISBN 9780521831109. [Google Scholar]
- Holderegger, R.; Balkenhol, N.; Bolliger, J.; Engler, J.O.; Gugerli, F.; Hochkirch, A.; Nowak, C.; Segelbacher, G.; Widmer, A.; Zachos, F.E. Conservation genetics: Linking science with practice. Mol. Ecol. 2019, 28, 3848–3856. [Google Scholar] [CrossRef] [Green Version]
- Habel, J.C.; Assmann, T.; Schmitt, T.; Avise, J.C. Relict Species: From Past to Future. In Relict Species; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–5. [Google Scholar]
- Ryder, O.A. Species conservation and systematics: The dilemma of subspecies. Trends Ecol. Evol. 1986, 1, 9–10. [Google Scholar] [CrossRef]
- Moritz, C. Defining “Evolutionarily Significant Units”. Tree Genet. Genomes 1994, 9, 373–375. [Google Scholar]
- Pérez-Collazos, E.; Segarra-Moragues, J.G.; Catalán, P. Two approaches for the selection of Relevant Genetic Units for Conservation in the narrow European endemic steppe plant Boleum asperum (Brassicaceae). Biol. J. Linn. Soc. 2008, 94, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Médail, F.; Baumel, A. Using phylogeography to define conservation priorities: The case of narrow endemic plants in the Mediterranean Basin hotspot. Biol. Conserv. 2018, 224, 258–266. [Google Scholar] [CrossRef]
- Salmerón-Sánchez, E.; Mendoza-Fernández, A.J.; Lorite, J.; Mota, J.F.; Peñas, J. Plant conservation in Mediterranean-type ecosystems. Mediterr. Bot. 2021, 42, e71333. [Google Scholar] [CrossRef]
- Medail, F.; Quezel, P. Hot-Spots Analysis for Conservation of Plant Biodiversity in the Mediterranean Basin. Ann. Missouri Bot. Gard. 1997, 84, 112. [Google Scholar] [CrossRef]
- Medail, F.; Quezel, P. Biodiversity hotspots in the Mediterranean Basin: Setting global conservation priorities. Conserv. Biol. 1999, 13, 1510–1513. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.; Koenig, K.; Bunting, G.; Costanza, J.; Williams, K.J. Biodiversity Hotspots (version 2016.1) (2016.1) [Data set]. Zenodo 2016. [Google Scholar] [CrossRef]
- Esler, K.J.; Jacobsen, A.L.; Pratt, R.B. The Biology of Mediterranean-Type Ecosystems; Oxford University Press: Oxford, UK, 2018; Volume 1, ISBN 9780198739135. [Google Scholar]
- Hopper, S.D.; Gioia, P. The Southwest Australian Floristic Region: Evolution and Conservation of a Global Hot Spot of Biodiversity. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 623–650. [Google Scholar] [CrossRef]
- Graham, N.E.; Ammann, C.M.; Fleitmann, D.; Cobb, K.M.; Luterbacher, J. Support for global climate reorganization during the “Medieval Climate Anomaly”. Clim. Dyn. 2011, 37, 1217–1245. [Google Scholar] [CrossRef] [Green Version]
- Underwood, E.C.; Viers, J.H.; Klausmeyer, K.R.; Cox, R.L.; Shaw, M.R. Threats and biodiversity in the mediterranean biome. Divers. Distrib. 2009, 15, 188–197. [Google Scholar] [CrossRef]
- Archibold, O.W. Mediterranean Ecosystems: Ecology of World Vegetation; Chapman Hall: London, UK, 1995; ISBN 0412442906. [Google Scholar]
- Schultz, J. The Ecozones of the World. The Ecological Divisions of the Geosphere; Springer: Berlin/Heidelberg, Germany, 1995; ISBN 9783662031612. [Google Scholar]
- Cowling, R.M.; Ojeda, F.; Lamont, B.B.; Rundel, P.W.; Lechmere-Oertel, R. Rainfall reliability, a neglected factor in explaining convergence and divergence of plant traits in fire-prone mediterranean-climate ecosystems. Glob. Ecol. Biogeogr. 2005, 14, 509–519. [Google Scholar] [CrossRef]
- Cowling, R.M.; MacDonald, I.A.W.; Simmons, M.T. The Cape Peninsula, South Africa: Physiographical, biological and historical background to an extraordinary hot-spot of biodiversity. Biodivers. Conserv. 1996, 5, 527–550. [Google Scholar] [CrossRef]
- Linder, H.P. The radiation of the Cape flora, southern Africa. Biol. Rev. 2003, 78, S1464793103006171. [Google Scholar] [CrossRef]
- Mendoza-Fernández, A.J.; Martínez-Hernández, F.; Salmerón-Sánchez, E.; Pérez-García, F.J.; Teruel, B.; Merlo, M.E.; Mota, J.F. The Relict Ecosystem of Maytenus senegalensis subsp. europaea in an Agricultural Landscape: Past, Present and Future Scenarios. Land 2020, 10, 1. [Google Scholar] [CrossRef]
- Vargas, P.; Nogales, M.; Jaramillo, P.; Olesen, J.M.; Traveset, A.; Heleno, R. Plant colonization across the Galápagos Islands: Success of the sea dispersal syndrome. Bot. J. Linn. Soc. 2014, 174, 349–358. [Google Scholar] [CrossRef]
- Verdú, M.; Dávila, P.; García-Fayos, P.; Flores-Hernández, N.; Valiente-Banuet, A. ‘Convergent’ traits of mediterranean woody plants belong to pre-mediterranean lineages. Biol. J. Linn. Soc. 2003, 78, 415–427. [Google Scholar] [CrossRef] [Green Version]
- Valente, L.M.; Vargas, P. Contrasting evolutionary hypotheses between two mediterranean-climate floristic hotspots: The Cape of southern Africa and the Mediterranean Basin. J. Biogeogr. 2013, 40, 2032–2046. [Google Scholar] [CrossRef]
- Rundel, P.W.; Arroyo, M.T.K.; Cowling, R.M.; Keeley, J.E.; Lamont, B.B.; Vargas, P. Mediterranean Biomes: Evolution of Their Vegetation, Floras, and Climate. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 383–407. [Google Scholar] [CrossRef]
- Mittermeier, R.A. Biodiversity Hotspots and Major Tropical Wilderness Areas: Approaches to Setting Conservation Priorities. Conserv. Biol. 1998, 12, 516–520. [Google Scholar] [CrossRef]
- Harrison, S.; Noss, R. Endemism hotspots are linked to stable climatic refugia. Ann. Bot. 2017, 119, 207–214. [Google Scholar] [CrossRef]
- Trew, B.T.; Maclean, I.M.D. Vulnerability of global biodiversity hotspots to climate change. Glob. Ecol. Biogeogr. 2021, 30, 768–783. [Google Scholar] [CrossRef]
- Esler, D.; Ballachey, B.E.; Matkin, C.; Cushing, D.; Kaler, R.; Bodkin, J.; Monson, D.; Esslinger, G.; Kloecker, K. Timelines and mechanisms of wildlife population recovery following the Exxon Valdez oil spill. Deep Sea Res. Part II Top. Stud. Oceanogr. 2018, 147, 36–42. [Google Scholar] [CrossRef]
- Sala, O.E.; Stuart Chapin, F., III; Armesto, J.J.; Berlow, E.; Bloomfield, J.; Dirzo, R.; Huber-Sanwald, E.; Huenneke, L.F.; Jackson, R.B.; Kinzig, A.; et al. Global Biodiversity Scenarios for the Year 2100. Science 2000, 287, 1770–1774. [Google Scholar] [CrossRef]
- Lozano, F.D.; Atkins, K.J.; Moreno Sáiz, J.C.; Sims, A.E.; Dixon, K. The nature of threat category changes in three Mediterranean biodiversity hotspots. Biol. Conserv. 2013, 157, 21–30. [Google Scholar] [CrossRef]
- Mendoza-Fernández, A.J.; Salmerón-Sánchez, E.; Lorite, J.; Mota, J.F.; Peñas, J. Plant Conservation Biology: A view from the Mediterranean ecoregions. Mediterr. Bot. 2021, 42, e71209. [Google Scholar] [CrossRef]
- Moreira, F.; Allsopp, N.; Esler, K.J.; Wardell-Johnson, G.; Ancillotto, L.; Arianoutsou, M.; Clary, J.; Brotons, L.; Clavero, M.; Dimitrakopoulos, P.G.; et al. Priority questions for biodiversity conservation in the Mediterranean biome: Heterogeneous perspectives across continents and stakeholders. Conserv. Sci. Pract. 2019, 1, e118. [Google Scholar] [CrossRef]
- Brooks, T.M.; Mittermeier, R.A.; da Fonseca, G.A.B.; Gerlach, J.; Hoffmann, M.; Lamoreux, J.F.; Mittermeier, C.G.; Pilgrim, J.D.; Rodrigues, A.S.L. Global Biodiversity Conservation Priorities. Science 2006, 313, 58–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, R.L.; Underwood, E.C. The Importance of Conserving Biodiversity Outside of Protected Areas in Mediterranean Ecosystems. PLoS ONE 2011, 6, e14508. [Google Scholar] [CrossRef]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.G.; Cosson, J.F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef]
- Comes, H.P.; Abbott, R.J. The relative importance of historical events and gene flow on the population structure of a mediterranean ragwort, Senecio gallicus (Asteraceae). Evolution 1998, 52, 355–367. [Google Scholar] [CrossRef]
- Fois, M.; Cuena-Lombraña, A.; Bacchetta, G. Knowledge gaps and challenges for conservation of Mediterranean wetlands: Evidence from a comprehensive inventory and literature analysis for Sardinia. Aquat. Conserv. Mar. Freshw. Ecosyst. 2021, 31, 2621–2631. [Google Scholar] [CrossRef]
- Massó, S.; López-Pujol, J.; Vilatersana, R. Reinterpretation of an endangered taxon based on integrative taxonomy: The case of Cynara baetica (Compositae). PLoS ONE 2018, 13, e0207094. [Google Scholar] [CrossRef] [PubMed]
- Baumel, A.; Médail, F.; Juin, M.; Paquier, T.; Clares, M.; Laffargue, P.; Lutard, H.; Dixon, L.; Pires, M. Population genetic structure and management perspectives for Armeria belgenciencis, a narrow endemic plant from Provence (France). Plant Ecol. Evol. 2020, 153, 219–228. [Google Scholar] [CrossRef]
- Jiang, T.; Pan, J.; Pu, X.-M.; Wang, B.; Pan, J.-J. Current status of coastal wetlands in China: Degradation, restoration, and future management. Estuar. Coast. Shelf Sci. 2015, 164, 265–275. [Google Scholar] [CrossRef]
- Bobo-Pinilla, J.; Barrios de León, S.B.; Seguí Colomar, J.; Fenu, G.; Bacchetta, G.; Peñas, J.; Martínez-Ortega, M.M. Phylogeography of Arenaria balearica L. (Caryophyllaceae): Evolutionary history of a disjunct endemic from the Western Mediterranean continental islands. PeerJ 2016, 4, e2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alipour, S.; Yousefzadeh, H.; Badehian, Z.; Asadi, F.; Espahbodi, K.; Dering, M. Genetic diversity and structure of the endemic and critically endangered Populus caspica in the Hyrcanian forests. Tree Genet. Genomes 2021, 17, 19. [Google Scholar] [CrossRef]
- Engelhardt, K.A.M.; Lloyd, M.W.; Neel, M.C. Effects of genetic diversity on conservation and restoration potential at individual, population, and regional scales. Biol. Conserv. 2014, 179, 6–16. [Google Scholar] [CrossRef]
- Thompson, J.D. Plant Evolution in the Mediterranean; Oxford University Press: Oxford, UK, 2005; ISBN 9780198515340. [Google Scholar]
- Habel, J.C.; Zachos, F.E.; Dapporto, L.; Rödder, D.; Radespiel, U.; Tellier, A.; Schmitt, T. Population genetics revisited-towards a multidisciplinary research field. Biol. J. Linn. Soc. 2015, 115, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Diniz-Filho, J.A.F.; Bini, L.M. Geographical Patterns in Biodiversity: Towards an Integration of Concepts and Methods from Genes to Species Diversity. Nat. Conserv. 2011, 9, 179–187. [Google Scholar] [CrossRef]
- Avise, J.C.; Bowen, B.W.; Ayala, F.J. In the light of evolution X: Comparative phylogeography. Proc. Natl. Acad. Sci. USA 2016, 113, 7957–7961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lexer, C.; Mangili, S.; Bossolini, E.; Forest, F.; Stölting, K.N.; Pearman, P.B.; Zimmermann, N.E.; Salamin, N. ‘Next generation’ biogeography: Towards understanding the drivers of species diversification and persistence. J. Biogeogr. 2013, 40, 1013–1022. [Google Scholar] [CrossRef]
- Prentice, H.C.; Ursula Malm, J.; Mateu-Andrés, I. Allozyme and chloroplast DNA variation in island and mainland populations of the rare Spanish endemic, Silene hifacensis (Caryophyllaceae). Conserv. Genet. 2003, 4, 543–555. [Google Scholar] [CrossRef]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Mota, J.F.; Sola, A.J.; Jiménez-Sánchez, M.L.; Pérez-García, F.; Merlo, M.E. Gypsicolous flora, conservation and restoration of quarries in the southeast of the Iberian Peninsula. Biodivers. Conserv. 2004, 13, 1797–1808. [Google Scholar] [CrossRef]
- Aitken, S.N.; Whitlock, M.C. Assisted Gene Flow to Facilitate Local Adaptation to Climate Change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367–388. [Google Scholar] [CrossRef]
- Breed, M.F.; Stead, M.G.; Ottewell, K.M.; Gardner, M.G.; Lowe, A.J. Which provenance and where? Seed sourcing strategies for revegetation in a changing environment. Conserv. Genet. 2013, 14, 1–10. [Google Scholar] [CrossRef]
- Hausdorf, B. Progress toward a general species concept. Evolution 2011, 65, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Duminil, J.; Di Michele, M. Plant species delimitation: A comparison of morphological and molecular markers. Plant Biosyst. 2009, 143, 528–542. [Google Scholar] [CrossRef]
- Mace, G.M. The role of taxonomy in species conservation. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2004, 359, 711–719. [Google Scholar] [CrossRef] [Green Version]
- IUCN. Guidelines for Re-Introductions; IUCN: Gland, Switzerland; Cambridge, UK, 1998; ISBN 2831704480. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Dudash, M.R.; Eldridge, M.D.B.; Fenster, C.B.; Lacy, R.C.; Mendelson, J.R.; Porton, I.J.; Ralls, K.; Ryder, O.A. Implications of different species concepts for conserving biodiversity. Biol. Conserv. 2012, 153, 25–31. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Srivastava, D.S.; Cadotte, M.W.; MacDonald, A.A.M.; Marushia, R.G.; Mirotchnick, N. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 2012, 15, 637–648. [Google Scholar] [CrossRef]
- Winter, M.; Devictor, V.; Schweiger, O. Phylogenetic diversity and nature conservation: Where are we? Trends Ecol. Evol. 2013, 28, 199–204. [Google Scholar] [CrossRef]
- Rumeu, B.; Vargas, P.; Jaén-Molina, R.; Nogales, M.; Caujapé-Castells, J. Phylogeography and genetic structure of the threatened Canarian Juniperus cedrus (Cupressaceae). Bot. J. Linn. Soc. 2014, 175, 376–394. [Google Scholar] [CrossRef] [Green Version]
- Avise, J.C. Gene trees and organismal histories: A phylogenetic approach to population biology. Evolution 1989, 43, 1192–1208. [Google Scholar] [CrossRef]
- Casacci, L.P.; Barbero, F.; Balletto, E. The “Evolutionarily Significant Unit” concept and its applicability in biological conservation. Ital. J. Zool. 2014, 81, 182–193. [Google Scholar] [CrossRef] [Green Version]
- Avise, J.C. Molecular Markers, Natural History and Evolution; Springer: Boston, MA, USA, 1994; ISBN 978-0-412-03781-8. [Google Scholar]
- Fraser, D.J.; Bernatchez, L. Adaptive evolutionary conservation: Towards a unified concept for defining conservation units. Mol. Ecol. 2001, 10, 2741–2752. [Google Scholar] [CrossRef] [PubMed]
- Dizon, A.E.; Lockyer, C.; Perrin, W.F.; Demaster, D.P.; Sisson, J. Rethinking the Stock Concept: A Phylogeographic Approach. Conserv. Biol. 1992, 6, 24–36. [Google Scholar] [CrossRef]
- Bowen, B.W.; Clark, A.M.; Abreu-Grobois, F.A.; Chaves, A.; Reichart, H.A.; Ferl, R.J. Global phylogeography of the ridley sea turtles (Lepidochelys spp.) as inferred from mitochondrial DNA sequences. Genetica 1998, 101, 179–189. [Google Scholar] [CrossRef]
- Crandall, K.A.; Bininda-Emonds, O.R.P.; Mace, G.M.; Wayne, R.K. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 2000, 15, 290–295. [Google Scholar] [CrossRef]
- Vogler, A.P.; Desalle, R. Diagnosing Units of Conservation Management. Conserv. Biol. 1994, 8, 354–363. [Google Scholar] [CrossRef]
- Waples, R.S. Pacific salmon, Oncorhynchus spp., and the definition of “species” under the Endangered Species Act. Mar. Fish. Rev. 1991, 53, 11. [Google Scholar]
- De Guia, A.P.O.; Saitoh, T. The gap between the concept and definitions in the Evolutionarily Significant Unit: The need to integrate neutral genetic variation and adaptive variation. Ecol. Res. 2007, 22, 604–612. [Google Scholar] [CrossRef]
- Doadrio, I.; Perdices, A.; Machordom, A. Allozymic variation of the endangered killifish Aphanius iberus and its application to conservation. Environ. Biol. Fishes 1996, 45, 259–271. [Google Scholar] [CrossRef]
- Corral-Lou, A.; Perea, S.; Doadrio, I. High genetic differentiation in the endemic and endangered freshwater fish Achondrostoma salmantinum. Conserv. Genet. 2021, 22, 585–600. [Google Scholar] [CrossRef]
- Eizirik, E.; Kim, J.-H.; Menotti-Raymond, M.; Crawshaw, P.G., Jr.; O’Brien, S.J.; Johnson, W.E. Phylogeography, population history and conservation genetics of jaguars (Panthera onca, Mammalia, Felidae). Mol. Ecol. 2001, 10, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Sarasola-Puente, V.; Madeira, M.J.; Gosá, A.; Lizana, M.; Gómez-Moliner, B. Population structure and genetic diversity of Rana dalmatina in the Iberian Peninsula. Conserv. Genet. 2012, 13, 197–209. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010; ISBN 9780521702713. [Google Scholar]
- Bengtsson, B.O.; Weibull, P.; Ghatnekar, L. The loss of alleles by sampling: A study of the common outbreeding grass Festuca ovina over three geographic scales. Hereditas 1995, 122, 221–238. [Google Scholar] [CrossRef]
- Lopez, S.; Rousset, F.Ç.; Shaw, F.H.; Shaw, R.G.; Ronce, O. Joint effects of inbreeding and local adaptation on the evolution of genetic load after fragmentation. Conserv. Biol. 2009, 23, 1618–1627. [Google Scholar] [CrossRef]
- Shaw, R.G.; Etterson, J.R. Rapid climate change and the rate of adaptation: Insight from experimental quantitative genetics. New Phytol. 2012, 195, 752–765. [Google Scholar] [CrossRef]
- Mee, J.A.; Bernatchez, L.; Reist, J.D.; Rogers, S.M.; Taylor, E.B. Identifying designatable units for intraspecific conservation prioritization: A hierarchical approach applied to the lake whitefish species complex (Coregonus spp.). Evol. Appl. 2015, 8, 423–441. [Google Scholar] [CrossRef]
- Semaan, M.T.; Dodd, R.S. Genetic variability and structure of the remnant natural populations of Cedrus libani (Pinaceae) of Lebanon. Tree Genet. Genomes 2008, 4, 757–766. [Google Scholar] [CrossRef]
- Szövényi, P.; Ricca, M.; Shaw, A.J. Multiple paternity and sporophytic inbreeding depression in a dioicous moss species. Heredity 2009, 103, 394–403. [Google Scholar] [CrossRef] [Green Version]
- Wahid, N.; Naydenov, K.D.; Kamari, S.; Boulli, A.; Tremblay, F. Genetic structure of Pinus pinaster Ait. populations in Morocco revealed by nuclear microsatellites. Biochem. Syst. Ecol. 2010, 38, 73–82. [Google Scholar] [CrossRef]
- Boulila, A.; Béjaoui, A.; Messaoud, C.; Boussaid, M. Genetic Diversity and Population Structure of Teucrium polium (Lamiaceae) in Tunisia. Biochem. Genet. 2010, 48, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Belletti, P.; Ferrazzini, D.; Piotti, A.; Monteleone, I.; Ducci, F. Genetic variation and divergence in Scots pine (Pinus sylvestris L.) within its natural range in Italy. Eur. J. For. Res. 2012, 131, 1127–1138. [Google Scholar] [CrossRef] [Green Version]
- Pellegrino, G.; Bellusci, F. Effects of human disturbance on reproductive success and population viability of Serapias cordigera (Orchidaceae). Bot. J. Linn. Soc. 2014, 176, 408–420. [Google Scholar] [CrossRef] [Green Version]
- Mucciarelli, M.; Ferrazzini, D.; Belletti, P. Genetic Variability and Population Divergence in the Rare Fritillaria tubiformis subsp. moggridgei Rix (Liliaceae) as Revealed by RAPD Analysis. PLoS ONE 2014, 9, e101967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardion, L.; Verlaque, R.; Saltonstall, K.; Leriche, A.; Vila, B. Origin of the invasive Arundo donax (Poaceae): A trans-Asian expedition in herbaria. Ann. Bot. 2014, 114, 455–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacchetta, G.; Bueno Sánchez, A.; Fenu, G.; Jiménez-Alfaro, B.; Mattana, E.; Piotto, B.; Virevaire, M. (Eds.) Conservación Ex Situ de Plantas Silvestres; Principado de Asturias/La Caixa: Gijón, Spain, 2008. [Google Scholar]
- Sarasan, V.; Cripps, R.; Ramsay, M.M.; Atherton, C.; McMichen, M.; Prendergast, G.; Rowntree, J.K. Conservation In vitro of threatened plants—Progress in the past decade. Vitr. Cell. Dev. Biol.-Plant 2006, 42, 206–214. [Google Scholar] [CrossRef]
- Hufford, K.M.; Mazer, S.J. Plant ecotypes: Genetic differentiation in the age of ecological restoration. Trends Ecol. Evol. 2003, 18, 147–155. [Google Scholar] [CrossRef]
- Godefroid, S.; Piazza, C.; Rossi, G.; Buord, S.; Stevens, A.-D.; Aguraiuja, R.; Cowell, C.; Weekley, C.W.; Vogg, G.; Iriondo, J.M.; et al. How successful are plant species reintroductions? Biol. Conserv. 2011, 144, 672–682. [Google Scholar] [CrossRef]
- Volis, S. Conservation meets restoration–rescuing threatened plant species by restoring their environments and restoring environments using threatened plant species. Isr. J. Plant Sci. 2016, 63, 262–275. [Google Scholar] [CrossRef]
- Mistretta, O. Genetics of species re-introductions: Applications of genetic analysis. Biodivers. Conserv. 1994, 3, 184–190. [Google Scholar] [CrossRef]
- McKay, J.K.; Christian, C.E.; Harrison, S.; Rice, K.J. “How Local Is Local?”—A Review of Practical and Conceptual Issues in the Genetics of Restoration. Restor. Ecol. 2005, 13, 432–440. [Google Scholar] [CrossRef]
- Falk, D.A. Process-centred restoration in a fire-adapted ponderosa pine forest. J. Nat. Conserv. 2006, 14, 140–151. [Google Scholar] [CrossRef]
- Vander Mijnsbrugge, K.; Bischoff, A.; Smith, B. A question of origin: Where and how to collect seed for ecological restoration. Basic Appl. Ecol. 2010, 11, 300–311. [Google Scholar] [CrossRef] [Green Version]
- Fenu, G.; Bacchetta, G.; Charalambos, S.C.; Fournaraki, C.; del Galdo, G.P.G.; Gotsiou, P.; Kyratzis, A.; Piazza, C.; Vicens, M.; Pinna, M.S.; et al. An early evaluation of translocation actions for endangered plant species on Mediterranean islands. Plant Divers. 2019, 41, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Caujapé-Castells, J.; Pedrola-Monfort, J. Designing ex-situ conservation strategies through the assessment of neutral genetic markers: Application to the endangered Androcymbium gramineum. Conserv. Genet. 2004, 5, 131–144. [Google Scholar] [CrossRef]
- Batista, F.; Bañares, A.; Caujapé-Castells, J.; Carqué, E.; Marrero-Gómez, M.; Sosa, P.A. Allozyme diversity in three endemic species of Cistus (Cistaceae) from the Canary Islands: Intraspecific and interspecific comparisons and implications for genetic conservation. Am. J. Bot. 2001, 88, 1582–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, H.; Freeland, J.R. Applications and Implications of Neutral versus Non-neutral Markers in Molecular Ecology. Int. J. Mol. Sci. 2011, 12, 3966–3988. [Google Scholar] [CrossRef] [Green Version]
- Ciofi, C.; Bruford, M.W. Genetic structure and gene flow among Komodo dragon populations inferredby microsatellite loci analysis. Mol. Ecol. 1999, 8, S17–S30. [Google Scholar] [CrossRef]
- Knapp, E.; Rice, K. Starting from Seed: Genetic Issues in Using Native Grasses for Restoration. Ecol. Restor. 1994, 12, 40–45. [Google Scholar] [CrossRef]
- Chung, M.Y.; Son, S.; Mao, K.; López-Pujol, J.; Chung, M.G. Seed collection strategies for plant restoration with the aid of neutral genetic diversity. Korean J. Plant Taxon. 2019, 49, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Krauss, S.L.; Koch, J.M. Methodological insights: Rapid genetic delineation of provenance for plant community restoration. J. Appl. Ecol. 2004, 41, 1162–1173. [Google Scholar] [CrossRef]
- Krauss, S.L.; He, T.H. Rapid genetic identification of local provenance seed collection zones for ecological restoration and biodiversity conservation. J. Nat. Conserv. 2006, 14, 190–199. [Google Scholar] [CrossRef]
- Hubert, J.; Cottrell, J. The Role of Forest Genetic Resources in Helping British Forests; Forestry Commission: Edinburgh, Scotland, 2004. [Google Scholar]
- Bobo-Pinilla, J.; Salmerón-Sánchez, E.; Mota, J.F.; Peñas, J. Genetic conservation strategies of endemic plants from edaphic habitat islands: The case of Jacobaea auricula (Asteraceae). J. Nat. Conserv. 2021, 61, 126004. [Google Scholar] [CrossRef]
- Bobo Pinilla, J.; López-González, N.; Caballero, A.; Peñas de Giles, J. Looking for a successful translocation: The case of Astragalus edulis. Mediterr. Bot. 2021, 42, e68048. [Google Scholar] [CrossRef]
- Friar, E.A.; Ladoux, T.; Roalson, E.H.; Robichaux, R.H. Microsatellite analysis of a population crash and bottleneck in the Mauna Kea silversword, Argyroxiphium sandwicense ssp. sandwicense (Asteraceae), and its implications for reintroduction. Mol. Ecol. 2000, 9, 2027–2034. [Google Scholar] [CrossRef]
- Armstrong, D.P.; Seddon, P.J. Directions in reintroduction biology. Trends Ecol. Evol. 2008, 23, 20–25. [Google Scholar] [CrossRef]
- Barrett, S.C.H.; Kohn, J.R. Genetic and evolutionary consequences of small population sizes in plants: Implications for conservation. In Genetics and Conservation of Rare Plants; Falk, D.A., Holsinger, K.A., Eds.; Oxford University Press: New York, NY, USA, 1991; pp. 3–30. ISBN 9780195064292. [Google Scholar]
- Fenster, C.B.; Galloway, L.F. Inbreeding and outbreeding depression in natural populations of Chamaecrista fasciculata (Fabaceae). Conserv. Biol. 2000, 14, 1406–1412. [Google Scholar] [CrossRef] [Green Version]
- Young, T.P.; Petersen, D.A.; Clary, J.J. The ecology of restoration: Historical links, emerging issues and unexplored realms. Ecol. Lett. 2005, 8, 662–673. [Google Scholar] [CrossRef]
- Young, A.G.; Clarke, G.M. Genetics, Demography and Viability of Fragmented Populations; Cambridge University Press: London, UK, 2000; ISBN 9780521782074. [Google Scholar]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002; ISBN 9780511808999. [Google Scholar]
- Broadhurst, L.M.; Lowe, A.; Coates, D.J.; Cunningham, S.A.; McDonald, M.; Vesk, P.A.; Yates, C. Seed supply for broadscale restoration: Maximizing evolutionary potential. Evol. Appl. 2008, 1, 587–597. [Google Scholar] [CrossRef]
- Nieto Feliner, G. Patterns and processes in plant phylogeography in the Mediterranean Basin. A review. Perspect. Plant Ecol. Evol. Syst. 2014, 16, 265–278. [Google Scholar] [CrossRef]
- Segarra-Moragues, J.G.; Palop-Esteban, M.; González-Candelas, F.; Catalán, P. On the verge of extinction: Genetics of the critically endangered Iberian plant species, Borderea chouardii (Dioscoreaceae) and implications for conservation management. Mol. Ecol. 2005, 14, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.G.; Wolfenbarger, L.L. AFLP genotyping and fingerprinting. Trends Ecol. Evol. 1999, 14, 389–394. [Google Scholar] [CrossRef]
- Agrimonti, C.; Bianchi, R.; Bianchi, A.; Ballero, M.; Poli, F.; Marmiroli, N. Understanding biological conservation strategies: A molecular-genetic approach to the case of myrtle (Myrtus communis L.) in two Italian regions: Sardinia and Calabria. Conserv. Genet. 2007, 8, 385–396. [Google Scholar] [CrossRef]
- Segarra-Moragues, J.G.; Catalán, P. The fewer and the better: Prioritization of populations for conservation under limited resources, a genetic study with Borderea pyrenaica (Dioscoreaceae) in the Pyrenean National Park. Genetica 2010, 138, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, A.; Chen, K.; Wan, D.; Liu, J. Genetic diversity and population structure of the endangered and medically important Rheum tanguticum (Polygonaceae) revealed by SSR Markers. Biochem. Syst. Ecol. 2009, 37, 613–621. [Google Scholar] [CrossRef]
- Garcia-Jacas, N.; Requena, J.; Massó, S.; Vilatersana, R.; Blanché, C.; López-Pujol, J. Genetic diversity and structure of the narrow endemic Seseli farrenyi (Apiaceae): Implications for translocation. PeerJ 2021, 9, e10521. [Google Scholar] [CrossRef]
- Juan, A.; Fay, M.F.; Pastor, J.; Juan, R.; Fernández, I.; Crespo, M.B. Genetic structure and phylogeography in Juniperus oxycedrus subsp. macrocarpa around the Mediterranean and Atlantic coasts of the Iberian Peninsula, based on AFLP and plastid markers. Eur. J. For. Res. 2012, 131, 845–856. [Google Scholar] [CrossRef]
- Salmerón-Sánchez, E.; Merlo, M.E.; Medina-Cazorla, J.M.; Pérez-García, F.J.; Martínez-Hernández, F.; Garrido-Becerra, J.A.; Mendoza-Fernández, A.J.; Valle, F.; Mota, J.F. Variability, genetic structure and phylogeography of the dolomitophilous species Convolvulus boissieri (Convolvulaceae) in the Baetic ranges, inferred from AFLPs, plastid DNA and ITS sequences. Bot. J. Linn. Soc. 2014, 176, 506–523. [Google Scholar] [CrossRef] [Green Version]
- Salmerón-Sánchez, E.; Martínez-Ortega, M.M.; Mota, J.F.; Peñas, J. A complex history of edaphic habitat islands in the Iberian Peninsula: Phylogeography of the halo-gypsophyte Jacobaea auricula (Asteraceae). Bot. J. Linn. Soc. 2017, 185, 376–392. [Google Scholar] [CrossRef]
- Gutierrez Larena, B.; Fuertes Aguilar, J.; Nieto Feliner, G. Glacial-induced altitudinal migrations in Armeria (Plumbaginaceae) inferred from patterns of chloroplast DNA haplotype sharing. Mol. Ecol. 2002, 11, 1965–1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropf, M.; Kadereit, J.W.; Comes, H.P. Differential cycles of range contraction and expansion in European high mountain plants during the Late Quaternary: Insights from Pritzelago alpina (L.) O. Kuntze (Brassicaceae). Mol. Ecol. 2003, 12, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, J.F.; Rosselló, J.A.; Feliner, G.N. Molecular evidence for the compilospecies model of reticulate evolution in Armeria (Plumbaginaceae). Syst. Biol. 1999, 48, 735–754. [Google Scholar] [CrossRef]
- Emerson, K.J.; Merz, C.R.; Catchen, J.M.; Hohenlohe, P.A.; Cresko, W.A.; Bradshaw, W.E.; Holzapfel, C.M. Resolving postglacial phylogeography using high-throughput sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 16196–16200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirschner, P.; Arthofer, W.; Pfeifenberger, S.; Záveská, E.; Schönswetter, P.; Frajman, B.; Gamisch, A.; Hilpold, A.; Paun, O.; Sanmartín, I.; et al. Performance comparison of two reduced-representation based genome-wide marker-discovery strategies in a multi-taxon phylogeographic framework. Sci. Rep. 2021, 11, 3978. [Google Scholar] [CrossRef] [PubMed]
- Onley, I.R.; Moseby, K.E.; Austin, J.J. Genomic approaches for conservation management in australia under climate change. Life 2021, 11, 653. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic diversity and conservation units: Dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef] [Green Version]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Jörger, K.M.; Schrödl, M. How to describe a cryptic species? Practical challenges of molecular taxonomy. Front. Zool. 2013, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Struck, T.H.; Feder, J.L.; Bendiksby, M.; Birkeland, S.; Cerca, J.; Gusarov, V.I.; Kistenich, S.; Larsson, K.-H.; Liow, L.H.; Nowak, M.D.; et al. Finding Evolutionary Processes Hidden in Cryptic Species. Trends Ecol. Evol. 2018, 33, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H.; Willis, J.H. Plant Speciation. Science 2007, 317, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.L.; Kunte, K. Adaptive Genetic Exchange: A Tangled History of Admixture and Evolutionary Innovation. Trends Ecol. Evol. 2017, 32, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Ottenlips, M.V.; Mansfield, D.H.; Buerki, S.; Feist, M.A.E.; Downie, S.R.; Dodsworth, S.; Forest, F.; Plunkett, G.M.; Smith, J.F. Resolving species boundaries in a recent radiation with the Angiosperms353 probe set: The Lomatium packardiae/L. anomalum clade of the L. triternatum (Apiaceae) complex. Am. J. Bot. 2021, 108, 1217–1233. [Google Scholar] [CrossRef]
- Milano, E.R.; Mulligan, M.R.; Rebman, J.P.; Vandergast, A.G. High-throughput sequencing reveals distinct regional genetic structure among remaining populations of an endangered salt marsh plant in California. Conserv. Genet. 2020, 21, 547–559. [Google Scholar] [CrossRef]
- Burge, D.O.; Rebman, J.P.; Mulligan, M.R.; Wilken, D.H. Three Edaphic-Endemic Ceanothus (Rhamnaceae) New to Science. Syst. Bot. 2017, 42, 529–542. [Google Scholar] [CrossRef]
- Fuentes, G.; González, F.; Saavedra, J.; López-Sepúlveda, P.; Victoriano, P.F.; Stuessy, T.F.; Ruiz-Ponce, E. Assessing signals of selection and historical demography to develop conservation strategies in the Chilean emblematic Araucaria araucana. Sci. Rep. 2021, 11, 20504. [Google Scholar] [CrossRef]
- Wood, G.; Marzinelli, E.M.; Campbell, A.H.; Steinberg, P.D.; Vergés, A.; Coleman, M.A. Genomic vulnerability of a dominant seaweed points to future-proofing pathways for Australia’s underwater forests. Glob. Chang. Biol. 2021, 27, 2200–2212. [Google Scholar] [CrossRef]
- Lim, S.L.; D’Agui, H.M.; Enright, N.J.; He, T. Characterization of Leaf Transcriptome in Banksia hookeriana. Genomics. Proteom. Bioinform. 2017, 15, 49–56. [Google Scholar] [CrossRef]
- Steane, D.A.; Potts, B.M.; McLean, E.H.; Collins, L.; Holland, B.R.; Prober, S.M.; Stock, W.D.; Vaillancourt, R.E.; Byrne, M. Genomic Scans across Three Eucalypts Suggest that Adaptation to Aridity is a Genome-Wide Phenomenon. Genome Biol. Evol. 2017, 9, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Supple, M.A.; Bragg, J.G.; Broadhurst, L.M.; Nicotra, A.B.; Byrne, M.; Andrew, R.L.; Widdup, A.; Aitken, N.C.; Borevitz, J.O. Landscape genomic prediction for restoration of a Eucalyptus foundation species under climate change. eLife 2018, 7, e31835. [Google Scholar] [CrossRef] [PubMed]
- Pickup, M.; Field, D.L.; Rowell, D.M.; Young, A.G. Source population characteristics affect heterosis following genetic rescue of fragmented plant populations. Proc. R. Soc. B Biol. Sci. 2013, 280. [Google Scholar] [CrossRef] [PubMed]
- Rey, O.; Eizaguirre, C.; Angers, B.; Baltazar-Soares, M.; Sagonas, K.; Prunier, J.G.; Blanchet, S. Linking epigenetics and biological conservation: Towards a conservation epigenetics perspective. Funct. Ecol. 2020, 34, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, B.G. Fine-Scale to Flora-Wide Phylogenetic Perspectives on Californian Plant Diversity, Endemism, and Conservation. Ann. Missouri Bot. Gard. 2019, 104, 429–440. [Google Scholar] [CrossRef]
- Kling, M.M.; Mishler, B.D.; Thornhill, A.H.; Baldwin, B.G.; Ackerly, D.D. Facets of phylodiversity: Evolutionary diversification, divergence and survival as conservation targets. Philos. Trans. R. Soc. B 2019, 374, 20170397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- López-González, N.; Bobo-Pinilla, J.; Gutiérrez-Larruscain, D.; Montserrat Martínez-Ortega, M.; Rojas-Andrés, B.M. Hybridization as a biodiversity driver: The case of Veronica × gundisalvi. Mediterr. Bot. 2021, 42, e67901. [Google Scholar] [CrossRef]
- Menon, S.; Choudhury, B.; Khan, M.; Townsend Peterson, A. Ecological niche modeling and local knowledge predict new populations of Gymnocladus assamicus a critically endangered tree species. Endanger. Species Res. 2010, 11, 175–181. [Google Scholar] [CrossRef]
- Sarkinen, T.; Gonzáles, P.; Knapp, S. Distribution models and species discovery: The story of a new Solanum species from the Peruvian Andes. PhytoKeys 2013, 31, 1. [Google Scholar] [CrossRef] [Green Version]
- Fois, M.; Cuena-Lombraña, A.; Fenu, G.; Bacchetta, G. Using species distribution models at local scale to guide the search of poorly known species: Review, methodological issues and future directions. Ecol. Modell. 2018, 385, 124–132. [Google Scholar] [CrossRef] [Green Version]
- Park, D.S.; Ellison, A.M.; Davis, C.C. Mating system does not predict niche breath. Glob. Ecol. Biogeogr. 2018, 27, 804–813. [Google Scholar] [CrossRef]
- Pearman, P.B.; D’Amen, M.; Graham, C.H.; Thuiller, W.; Zimmermann, N.E. Within-taxon niche structure: Niche conservatism, divergence and predicted effects of climate change. Ecography 2010, 33, 990–1003. [Google Scholar] [CrossRef]
- Bálint, M.; Domisch, S.; Engelhardt, C.H.M.; Haase, P.; Lehrian, S.; Sauer, J.; Theissinger, K.; Pauls, S.U.; Nowak, C. Cryptic biodiversity loss linked to global climate change. Nat. Clim. Chang. 2011, 1, 313–318. [Google Scholar] [CrossRef]
- Bendiksby, M.; Mazzoni, S.; Jørgensen, M.H.; Halvorsen, R.; Holien, H. Combining genetic analyses of archived specimens with distribution modelling to explain the anomalous distribution of the rare lichen Staurolemma omphalarioides: Long-distance dispersal or vicariance? J. Biogeogr. 2014, 41, 2020–2031. [Google Scholar] [CrossRef]
- Chen, G.; Kéry, M.; Plattner, M.; Ma, K.; Gardner, B. Imperfect detection is the rule rather than the exception in plant distribution studies. J. Ecol. 2013, 101, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Winkler, J.A.; Viña, A.; Liu, J.; Zhang, Y.; Zhang, X.; Li, X.; Wang, F.; Zhang, J.; Zhao, Z. Uncertainty of future projections of species distributions in mountainous regions. PLoS ONE 2018, 13, e0189496. [Google Scholar] [CrossRef] [Green Version]
- Cho, N.; Kim, E.; Lee, B.; Lim, J.; Kang, S. Predicting the Potential Distribution of Pinus densiflora and Analyzing the Relationship with Environmental Variable Using MaxEnt Model. Korean J. Agric. For. Meteorol. 2020, 22, 47–56. [Google Scholar] [CrossRef]
- Han, E.K.; Cho, W.B.; Park, J.S.; Choi, I.S.; Kwak, M.; Kim, B.Y.; Lee, J.H. A Disjunctive Marginal Edge of Evergreen Broad-Leaved Oak (Quercus gilva) in East Asia: The High Genetic Distinctiveness and Unusual Diversity of Jeju Island Populations and Insight into a Massive, Independent Postglacial Colonization. Genes 2020, 11, 1114. [Google Scholar] [CrossRef]
- Nualart, N.; Herrando-Moraira, S.; Cires, E.; Guardiola, M.; Laguna, E.; Pérez-Prieto, D.; Sáez, L.; López-Pujol, J. Reusing old and producing new data is useful for species delimitation in the taxonomically controversial iberian endemic pair Petrocoptis montsicciana/ P. pardoi (caryophyllaceae). Diversity 2021, 13, 205. [Google Scholar] [CrossRef]
- Nygaard, M.; Kemppainen, P.; Speed, J.D.M.; Elven, R.; Flatberg, K.I.; Galten, L.P.; Yousefi, N.; Solstad, H.; Bendiksby, M. Combining population genomics and ecological niche modeling to assess taxon limits between Carex jemtlandica and C. lepidocarpa. J. Syst. Evol. 2021, 59, 627–641. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobo-Pinilla, J.; Salmerón-Sánchez, E.; Mendoza-Fernández, A.J.; Mota, J.F.; Peñas, J. Conservation and Phylogeography of Plants: From the Mediterranean to the Rest of the World. Diversity 2022, 14, 78. https://doi.org/10.3390/d14020078
Bobo-Pinilla J, Salmerón-Sánchez E, Mendoza-Fernández AJ, Mota JF, Peñas J. Conservation and Phylogeography of Plants: From the Mediterranean to the Rest of the World. Diversity. 2022; 14(2):78. https://doi.org/10.3390/d14020078
Chicago/Turabian StyleBobo-Pinilla, Javier, Esteban Salmerón-Sánchez, Antonio J. Mendoza-Fernández, Juan F. Mota, and Julio Peñas. 2022. "Conservation and Phylogeography of Plants: From the Mediterranean to the Rest of the World" Diversity 14, no. 2: 78. https://doi.org/10.3390/d14020078








