Diet Metabarcoding Reveals Extensive Dietary Overlap between Two Benthic Stream Fishes (Zingel asper and Cottus gobio) and Provides Insights into Their Coexistence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Macroinvertebrate Community Sampling
2.3. Taxonomic Assignment of Macroinvertebrates
2.4. Dietary Data Sampling
2.5. Metabarcoding Protocol
2.6. Statistical Analyses
2.6.1. Macroinvertebrate Community Composition
2.6.2. Electivity
2.6.3. Trophic Niche Traits
2.6.4. Dietary Overlap
3. Results
3.1. Macroinvertebrate Community Composition
3.2. Z. asper and C. gobio Diet Composition
3.3. Prey Preferences
3.4. Comparison of Z. asper and C. gobio Trophic Traits
3.5. Trophic Niche Overlap
4. Discussion
4.1. Dietary Overlap and Coexistence
4.2. Conservation Implications for the Critically Endangered Z. asper
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tilman, D. The Importance of the Mechanisms of Interspecific Competition. Am. Nat. 1987, 129, 769–774. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Amarasekare, P.; Araújo, M.S.; Bürger, R.; Levine, J.M.; Novak, M.; Rudolf, V.H.W.; Schreiber, S.J.; Urban, M.C.; Vasseur, D.A. Why Intraspecific Trait Variation Matters in Community Ecology. Trends Ecol. Evol. 2011, 26, 183–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoday, J.M. Evolution of Niche Width. Am. Nat. 1974, 108, 142–143. [Google Scholar] [CrossRef]
- Schoener, T.W. Theory of Feeding Strategies. Annu. Rev. Ecol. Syst. 1971, 2, 369–404. [Google Scholar] [CrossRef] [Green Version]
- Chesson, P. Mechanisms of Maintenance of Species Diversity. Annu. Rev. Ecol. Syst. 2000, 31, 343–366. [Google Scholar] [CrossRef] [Green Version]
- Toft, C.A. Resource Partitioning in Amphibians and Reptiles. Copeia 1985, 1985, 1–21. [Google Scholar] [CrossRef]
- Ross, S.T. Resource Partitioning in Fish Assemblages: A Review of Field Studies. Copeia 1986, 1986, 352–388. [Google Scholar] [CrossRef]
- Worischka, S.; Schmidt, S.I.; Hellmann, C.; Winkelmann, C. Selective Predation by Benthivorous Fish on Stream Macroinvertebrates—The Role of Prey Traits and Prey Abundance. Limnologica 2015, 52, 41–50. [Google Scholar] [CrossRef]
- Sagar, P.M.; Glova, G.J. Food Partitioning by Small Fish in a Coastal New Zealand Stream. N. Z. J. Mar. Freshw. Res. 1994, 28, 429–436. [Google Scholar] [CrossRef]
- Piria, M.; Jakšić, G.; Jakovlić, I.; Treer, T. Dietary Habits of Invasive Ponto-Caspian Gobies in the Croatian Part of the Danube River Basin and Their Potential Impact on Benthic Fish Communities. Sci. Total Environ. 2016, 540, 386–395. [Google Scholar] [CrossRef]
- MacArthur, R.; Levins, R. The Limiting Similarity, Convergence, and Divergence of Coexisting Species. Am. Nat. 1967, 101, 377–385. [Google Scholar] [CrossRef]
- Schmitt, R.J.; Holbrook, S.J. Seasonally Fluctuating Resources and Temporal Variability of Interspecific Competition. Oecologia 1986, 69, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Fausch, K.D.; Kitano, S. Flexible Niche Partitioning via a Foraging Mode Shift: A Proposed Mechanism for Coexistence in Stream-Dwelling Charts. J. Anim. Ecol. 1999, 68, 1079–1092. [Google Scholar] [CrossRef]
- Prejs, A.; Prejs, K. Feeding of Tropical Freshwater Fishes: Seasonality in Resource Availability and Resource Use. Oecologia 1987, 71, 397–404. [Google Scholar] [CrossRef]
- Keith, P.; Poulet, N.; Denys, G.; Changeux, T.; Feunteun, É.; Persat, H. Les Poissons D’eau Douce de France, 2nd ed.; Biotope éditions: Mèze, France, 2020. [Google Scholar]
- Georget, M. Retours Sur Deux Programmes Life et Un Plan National d’action En Faveur de l’apron Du Rhône. Sci. Eaux Territ. 2019, IV, 2–5. [Google Scholar] [CrossRef]
- Cavalli, L.; Pech, N.; Chappaz, R. Diet and Growth of the Endangered Zingel asper in the Durance River. J. Fish Biol. 2003, 63, 460–471. [Google Scholar] [CrossRef]
- Labonne, J.; Gaudin, P. Exploring Population Dynamics Patterns in a Rare Fish, Zingel asper, through Capture-Mark-Recapture Methods. Conserv. Biol. 2005, 19, 463–472. [Google Scholar] [CrossRef]
- Danancher, D.; Labonne, J.; Gaudin, P.; Joly, P. Scale Measurements as a Conservation Tool in Endangered Zingel asper (Linnaeus, 1758). Aquat. Conserv. Mar. Freshw. Ecosyst. 2007, 17, 712–723. [Google Scholar] [CrossRef]
- Monnet, G.; Corse, E.; Archambaud-Suard, G.; Grenier, R.; Chappaz, R.; Dubut, V. Growth Variation in the Endangered Fish Zingel asper: Contribution of Substrate Quality, Hydraulics, Prey Abundance and Water Temperature. Aquat. Conserv. Mar. Freshw. Ecosyst. 2022, in press. [CrossRef]
- Cavalli, L.; Knight, C.M.; Durbec, M.; Chappaz, R.; Gozlan, R.E. Twenty-Four Hours in the Life of Zingel asper. J. Fish Biol. 2009, 75, 723–727. [Google Scholar] [CrossRef]
- Labonne, J.; Allouche, S.; Gaudin, P. Use of a Generalised Linear Model to Test Habitat Preferences: The Example of Zingel asper, an Endemic Endangered Percid of the River Rhône. Freshw. Biol. 2003, 48, 687–697. [Google Scholar] [CrossRef]
- Villsen, K.; Corse, E.; Meglécz, E.; Archambaud-Suard, G.; Vignes, H.; Ereskovsky, A.V.; Chappaz, R.; Dubut, V. DNA Metabarcoding Reveals Adaptive Seasonal Variation of Individual Trophic Traits in a Critically Endangered Fish. bioRxiv 2021, 2021.01.25.428043. [Google Scholar] [CrossRef]
- Vandelannoote, A.; Yseboodt, R.; Bruylants, B.; Verheyen, R.; Coeck, J.; Belpaire, C.; Van Thuyne, G.; Denayer, B.; Beyens, J.; De Charleroy, D. Atlas van de Vlaamse Beek-En Riviervissen; Water-Energik-Vlario: Wijnegem, The Netherlands, 1998. [Google Scholar]
- Zaugg, B. Fauna Helvetica: Pisces Atlas, 2nd ed.; Centre Suisse de Cartographie de la Faune: Neuchâtel, Switzerland, 2018; ISBN 978-2-88414-020-1. [Google Scholar]
- Lusk, S.; Hanel, L.; Lusková, V. Red List of the Ichthyofauna of the Czech Republic: Development and Present Status. Folia Zool. 2004, 53, 215–226. [Google Scholar]
- Mills, C.A.; Mann, R.H.K. The Bullhead Cottus gobio, a Versatile and Successful Fish. Annnual Rep. Freshw. Biol. Assoc. 1983, 51, 76–88. [Google Scholar]
- Davey, A.J.H.; Turner, G.F.; Hawkins, S.J.; Doncaster, C.P. Mechanisms of Density Dependence in Stream Fish: Exploitation Competition for Food Reduces Growth of Adult European Bullheads (Cottus gobio). Can. J. Fish. Aquat. Sci. 2006, 63, 597–606. [Google Scholar] [CrossRef]
- Števove, B.; Babeľová, M.; Haruštiaková, D.; Kováč, V. When a River Is Affected by a Reservoir: Trophic Interactions and Flexibility in Feeding Strategies of Alpine Bullhead (Cottus poecilopus) and European Bullhead (Cottus gobio). Sci. Total Environ. 2019, 651, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Englund, G.; Olsson, T.; Englund, G. Treatment Effects in a Stream Fish Enclosure Experiment: Influence of Predation Rate and Prey Movements. Oikos 1996, 77, 519. [Google Scholar] [CrossRef]
- Dahl, J. Effects of a Benthivorous and a Drift-Feeding Fish on a Benthic Stream Assemblage. Oecologia 1998, 116, 426–432. [Google Scholar] [CrossRef]
- Crivelli, A.J. Zingel asper. The IUCN Red List of Threatened Species 2006: E.T23207A9427850; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland, 2006. [Google Scholar] [CrossRef]
- Okuyama, T. Individual Variation in Prey Choice in a Predator–Prey Community. Theor. Popul. Biol. 2011, 79, 64–69. [Google Scholar] [CrossRef]
- Greenberg, L.A.; Giller, P.S. Individual Variation in Habitat Use and Growth of Male and Female Brown Trout. Ecography. 2001, 24, 212–224. [Google Scholar] [CrossRef]
- Svanbäck, R.; Bolnick, D.I. Intraspecific Competition Drives Increased Resource Use Diversity within a Natural Population. Proc. R. Soc. B Biol. Sci. 2007, 274, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Bolnick, D.I.; Ingram, T.; Stutz, W.E.; Snowberg, L.K.; Lau, O.L.; Pauli, J.S. Ecological Release from Interspecific Competition Leads to Decoupled Changes in Population and Individual Niche Width. Proc. R. Soc. B Biol. Sci. 2010, 277, 1789–1797. [Google Scholar] [CrossRef]
- Sjödin, H.; Ripa, J.; Lundberg, P. Principles of Niche Expansion. Proc. R. Soc. B Biol. Sci. 2018, 285, 20182603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolnick, D.I.; Yang, L.H.; Fordyce, J.A.; Davis, J.M.; Svanbäck, R. Measuring Individual-Level Resource Specialization. Ecology 2002, 83, 2936–2941. [Google Scholar] [CrossRef]
- Barbini, S.A.; Lucifora, L.O.; Sabadin, D.E.; Figueroa, D.E. Ecological Specialization Is Associated with High Conservation Concern in Skates (Chondrichthyes, Rajiformes). Anim. Conserv. 2020, 23, 222–228. [Google Scholar] [CrossRef]
- Costa-Pereira, R.; Araújo, M.S.; Souza, F.L.; Ingram, T. Competition and Resource Breadth Shape Niche Variation and Overlap in Multiple Trophic Dimensions. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falke, L.P.; Henderson, J.S.; Novak, M.; Preston, D.L. Temporal Shifts in Intraspecific and Interspecific Diet Variation among 3 Stream Predators. Freshw. Sci. 2020, 39, 115–125. [Google Scholar] [CrossRef]
- Alberdi, A.; Aizpurua, O.; Bohmann, K.; Gopalakrishnan, S.; Lynggaard, C.; Nielsen, M.; Gilbert, M.T.P. Promises and Pitfalls of Using High-Throughput Sequencing for Diet Analysis. Mol. Ecol. Resour. 2019, 19, 327–348. [Google Scholar] [CrossRef]
- de Sousa, L.L.; Silva, S.M.; Xavier, R. DNA Metabarcoding in Diet Studies: Unveiling Ecological Aspects in Aquatic and Terrestrial Ecosystems. Environ. DNA 2019, 1, 199–214. [Google Scholar] [CrossRef] [Green Version]
- Hoenig, B.D.; Trevelline, B.K.; Nuttle, T.; Porter, B.A. Dietary DNA Metabarcoding Reveals Seasonal Trophic Changes among Three Syntopic Freshwater Trout Species. Freshw. Biol. 2021, 66, 509–523. [Google Scholar] [CrossRef]
- Corse, E.; Valladares, S.; Planas, M.; Chamorro, A.; Pintado, J. Analysis of the Diet of the Long-Snouted Seahorse Hippocampus guttulatus by 18SrDNA Amplification of Prey in Faeces. Aquac. Nutr. 2015, 21, 528–540. [Google Scholar] [CrossRef]
- MacColl, A.D.C. The Ecological Causes of Evolution. Trends Ecol. Evol. 2011, 26, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Warner, R.F. Gross Channel Changes along the Durance River, Southern France, Over the Last 100 Years Using Cartographic Data. Regul. River 2000, 16, 141–157. [Google Scholar] [CrossRef]
- Clifford, H. Life Cycles of Mayflies (Ephemeroptera), with Special Reference to Voltinism. Quaest. Entomol. 1982, 18, 15–90. [Google Scholar]
- Tachet, H.; Richoux, P.; Bournaud, M.; Usseglio-Polatera, P. Invertébrés d’Eau Douce: Systématique, Biologie, Écologie; CNRS éditions: Paris, France, 2010; ISBN 978-2-271-06945-0. [Google Scholar]
- Corse, E.; Meglécz, E.; Archambaud, G.; Ardisson, M.; Martin, J.F.; Tougard, C.; Chappaz, R.; Dubut, V. A From-Benchtop-to-Desktop Workflow for Validating HTS Data and for Taxonomic Identification in Diet Metabarcoding Studies. Mol. Ecol. Resour. 2017, 17, e146–e159. [Google Scholar] [CrossRef]
- Corse, E.; Tougard, C.; Archambaud-Suard, G.; Agnèse, J.F.; Messu Mandeng, F.D.; Bilong Bilong, C.F.; Duneau, D.; Zinger, L.; Chappaz, R.; Xu, C.C.Y.; et al. One-Locus-Several-Primers: A Strategy to Improve the Taxonomic and Haplotypic Coverage in Diet Metabarcoding Studies. Ecol. Evol. 2019, 9, 4603–4620. [Google Scholar] [CrossRef]
- Meusnier, I.; Singer, G.A.C.; Landry, J.-F.; Hickey, D.A.; Hebert, P.D.N.; Hajibabaei, M. A Universal DNA Mini-Barcode for Biodiversity Analysis. BMC Genomics 2008, 9, 214. [Google Scholar] [CrossRef] [Green Version]
- Zeale, M.R.K.; Butlin, R.K.; Barker, G.L.A.; Lees, D.C.; Jones, G. Taxon-specific PCR for DNA Barcoding Arthropod Prey in Bat Faeces. Mol. Ecol. Resour. 2011, 11, 236–244. [Google Scholar] [CrossRef]
- González, A.; Dubut, V.; Corse, E.; Mekdad, R.; Dechatre, T.; Meglécz, E. VTAM: A Robust Pipeline for Validating Metabarcoding Data Using Internal Controls. bioRxiv 2020, 2020.11.06.371187. [Google Scholar] [CrossRef]
- Raveret-Wattel, C. Atlas de Poche des Poissons D’eau Douce de La France de La Suisse Romande et de La Belgique; Paul Klincksieck: Paris, France, 1900. [Google Scholar]
- White, T.E. A Method of Calculating the Dietary Percentage of Various Food Animals Utilized by Aboriginal Peoples. Am. Antiq. 1953, 18, 396–398. [Google Scholar] [CrossRef]
- R Core Development Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/ (accessed on 15 October 2021).
- Hsieh, T.C.; Ma, K.H.; Chao, A. INEXT: An R Package for Rarefaction and Extrapolation of Species Diversity (Hill Numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.H.; Jost, L. Unifying Species Diversity, Phylogenetic Diversity, Functional Diversity, and Related Similarity and Differentiation Measures through Hill Numbers. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 297–324. [Google Scholar] [CrossRef] [Green Version]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, I.P.; Gotelli, N.J.; Memmott, J.; Pearson, C.E.; Woodward, G.; Symondson, W.O.C. Econullnetr: An R Package Using Null Models to Analyse the Structure of Ecological Networks and Identify Resource Selection. Methods Ecol. Evol. 2018, 9, 728–733. [Google Scholar] [CrossRef] [Green Version]
- Hill, M.O. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef] [Green Version]
- Ohlmann, M.; Miele, V.; Dray, S.; Chalmandrier, L.; O’Connor, L.; Thuiller, W. Diversity Indices for Ecological Networks: A Unifying Framework Using Hill Numbers. Ecol. Lett. 2019, 22, 737–747. [Google Scholar] [CrossRef]
- Alberdi, A.; Gilbert, M.T.P. A Guide to the Application of Hill Numbers to DNA-Based Diversity Analyses. Mol. Ecol. Resour. 2019, 19, 804–817. [Google Scholar] [CrossRef]
- Alberdi, A.; Gilbert, M.T.P. Hilldiv: An R Package for the Integral Analysis of Diversity Based on Hill Numbers. bioRxiv 2019, 545665. [Google Scholar] [CrossRef] [Green Version]
- Chao, A.; Jost, L. Coverage-Based Rarefaction and Extrapolation: Standardizing Samples by Completeness Rather than Size. Ecology 2012, 93, 2533–2547. [Google Scholar] [CrossRef]
- Pianka, E.R. Niche Overlap and Diffuse Competition. Proc. Natl. Acad. Sci. USA 1974, 71, 2141–2145. [Google Scholar] [CrossRef] [Green Version]
- Gotelli, N.J.; Ellison, A.M. EcoSimR 1.00. Available online: http://www.uvm.edu/~ngotelli/EcoSim/EcoSim.html (accessed on 15 October 2021).
- Angermeier, P.L. Resource Seasonality and Fish Diets in an Illinois Stream. Environ. Biol. Fishes 1982, 7, 251–264. [Google Scholar] [CrossRef]
- Sánchez-Hernández, J.; Finstad, A.G.; Arnekleiv, J.V.; Kjærstad, G.; Amundsen, P.A. Beyond Ecological Opportunity: Prey Diversity Rather than Abundance Shapes Predator Niche Variation. Freshw. Biol. 2021, 66, 44–61. [Google Scholar] [CrossRef]
- Costa-Pereira, R.; Tavares, L.E.R.; de Camargo, P.B.; Araújo, M.S. Seasonal Population and Individual Niche Dynamics in a Tetra Fish in the Pantanal Wetlands. Biotropica 2017, 49, 531–538. [Google Scholar] [CrossRef]
- Murdoch, W.W. Switching in General Predators: Experiments on Predator Specificity and Stability of Prey Populations. Ecol. Monogr. 1969, 39, 335–354. [Google Scholar] [CrossRef]
- Ivlev, V.S. Experimental Ecology of the Feeding of Fishes; Yale University Press: New Haven, CT, USA, 1961. [Google Scholar]
- Manly, B.F.J.; Miller, P.; Cook, L.M. Analysis of a Selective Predation Experiment. Am. Nat. 1972, 106, 719–736. [Google Scholar] [CrossRef]
- Werner, E.E.; Hall, D.J. Optimal Foraging and the Size Selection of Prey by the Bluegill Sunfish (Lepomis macrochirus). Ecology 1974, 55, 1042–1052. [Google Scholar] [CrossRef]
- Perry, G.; Pianka, E.R. Animal Foraging: Past, Present and Future. Trends Ecol. Evol. 1997, 12, 360–364. [Google Scholar] [CrossRef]
- Welton, J.S.; Mill, C.A.; Pygott, J.R. The Effect of Interaction between the Stone Loach Noemacheilus barbatulus (L.) and the Bullhead Cottus gobio (L.) on Prey and Habitat Selection. Hydrobiologia 1991, 220, 1–7. [Google Scholar] [CrossRef]
- Pulliam, H.R. Niche Expansion and Contraction in a Variable Environment. Integr. Comp. Biol. 1986, 26, 71–79. [Google Scholar] [CrossRef]
- Neves, M.P.; Costa-Pereira, R.; Delariva, R.L.; Fialho, C.B. Seasonality and Interspecific Competition Shape Individual Niche Variation in Co-occurring Tetra Fish in Neotropical Streams. Biotropica 2021, 53, 329–338. [Google Scholar] [CrossRef]
- Liu, F.; Wang, J.; Liu, H. Seasonal Variations in Food Resource Partitioning among Four Sympatric Gudgeon Species in the Upper Yangtze River. Ecol. Evol. 2019, 9, 7227–7236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svanbäck, R.; Bolnick, D.I. Intraspecific Competition Affects the Strength of Individual Specialization: An Optimal Diet Theory Method. Evol. Ecol. Res. 2005, 7, 993–1012. [Google Scholar]
- Araújo, M.S.; Bolnick, D.I.; Layman, C.A. The Ecological Causes of Individual Specialisation. Ecol. Lett. 2011, 14, 948–958. [Google Scholar] [CrossRef] [PubMed]
- Van Valen, L. Morphological Variation and Width of Ecological Niche. Am. Nat. 1965, 99, 377–390. [Google Scholar] [CrossRef]
- Nunn, A.D.; Tewson, L.H.; Cowx, I.G. The Foraging Ecology of Larval and Juvenile Fishes. Rev. Fish Biol. Fish. 2012, 22, 377–408. [Google Scholar] [CrossRef]
- Svanbäck, R.; Eklöv, P.; Fransson, R.; Holmgren, K. Intraspecific Competition Drives Multiple Species Resource Polymorphism in Fish Communities. Oikos 2008, 117, 114–124. [Google Scholar] [CrossRef]
- Gabler, H.-M.; Amundsen, P.-A. Feeding Strategies, Resource Utilisation and Potential Mechanisms for Competitive Coexistence of Atlantic Salmon and Alpine Bullhead in a Sub-Arctic River. Aquat. Ecol. 2010, 44, 325–336. [Google Scholar] [CrossRef]
- Osborne, L.L.; Herricks, E.E. Microhabitat Characteristics of Hydropsyche (Trichoptera: Hydropsychidae) and the Importance of Body Size. J. N. Am. Benthol. Soc. 1987, 6, 115–124. [Google Scholar] [CrossRef]
- Pirvu, M.; Zaharia, C.; Satmari, A.; Parvulescu, L. Spatial Ecology of Hydropsyche incognita (Trichoptera: Hydropsychidae) in the Carpathians. EJE 2015, 112, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Hernández, J.; Vieira-Lanero, R.; Servia, M.J.; Cobo, F. Feeding Habits of Four Sympatric Fish Species in the Iberian Peninsula: Keys to Understanding Coexistence Using Prey Traits. Hydrobiologia 2011, 667, 119–132. [Google Scholar] [CrossRef]
- Schoener, T.W. Resource Partitioning in Ecological Communities. Science 1974, 185, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Gjelland, K.Ø.; Bøhn, T.; Amundsen, P. Is Coexistence Mediated by Microhabitat Segregation? An In-depth Exploration of a Fish Invasion. J. Fish Biol. 2007, 71, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Ewen, J.G.; Armstrong, D.P. Strategic Monitoring of Reintroductions in Ecological Restoration Programmes. Ecoscience 2007, 14, 401–409. [Google Scholar] [CrossRef]
- Jachowski, D.; Millspaugh, J.J.; Angermeier, P.L.; Slotow, R.H. Reintroduction of Fish and Wildlife Populations; University of California Press: Oakland, CA, USA, 2016; ISBN 0520960386. [Google Scholar]
- Stamps, J.A.; Swaisgood, R.R. Someplace like Home: Experience, Habitat Selection and Conservation Biology. Appl. Anim. Behav. Sci. 2007, 102, 392–409. [Google Scholar] [CrossRef]
- Mönkkönen, M.; Härdling, R.; Forsman, J.T.; Tuomi, J. Evolution of Heterospecific Attraction: Using Other Species as Cues in Habitat Selection. Evol. Ecol. 1999, 13, 91–104. [Google Scholar] [CrossRef]
- Parejo, D.; Danchin, E.; Avilés, J.M. The Heterospecific Habitat Copying Hypothesis: Can Competitors Indicate Habitat Quality? Behav. Ecol. 2005, 16, 96–105. [Google Scholar] [CrossRef]
- Thomson, R.L.; Forsman, J.T.; Mönkkönen, M. Positive Interactions between Migrant and Resident Birds: Testing the Heterospecific Attraction Hypothesis. Oecologia 2003, 134, 431–438. [Google Scholar] [CrossRef]
- Hattori, A. Small and Large Anemonefishes Can Coexist Using the Same Patchy Resources on a Coral Reef, before Habitat Destruction. J. Anim. Ecol. 2002, 71, 824–831. [Google Scholar] [CrossRef] [Green Version]
- Kokko, H.; Sutherland, W.J. Ecological Traps in Changing Environments: Ecological and Evolutionary Consequences of a Behaviourally Mediated Allee Effect. Evol. Ecol. Res. 2001, 3, 537–551. [Google Scholar]
- Kristan, W.B. The Role of Habitat Selection Behavior in Population Dynamics: Source-Sink Systems and Ecological Traps. Oikos 2003, 103, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Battin, J. When Good Animals Love Bad Habitats: Ecological Traps and the Conservation of Animal Populations. Conserv. Biol. 2004, 18, 1482–1491. [Google Scholar] [CrossRef]
- Zarzoso-Lacoste, D.; Bonnaud, E.; Corse, E.; Dubut, V.; Lorvelec, O.; De Meringo, H.; Santelli, C.; Meunier, J.Y.; Ghestemme, T.; Gouni, A.; et al. Stuck amongst introduced species: Trophic ecology reveals complex relationships between the critically endangered Niau kingfisher and introduced predators, competitors and prey. NeoBiota 2019, 53, 61–82. [Google Scholar] [CrossRef] [Green Version]
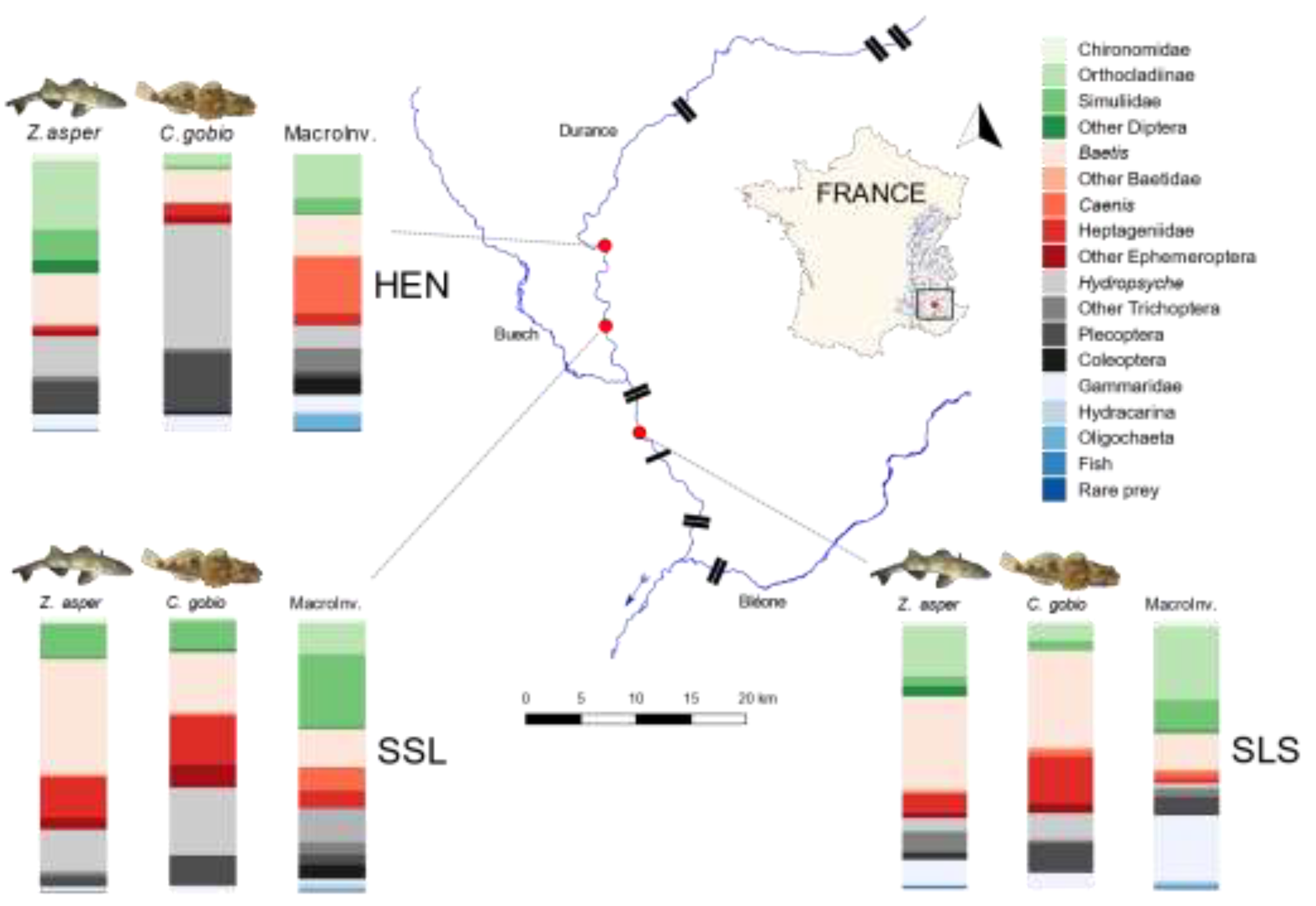

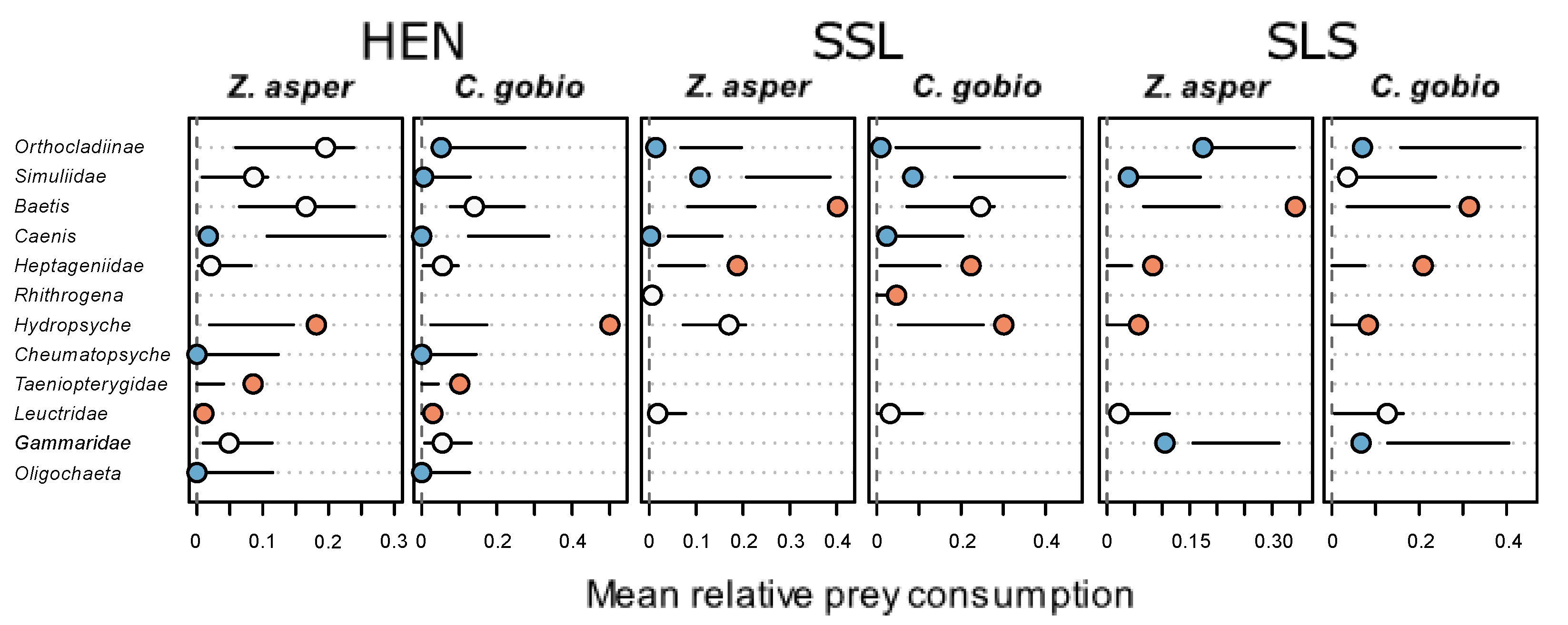



| Site | Coordinates | Date of Faeces Sampling | No. Faeces Z. asper | No. Faeces C. gobio | Date of Surber Sampling | No. Surber Samples |
|---|---|---|---|---|---|---|
| HEN | N 44° 18′ 46″ E 5° 55′ 29″ | 5 November 2015 | 29 | 27 | 6 November 2015 | 90 |
| SSL | N 44° 14′ 50″ E 5° 55′ 17″ | 22 September 2015 | 44 | 21 | 21 September 2015 | 60 |
| SLS | N 44° 9′ 32″ E 5° 57′ 15″ | 28 August 2015 | 43 | 16 | 27 August 2015 | 60 |
| Primer Set | Primer Name | Forward (F)/ Reverse (R) | Sequence (5’-3’) | Reference |
|---|---|---|---|---|
| MFZR | Uni-Minibar-F1 | F | TCCACTAATCACAARGATATTGGTAC | [52] |
| ZBJ-ArtR2c | R | WACTAATCAATTWCCAAATCCTCC | [53] | |
| ZFZR | ZBJ-ArtF1c | F | AGATATTGGAACWTTATATTTTATTTTTGG | [53] |
| ZBJ-ArtR2c | R | WACTAATCAATTWCCAAATCCTCC | [53] | |
| LFCR | LepLCO | F | RKTCAACMAATCATAAAGATATTGG | [51] |
| McoiR2 | R | CCBCCRATTAWAATKGGTATHAC | [51] |
| Z. asper | C. gobio | Macroinvertebrate Community | ||||||
|---|---|---|---|---|---|---|---|---|
| Site | Estimate | [95% CI] or ±SE | Estimate | [95% CI] or ±SE | Estimate | [95% CI] or ±SE | ||
| HEN | TNW | 28.82 | [25.44–32.21] | 20.63 * | [13.62–27.64] | 1D | 10.79 | [10.63–10.95] |
| INW | 4.42 | ±0.77 | 2.03 | ±0.26 | Density (inv.m−2) | 3659 | ±490 | |
| BIC | 0.88 | ±0.01 | 0.88 | ±0.02 | ||||
| SSL | TNW | 11.44 | [9.66–23.05] | 17.33 * | [11.61–23.05] | 1D | 10.11 | [9.89–10.32] |
| INW | 4.40 | ±0.28 | 3.39 | ±0.37 | Density (inv.m−2) | 3004 | ±397 | |
| BIC | 0.54 | ±0.01 | 0.67 | ±0.03 | ||||
| SLS | TNW | 20.99 | [18.60–23.38] | 15.27 | [12.57–17.98] | 1D | 7.49 | [7.37–7.62] |
| INW | 6.06 | ±0.45 | 4.66 | ±0.48 | Density (inv.m−2) | 5194 | ±774 | |
| BIC | 0.69 | ±0.02 | 0.68 | ±0.02 | ||||
| Site | Taxa | Size-Class | Density inv.m−2 (±SE) |
|---|---|---|---|
| HEN | Baetis | small (<5 mm) | 545 (±121) |
| large (>5 mm) | 5 (±1) | ||
| Heptageniidae | small (<5 mm) | 124 (±21) | |
| large (>5 mm) | 9 (±3) | ||
| SSL | Baetis | small (<5 mm) | 390 (±66) |
| large (>5 mm) | 30 (±9) | ||
| Heptageniidae | small (<5 mm) | 159 (±26) | |
| large (>5 mm) | 10 (±2) | ||
| SLS | Baetis | small (<5 mm) | 577 (±129) |
| large (>5 mm) | 121 (±34) | ||
| Heptageniidae | small (<5 mm) | 67 (±22) | |
| large (>5 mm) | 21 (±4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villsen, K.; Corse, E.; Archambaud-Suard, G.; Héran, K.; Meglécz, E.; Ereskovsky, A.V.; Chappaz, R.; Dubut, V. Diet Metabarcoding Reveals Extensive Dietary Overlap between Two Benthic Stream Fishes (Zingel asper and Cottus gobio) and Provides Insights into Their Coexistence. Diversity 2022, 14, 412. https://doi.org/10.3390/d14050412
Villsen K, Corse E, Archambaud-Suard G, Héran K, Meglécz E, Ereskovsky AV, Chappaz R, Dubut V. Diet Metabarcoding Reveals Extensive Dietary Overlap between Two Benthic Stream Fishes (Zingel asper and Cottus gobio) and Provides Insights into Their Coexistence. Diversity. 2022; 14(5):412. https://doi.org/10.3390/d14050412
Chicago/Turabian StyleVillsen, Kurt, Emmanuel Corse, Gaït Archambaud-Suard, Kévin Héran, Emese Meglécz, Alexander V. Ereskovsky, Rémi Chappaz, and Vincent Dubut. 2022. "Diet Metabarcoding Reveals Extensive Dietary Overlap between Two Benthic Stream Fishes (Zingel asper and Cottus gobio) and Provides Insights into Their Coexistence" Diversity 14, no. 5: 412. https://doi.org/10.3390/d14050412
APA StyleVillsen, K., Corse, E., Archambaud-Suard, G., Héran, K., Meglécz, E., Ereskovsky, A. V., Chappaz, R., & Dubut, V. (2022). Diet Metabarcoding Reveals Extensive Dietary Overlap between Two Benthic Stream Fishes (Zingel asper and Cottus gobio) and Provides Insights into Their Coexistence. Diversity, 14(5), 412. https://doi.org/10.3390/d14050412







