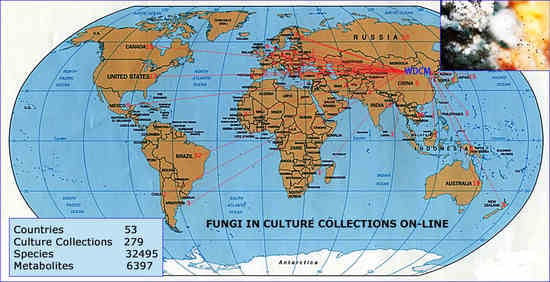
| | Country | Acronym | WDCM Number | Culture Collection Name | Number of Species |
| 1 | Argentina | BGIV | WDCM962 | Banco de Glomeromycota In Vitro (Bank of Glomeromycota In Vitro) | 8 |
| 2 | CCM | WDCM29 | Coleccion de Cultivos Microbianos | 28 |
| 3 | CEP | WDCM973 | Entomopathogenic Fungal Culture Collection of Argentina | 30 |
| 4 | Armenia | MDC | WDCM803 | Microbial Depository Center (National Microbial Culture Collection of the Republic of Armenia) | 305 |
| 5 | Australia | AMMRL | WDCM42 | Australian National Reference Laboratory in Medical Mycology | 326 |
| 6 | AWRI MCC | WDCM22 | AWRI Microorganism Culture Collection | 55 |
| 7 | CC | WDCM61 | CSIRO Canberra Rhizobium Collection | 1 |
| 8 | CS | WDCM532 | CSIRO Collection of Living Micro-algae | 1 |
| 9 | DE-CSIRO | WDCM70 | CSIRO Insect Pathogen Culture Collection | 14 |
| 10 | DMPMC | WDCM454 | Department of Microbiology | 20 |
| 11 | DFP | WDCM102 | DFP Culture Collection | 447 |
| 12 | FRR | WDCM18 | Food Science Australia, Ryde | 451 |
| 13 | WAITE | WDCM35 | Insect Pathology Pathogen Collection | 19 |
| 14 | JCT | WDCM387 | James Cook Townsville | 161 |
| 15 | KEMH | WDCM11 | KEMH/PMH Culture collection | 33 |
| 16 | ACH | WDCM47 | Mycology Culture Collection | 123 |
| 17 | WAC | WDCM77 | Plant Pathology Culture Collection | 347 |
| 18 | SBSFU | WDCM78 | School of Biological Sciences | 3 |
| 19 | SMTWA | WDCM90 | School of Medical Technology Western Australia | 1 |
| 20 | SAITP | WDCM569 | School of Pharmacy and Medical Sciences, University of South Australia | 1 |
| 21 | WACC | WDCM452 | Western Australian Culture Collection | 13 |
| 22 | WM | WDCM1205 | Westmead Medical Mycology Collection | 366 |
| 23 | DWT | WDCM36 | Wood Technology and Forest Research Division | 80 |
| 24 | Belarus | BIM | WDCM909 | Belarusian Collection of non-pathogenic microorganisms | 164 |
| 25 | Belgium | MUCL | WDCM308 | Agro-food and Environmental Fungal Collection | 4601 |
| 26 | BCCM/IHEM | WDCM642 | BCCM/IHEM—Fungi Collection: Human and Animal Health | 1855 |
| 27 | CRA-W | - | Fungi collection, Walloon Agricultural Research Centre | 11 |
| 28 | GINCO | - | Glomeromycota in vitro collection | 7 |
| 29 | LUC | - | Limburgs Universitair Centrum | 11 |
| 30 | CLO-Gent | - | Verticillium chlamydosporium (Fungi) strain collection | 1 |
| 31 | Brazil | IPT | WDCM721 | Agrupamento de Biotecnologia, Culture Collection of Microorganisms | 4 |
| 32 | ITAL | WDCM143 | Banco de Fermentos Lacticos | 2 |
| 33 | CBMAI | WDCM823 | Brazilian Collection of Microorganisms from the Environment and Industry | 111 |
| 34 | BCCCp | WDCM921 | Brazilian Culture collection of Crinipellis perniciosa | 1 |
| 35 | CRM-UNESP | WDCM1043 | Central de Recursos Microbianos do Instituto de Biociencias da UNESP | 60 |
| 36 | FTI | WDCM716 | Centro de Biotecnologia e Quimica-CEBIQ | 14 |
| 37 | CCB | WDCM713 | Colecao de Culturas de Basidiomicetos | 16 |
| 38 | CFAF | - | Colecao de Culturas de Fitopatogenos e Agentes de Controle Biologico de Fitopatogenos | 13 |
| 39 | Fiocruz/CCFF | WDCM720 | Colecao de Culturas de Fungos Filamentosos | 359 |
| 40 | CCT | WDCM885 | Colecao de Culturas Tropical | 692 |
| 41 | Fiocruz/CFAM | WDCM957 | Colecao de Fungos da Amazonia | 71 |
| 42 | CFEUnioeste | - | Colecao de Fungos Entomopatogenicos do Laboratorio de Biotecnologia Agricola | 5 |
| 43 | CFEOCA | - | Colecao de Fungos Entomopatogenicos Oldemar Cardim Abre | 19 |
| 44 | Fiocruz/CFP | WDCM951 | Colecao de Fungos Patogenicos | 20 |
| 45 | UFPEDA | WDCM114 | Colecao de Microrganismos UFPEDA | 127 |
| 46 | CICG | - | Colecao Internacional de Cultura de Glomeromycota | 24 |
| 47 | CCMA-UFLA | WDCM1083 | Culture Collection of Agricultural Microbiology | 59 |
| 48 | CCDCA | WDCM1081 | Culture Collection of Microorganisms from the Department of Food Science | 58 |
| 49 | UFRJIM | WDCM725 | Departamento de Microbiologia Medica | 2 |
| 50 | DPUA | WDCM715 | Departamento de Patologia/ICB | 81 |
| 51 | IZ | WDCM724 | Departamento de Tecnologia Rural | 344 |
| 52 | CG | WDCM712 | Embrapa Genetic Resources and Biotechnology Collection of Fungi of Interest to Biological Control | 42 |
| 53 | CCOC | WDCM575 | Fundacao Oswaldo Cruz-FIOCRUZ | 45 |
| 54 | CCT | WDCM711 | Fundacao Tropical de Pesquisas e Tecnologia “Andre Tosello” | 97 |
| 55 | INPA | WDCM719 | Laboratorio de Micologia Medica Divisao de Microbiologia e Nutricao | 93 |
| 56 | IALMIC | WDCM717 | Micoteca do Insituto Adolfo Lutz | 90 |
| 57 | IMT | WDCM718 | Micoteca do Instituto de Medicina Tropical de Sao Paulo | 254 |
| 58 | MGSS | - | Micoteca Prof. Gilson Soares da Silva | 61 |
| 59 | CMRP | WDCM1240 | Microbiological Collections of Parana Network | 344 |
| 60 | IAL | WDCM282 | Nucleo de Colecao de Micro-organismos | 3 |
| 61 | ITALSM | WDCM723 | Secao de Microbiologia | 19 |
| 62 | URM | WDCM604 | Universidade Federal de Pernambuco | 1144 |
| 63 | Bulgaria | BTCC | WDCM66 | Bulgarian Type Culture Collection | 77 |
| 64 | NBIMCC | WDCM135 | National Bank for Industrial Microorganisms and Cell Cultures | 213 |
| 65 | Canada | DAOMC | WDCM150 | Canadian Collection of Fungal Cultures | 2716 |
| 66 | CSCC | - | Cereal Smuts Cultures Collection, Winnipeg Research Centre Agriculture and Agri-Food | 6 |
| 67 | LSRRW | - | Department of Crop Sciences and Plant Ecology
University of Saskatchewan | 2 |
| 68 | MUL | WDCM250 | Department of Microbiology MUL-B 250 | 6 |
| 69 | UWO | WDCM91 | Department of Plant Sciences | 294 |
| 70 | HER | WDCM6 | Felix d’Herelle Reference Center for Bacterial Viruses | 1 |
| 71 | DFF | WDCM50 | Forest Pathology Culture Collection, Pacific Forest Research Centre | 164 |
| 72 | FSC | WDCM237 | Fredericton Stock Culture Collection | 128 |
| 73 | LYCC | WDCM634 | Lallemand Yeast Culture Collection | 1 |
| 74 | OCRC | - | Oat crown rust Collection, Winnipeg Research Centre Agriculture and Agri-Food | 1 |
| 75 | OSRC | - | Oat Stem Rust Collection, Winnipeg Research Centre Agriculture and Agri-Food | 1 |
| 76 | PFCWDCC | - | PFC Wood Decay Culture Collection, Pacific Forestry Centre | 8 |
| 77 | CCRCAF | - | Research Centre Culture Collection of Agriculture and Agri-Food | 2 |
| 78 | SGSC | WDCM338 | Salmonella Genetic Stock Centre | 21 |
| 79 | SCCM | WDCM920 | Sporometrics Culture Collection of Microorganisms | 30 |
| 80 | UAMH | WDCM73 | UAMH Center for Global Microfungal Biodiversity | 1721 |
| 81 | WLRC | - | Wheat Leaf Rust Collection, Winnipeg Research Centre Agriculture and Agri-Food | 1 |
| 82 | WSRC | - | Wheat Stem Rust Collection | 1 |
| 83 | Chile | CChRGM | WDCM1067 | Chilean Collection of Microbial Genetic Resources | 21 |
| 84 | China | CCTCC | WDCM611 | China Center for Type Culture Collection | 822 |
| 85 | ACCC | WDCM572 | Agricultural Culture Collection of China | 300 |
| 86 | CGMCC | WDCM550 | China General Microbiological Culture Collection Center | 1118 |
| 87 | CCDM | WDCM117 | Culture Collection of Department of Microbiology | 41 |
| 88 | CMCC(B) | WDCM123 | National Center for Medical Culture Collections | 34 |
| 89 | Czech Republic | RIBM | WDCM655 | Collection of Brewing Yeasts, Research Institute for Brewing and Malting | 28 |
| 90 | CMF ISB | - | Collection of Microscopic Fungi ISB (CMF ISB) | 280 |
| 91 | DBM | WDCM654 | Collection of Yeasts and Industrial Microorganisms, Institute of Chemical Technology, Department of Biochemistry and Microbiology | 103 |
| 92 | DMUP | WDCM658 | Collection of Yeasts, Department of Genetics and Microbiology, Faculty of Science, Charles University | 71 |
| 93 | CCBAS | WDCM558 | Culture Collection of Basidiomycetes | 288 |
| 94 | CCDM | - | Culture Collection of Dairy Microorganisms | 16 |
| 95 | CCF | WDCM182 | Culture Collection of Fungi | 615 |
| 96 | RIFIS | - | Culture Collection of Microorganisms with Application in the Fodder Industry, Food Research Institute | 30 |
| 97 | CCC | - | Czech Collection Clavicipitales | 31 |
| 98 | CCM | WDCM65 | Czech Collection of Microorganisms | 513 |
| 99 | CNCTC | WDCM130 | Czech National Collection of Type Cultures | 78 |
| 100 | Denmark | IBT | WDCM758 | IBT Culture Collection of Fungi | 103 |
| 101 | SSI | WDCM158 | The International Escherichia and Klebsiella Centre (WHO) | 1 |
| 102 | Finland | HAMBI | WDCM779 | HAMBI Culture Collection | 36 |
| 103 | VTTCC | WDCM139 | VTT Culture Collection | 139 |
| 104 | France | UMIP | WDCM344 | Collection de Champignons et Actinomycetes Pathogenes | 229 |
| 105 | CNCM | WDCM174 | Collection Nationale de Cultures de Microorganismes | 54 |
| 106 | LCP | WDCM659 | Fungal Strain Collection, Laboratory of Cryptogamy | 588 |
| 107 | UCLAF | WDCM552 | HMR/Romainville | 34 |
| 108 | Germany | BLWG | WDCM264 | Bayerische Landesanstalt fur Weinbau und Gartenbau | 41 |
| 109 | DSMZ | WDCM274 | DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH | 1603 |
| 110 | IFAM | WDCM145 | Institut fur Allgemeine Mikrobiologie | 2 |
| 111 | BBLF | WDCM204 | Institut fur Pflanzenschutz im Forst | 112 |
| 112 | Greece | ATHUM | WDCM650 | ATHens University Mycology | 203 |
| 113 | BPIC | WDCM610 | Benaki Phytopathological Institute Collection | 127 |
| 114 | NUA | WDCM281 | Department of Microbiology, National University of Athens | 1 |
| 115 | Hong Kong | CUHK | WDCM68 | Biology Department, Chinese University of Hong Kong | 30 |
| 116 | Hungary | DACT | WDCM496 | Dept. Agricult. Chem. Technol. | 256 |
| 117 | NCAIM | WDCM485 | National Collection of Agricultural and Industrial Microorganisms | 127 |
| 118 | India | MPKV | WDCM448 | Biological Nitrogen Fixation Project College of Agriculture | 13 |
| 119 | CCDMBI | WDCM119 | Culture Collection, Department of Microbiology | 78 |
| 120 | NTCCI | WDCM107 | Culture Collection, Microbiology and Cell Biology Laboratory | 48 |
| 121 | DUM | WDCM40 | Delhi University Mycological Herbarium | 1273 |
| 122 | DBV | WDCM173 | Division of Standardisation | 1 |
| 123 | DMSRDE | WDCM166 | DMSRDE Culture Collection | 150 |
| 124 | UMFFTD | WDCM562 | Food and Fermentation Technology Division, University of Mumbai | 5 |
| 125 | VPCI | WDCM497 | Fungal Culture Collection | 60 |
| 126 | GPCK | - | Germplasm Centre for Keratinophilic Fungi | 22 |
| 127 | ITCC | WDCM430 | Indian Type Culture Collection | 746 |
| 128 | MCM | WDCM561 | MACS Collection of Microorganisms | 8 |
| 129 | MTCC | WDCM773 | Microbial Type Culture Collection and Gene Bank | 1004 |
| 130 | NCIM | WDCM3 | National Collection of Industrial Microorganisms | 288 |
| 131 | Indonesia | FNCC | WDCM755 | Food and Nutrition Culture Collection | 92 |
| 132 | ICBB | WDCM842 | ICBB Culture Collection for Microorganisms and Cell Culture | 27 |
| 133 | ITBCC | WDCM44 | Institute of Technology Bandung Culture Collection | 57 |
| 134 | InaCC | WDCM769 | Lembaga Ilmu Pengetahuan Indonesia, Indonesian Institute for Sciences | 150 |
| 135 | Iran | IBRC | WDCM950 | Iranian Biological Resource Center | 288 |
| 136 | PTCC | WDCM124 | Persian Type Culture Collection | 50 |
| 137 | Ireland | IMD | WDCM227 | Industrial Microbiology Dublin | 104 |
| 138 | Italy | ITEM | - | Agro-Food Microbial Culture Collection | 92 |
| 139 | CSMA | WDCM147 | Centro di Studio dei Microorganismi Autotrofi—CNR | 1 |
| 140 | DBVPG | WDCM180 | Industrial Yeasts Collection | 288 |
| 141 | Japan | AHU | WDCM635 | AHU Culture Collection | 339 |
| 142 | OUT | WDCM748 | Department of Biotechnology | 220 |
| 143 | ATU | WDCM636 | Dept. of Biotechnology University of Tokyo | 4 |
| 144 | HUT | WDCM195 | HUT Culture Collection | 241 |
| 145 | IAM | WDCM190 | IAM Culture Collection | 509 |
| 146 | IFO | WDCM191 | Institute for Fermentation, Osaka | 3024 |
| 147 | RIFY | WDCM749 | Institute of Enology and Viticulture | 15 |
| 148 | TIMM | WDCM750 | Institute of Medical Mycology | 148 |
| 149 | JCM | WDCM567 | Japan Collection of Microorganisms | 1984 |
| 150 | TSY | WDCM67 | Laboratory of Mycology, Division of Microbiology | 5 |
| 151 | MAFF | WDCM637 | NARO Genebank, Microorganism Section | 777 |
| 152 | NIBH | WDCM746 | National Institute of Bioscience and Human-Technology | 15 |
| 153 | RIB | WDCM640 | National Research Institute of Brewing | 14 |
| 154 | NRIC | WDCM747 | Nodai Research Institute Culture Collection | 161 |
| 155 | IFM | WDCM60 | Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University | 394 |
| 156 | Malaysia | SKUK | WDCM565 | Simpanan Kultur Universiti Kebangsaan | 40 |
| 157 | UKKP | WDCM430 | Universiti Kebangsaan Kultur Perubatan | 21 |
| 158 | Mexico | CENACUMI | WDCM757 | Centro Nacional de Cultivos Microbianos (National Center For Microbial Cultures) | 277 |
| 159 | CFQ | WDCM100 | Cepario de la Facultad de Quimica | 65 |
| 160 | ITD | WDCM99 | Coleccion de Cepas Microbianas | 10 |
| 161 | ENCB-IPN | WDCM449 | Coleccion de Cultivos de la Escuela Nacional de Ciencias Biologicas | 97 |
| 162 | INIF | WDCM104 | Coleccion de Microhongos | 83 |
| 163 | LIH-UNAM | WDCM817 | CultureCollection of Histoplasma capsulatum Strains from the Fungal Immunology Laboratory of the Department of Microbiology and Parasitology, Faculty of Medicine, UNAM | 1 |
| 164 | IIBM-UNAM | WDCM48 | Industrial Culture Collection | 40 |
| 165 | CDBB | WDCM500 | Unidad de Servicios de la Coleccion Nacional de Cepas Microbianas y Cultivos Celulares | 123 |
| 166 | CISM | WDCM95 | Verticillium dahliae from cotton | 3 |
| 167 | The Netherlands | CBS | WDCM133 | Centraalbureau voor Schimmelcultures, Fungal and Yeast Collection | 18,346 |
| 168 | New Zealand | NZFS | WDCM62 | Forest Research Culture Collection | 100 |
| 169 | ICMP | WDCM589 | International Collection of Microorganisms from Plants | 969 |
| 170 | WARC | WDCM376 | New Zealand Reference Culture Collection | 1 |
| 171 | NZRD | WDCM318 | New Zealand Reference Culture Collection of Microorganisms, Dairy Section | 2 |
| 172 | NZRM | WDCM457 | New Zealand Reference Culture Collection, Medical Section | 5 |
| 173 | Pakistan | FCBP | WDCM859 | First fungal culture bank of Pakistan | 119 |
| 174 | Philippines | ITDI | WDCM503 | Industrial Technology Development Institute | 49 |
| 175 | UPCC | WDCM310 | Natural Sciences Research Institute Culture Collection | 214 |
| 176 | PNCM-BIOTECH | WDCM620 | Philippine National Collection of Microorganisms | 144 |
| 177 | Poland | LOCK | WDCM105 | Centre of Industrial Microorganisms Collection | 38 |
| 178 | IAFB | WDCM212 | Collection of Industrial Microorganisms | 159 |
| 179 | IAW | - | Research and Development Centre for Biotechnology Culture Collection | 17 |
| 180 | LCC | WDCM231 | University of Warmia and Mazury in Olsztyn | 34 |
| 181 | Portugal | MUM | WDCM816 | Micoteca da Universidade do Minho | 125 |
| 182 | Republic of Korea | EFCC | - | Entomopatogenic Fungal Culture Collection | 58 |
| 183 | Romania | ICCF | WDCM232 | Collection of Industrial Microorganisms | 21 |
| 184 | Russian Federation | VKM | WDCM342 | All-Russian Collection of Microorganisms | 2100 |
| 185 | VIZR | WDCM760 | Collection for plant protection, All-Russian Institute of Plant Protection | 14 |
| 186 | KMM | WDCM644 | Collection of Marine Microorganisms of the Pacific Institute of Biorganic Chemistry of the Far-Eastern Branch of the RAS | 48 |
| 187 | IPP | - | Collection of Monoxenic Cultures of Arbuscular Mycorrhizal Fungi of the Institute of Plant Physiology RAS | 4 |
| 188 | VNIISC | - | Culture Collection of the Institute of Agricultural Microbiology, Russian Academy of Agricultural Sciences | 143 |
| 189 | IBC | - | Institute of Cell Biology RAS | 13 |
| 190 | LE(BIN) | WDCM1015 | Komarov Botanical Institute Basidiomycetes Culture Collection | 672 |
| 191 | VKPM | WDCM588 | Russian National Collection of Industrial Microorganisms | 733 |
| 192 | DSB MSU | - | The Department of Soil Sciences Moscow State University | 35 |
| 193 | RIA | WDCM337 | The Russia Research Institute for Antibiotics Culture Collection | 43 |
| 194 | Senegal | MAO | WDCM53 | Mircen Afrique Ouest | 1 |
| 195 | Serbia | ISS | WDCM375 | Collection of Bacteria | 1 |
| 196 | Singapore | DBS | WDCM510 | Department of Biological Culture Collection | 98 |
| 197 | NUSDM | WDCM568 | Department of Microbiology | 24 |
| 198 | Slovak Republic | CCWDF | - | Culture Collection of Wood-destroying Fungi | 52 |
| 199 | CCY | WDCM333 | Culture Collection of Yeasts | 394 |
| 200 | RIVE | WDCM28 | Research Institute for Viticulture and Enology | 96 |
| 201 | Slovenia | MZKI | WDCM599 | Microbial Culture Collection of National Institute of Chemistry | 174 |
| 202 | ZIM | WDCM810 | ZIM Collection of Industrial Microorganisms | 112 |
| 203 | Spain | CECT | WDCM412 | Coleccion Espanola de Cultivos Tipo | 394 |
| 204 | CCMCU | WDCM599 | Culture Collection of Microorganisms | 182 |
| 205 | Sri Lanka | DMBUK | WDCM564 | Department of Microbiology | 44 |
| 206 | Sweden | CCUG | WDCM32 | Culture Collection University of Goteborg | 92 |
| 207 | FCUG | WDCM651 | Fungal Cultures University of Goteborg | 507 |
| 208 | UPSC | WDCM603 | Uppsala University Culture Collection of Fungi | 800 |
| 209 | Switzerland | CCTM | WDCM475 | Centre de Collection de Type Microbien | 19 |
| 210 | Taiwan | BCRC | WDCM59 | Bioresource Collection and Research Center | 1545 |
| 211 | Thailand | BSMB | WDCM491 | Bacteriology and Soil Microbiology Branch | 20 |
| 212 | BCC | WDCM783 | BIOTEC Culture Collection | 399 |
| 213 | NRPSU | WDCM679 | Department of Agro-industry, Faculty of Natural Resources | 36 |
| 214 | ABKMI | WDCM698 | Department of Applied Biology, Faculty of Science | 12 |
| 215 | DBKKU1 | WDCM687 | Department of Biology, Faculty of Science | 27 |
| 216 | SWU2 | WDCM697 | Department of Biology, Faculty of Science | 4 |
| 217 | DBMU2 | WDCM667 | Department of Biotechnology, Faculty of Science | 62 |
| 218 | FTCMU | WDCM690 | Department of Food Science and Technology, Faculty of Agriculture | 10 |
| 219 | DMST | WDCM707 | Department of Medical Sciences Culture Collection | 204 |
| 220 | MPSU | WDCM492 | Department of Microbiology | 2 |
| 221 | DMKKU1 | WDCM680 | Department of Microbiology, Faculty of Medicine | 2 |
| 222 | DMMU3 | WDCM668 | Department of Microbiology, Faculty of Medicine Siriraj Hospital | 66 |
| 223 | DMKU | WDCM669 | Department of Microbiology, Faculty of Science | 57 |
| 224 | NU | WDCM696 | Department of Microbiology, Faculty of Science | 2 |
| 225 | PPKU1 | WDCM670 | Department of Plant Pathology, Faculty of Agriculture | 5 |
| 226 | PPKU3 | WDCM672 | Department of Plant Pathology, Faculty of Agriculture | 19 |
| 227 | PPKU4 | WDCM673 | Department of Plant Pathology, Faculty of Agriculture | 18 |
| 228 | PPKU5 | WDCM674 | Department of Plant Pathology, Faculty of Agriculture | 10 |
| 229 | MLMJI | WDCM701 | Department of Plant Protection, Faculty of Agricultural Production | 7 |
| 230 | CMKKU | WDCM684 | Diagnostic Microbiology Unit Division of Clinical Laboratory Srinagarind Hospital, Faculty of Medicine | 9 |
| 231 | IFRPD | WDCM676 | Institute of Food Research and Product Development, Kasetsart University | 35 |
| 232 | KUFC | WDCM677 | Kasetsart University Fungus Collection, Department of Plant Pathology, Faculty of Agriculture | 18 |
| 233 | KKU | WDCM23 | MICKKU Culture Collection | 18 |
| 234 | MLLD | WDCM702 | Microbiological Research Laboratory, Soil and Water Section, Department of Land Development | 3 |
| 235 | CHULA | WDCM511 | Department of Microbiology, Faculty of Science | 34 |
| 236 | DMCU | WDCM663 | Microbiology Department, Faculty of Science | 24 |
| 237 | MLRU | WDCM695 | Microbiology Laboratory, Department of Biology, Faculty of Science | 7 |
| 238 | MSDS | WDCM494 | Microbiology Section, Biological Science Division, Department of Science Services | 9 |
| 239 | MSCMU | WDCM692 | Microbiology Section, Chiang Mai University (MSCMU) | 50 |
| 240 | MSPP | WDCM704 | Mycology Section, Plant Pathology and Microbiology Division, Department of Agricultural Science | 4 |
| 241 | NCSC | WDCM664 | National Center of Streptococcus Collection, Department of Microbiology, Faculty of Medical Science | 16 |
| 242 | PCU | WDCM662 | Pharmaceutical Sciences Chulalongkorn University Culture Collection | 10 |
| 243 | PPKM | WDCM699 | Plant Production Technology Department, Faculty of Agricultural Technology | 5 |
| 244 | ERAEP | WDCM706 | Radiation Ecology Section, Biological Science Division, Office of Atomic Energy for Peace | 5 |
| 245 | SSMJI | WDCM700 | Science Section, Department of General Education, Faculty of Agricultural Business | 9 |
| 246 | TISTR | WDCM383 | TISTR Culture Collection, Bangkok MIRCEN | 235 |
| 247 | Turkey | KUKENS | WDCM101 | Centre for Research and Application of Culture Collections of Microorganisms | 83 |
| 248 | RSKK | WDCM828 | Refik Saydam National Type Culture Collection-RSKK | 22 |
| 249 | UK | IMI | WDCM214 | CABI Bioscience Genetic Resource Collection | 3716 |
| 250 | BEG | WDCM777 | La Banque European des Glomales | 30 |
| 251 | NCPF | WDCM184 | National Collection of Pathogenic Fungi | 151 |
| 252 | NCTC | WDCM154 | National Collection of Type Cultures | 1 |
| 253 | NCWRF | WDCM134 | National Collection of Wood Rotting Fungi | 296 |
| 254 | NCYC | WDCM169 | National Collection of Yeast Cultures | 449 |
| 255 | PHBL | WDCM508 | Philip Harris Biological Ltd. | 27 |
| 256 | DMCCUS | WDCM478 | School of Biological Sciences Culture Collection | 17 |
| 257 | CCMF | WDCM766 | University of Portsmouth | 207 |
| 258 | Ukraine | IBK | WDCM1152 | Culture Collection of Mushrooms | 195 |
| 259 | UCM | WDCM1203 | Ukrainian Collection of Microorganisms | 195 |
| 260 | USA | NRRL | WDCM97 | Agricultural Research Service Culture Collection | 742 |
| 261 | ATCC | WDCM1 | American Type Culture Collection | 5585 |
| 262 | ARSEF | WDCM112 | ARS Collection of Entomopathogenic Fungi | 366 |
| 263 | LMS | WDCM530 | Carolina Biological Supply Company | 67 |
| 264 | FGSC | WDCM115 | Fungal Genetics Stock Center | 21 |
| 265 | INVAM | - | International Culture Collection of VA Mycorrhizal Fungi | 76 |
| 266 | MCC | - | Mushroom Culture Collection Department of Plant Pathology, The Pennsylvania State University, USA | 198 |
| 267 | DM | - | National Center for Agricultural Utilization Research | 54 |
| 268 | NCMA | WDCM2 | Provasoli-Guillard National Center for Marine Algae and Microbiota | 4 |
| 269 | BMP | - | Pryor Lab Culture Collection, The University of Arizona, USA | 119 |
| 270 | RMF | - | Rocky Mountain Fungus, Wyoming | 374 |
| 271 | DSC | WDCM849 | The Dicty Stock Center | 16 |
| 272 | UA | - | The University of Alabama | 4 |
| 273 | UBC | - | University of California at Berkeley | 1 |
| 274 | UM | - | University of Maine, Orono, Maine, USA | 14 |
| 275 | WVDH | WDCM411 | West Virginia Hygienic Laboratory | 13 |
| 276 | WSF | - | Wisconsin Soil Fungus | 183 |
| 277 | Uzbekistan | NCAM | WDCM808 | National Collection of Agricultural Microorganisms | 146 |
| 278 | Vietnam | CNTP | - | The Industrial Microorganisms Culture Collection | 39 |
| 279 | Zimbabwe | BDUZ | WDCM17 | Biological Sciences | 17 |