Runs of Homozygosity and Gene Identification in Pelibuey Sheep Using Genomic Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Genotyping and Data Quality Control
2.3. Linkage Disequilibrium
2.4. Genetic Structure of the Population, Inbreeding Coefficient, and Effective Population Size
2.5. Detection of Runs of Homozygosity
2.6. Annotation of Significant Genomic Regions
3. Results
3.1. Dataset
3.2. Linkage Disequilibrium (LD)
3.3. Genetic Diversity, Inbreeding Coefficient, and Effective Population Size
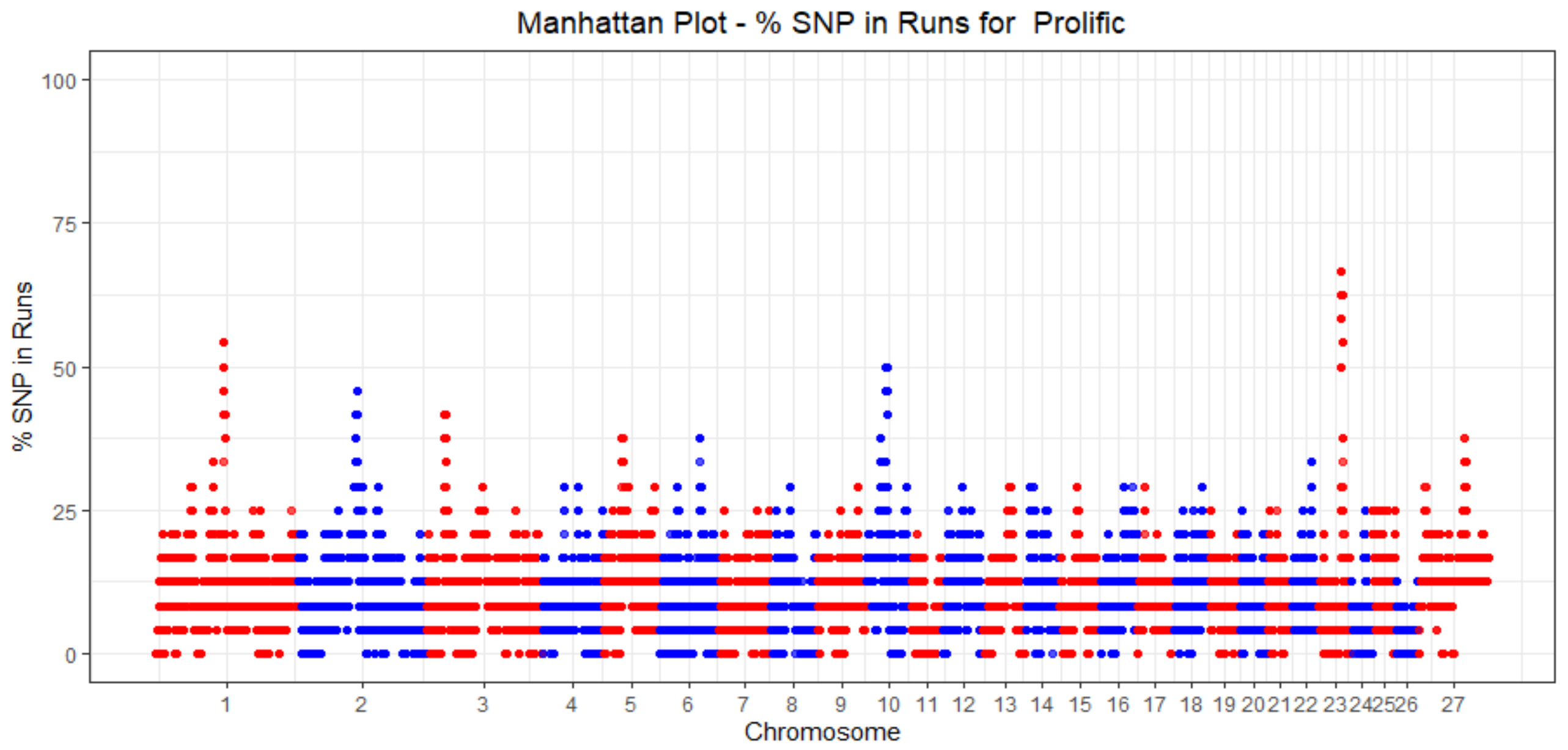
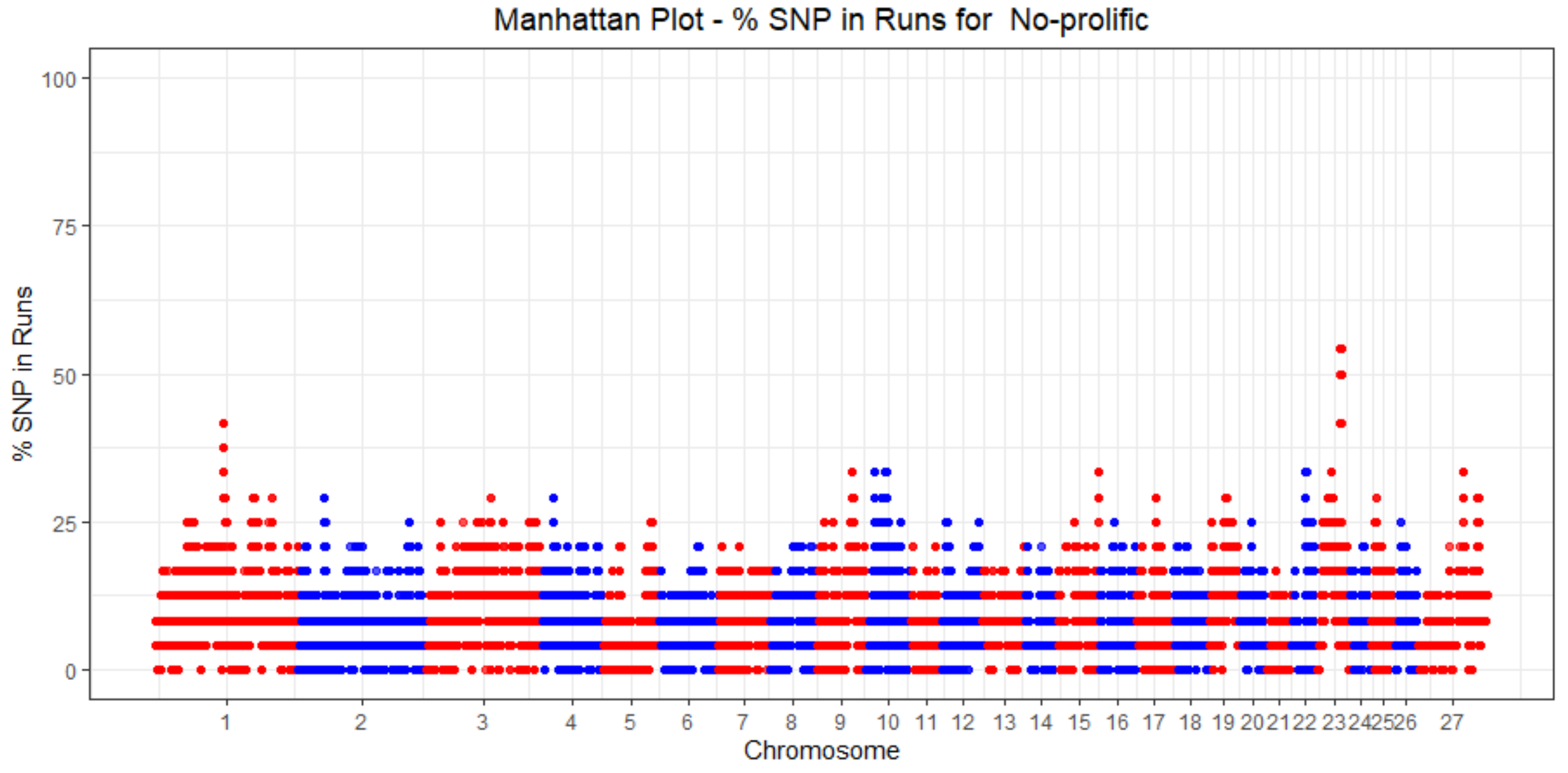
3.4. ROH Identification
3.5. Functional Annotation of ROH in Prolific Sheep
4. Discussion
4.1. Genetic Diversity, Inbreeding Coefficient, and Effective Population Size
4.2. Functional Annotation of ROH in Prolific Sheep
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.K.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals—A review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirkena, T.; Duguma, G.; Haile, A.; Tibbo, M.; Okeyo, A.M.; Wurzinger, M.; Sölkner, J. Genetics of adaptation in domestic farm animals: A review. Livest. Sci. 2010, 132, 1–12. [Google Scholar] [CrossRef]
- Diamond, J. Evolution, consequences and future of plant and animal domestication. Nature 2002, 418, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.J.; et al. Revealing the history of sheep domestication using retrovirus integrations. Science 2009, 324, 532–536. [Google Scholar] [CrossRef] [Green Version]
- Delgado, J.V.; Perezgrovas, R.; Camacho, M.E.; Fresno, M.; Barba, C. The Wool-Less Canary Sheep and their relationship with the present breeds in America. Anim. Genet. Resour. Inf. 2000, 28, 27–35. [Google Scholar] [CrossRef]
- Aguilar-Martinez, C.U.; Berruecos-Villalobos, J.M.; Espinoza-Gutiérrez, B.; Segura-Correa, J.C.; Valencia-Méndez, J.; Roldán-Roldán, A. Origen, historia y situacion actual de la oveja pelibuey en Mexico. Trop. Subtrop. Agroecosyst. 2017, 20, 429–439. [Google Scholar]
- Gutiérrez, J.; Rubio, M.S.; Méndez, R.D. Effects of crossbreeding Mexican Pelibuey sheep with Rambouillet and Suffolk on carcass traits. Meat Sci. 2005, 70, 1–5. [Google Scholar] [CrossRef]
- Abied, A.; Bagadi, A.; Bordbar, F.; Pu, Y.; Augustino, S.M.A.; Xue, X.; Xing, F.; Gebreselassie, G.; Han, J.-L.; Mwacharo, J.M.; et al. Genomic Diversity, Population Structure, and Signature of Selection in Five Chinese Native Sheep Breeds Adapted to Extreme Environments. Genes 2020, 11, 494. [Google Scholar] [CrossRef]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [Green Version]
- Miao, X.; Luo, Q. Genome-wide transcriptome analysis between Small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 2013, 145, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Jansson, T. Genes Involved in Ovulation Rate and Litter Size in Sheep. Bachelor’s Thesis, Department of Animal Breeding and Genetics, SLU, Uppsala, Sweden, 2014; pp. 1–14. [Google Scholar]
- Abdoli, R.; Zamani, P.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Nadri, S. A review on prolificacy genes in sheep. Reprod. Domest. Anim. 2016, 51, 631–637. [Google Scholar] [CrossRef]
- Gootwine, E. Invited review: Opportunities for genetic improvement toward higher prolificacy in sheep. Small Rumin. Res. 2020, 186, 106090. [Google Scholar] [CrossRef]
- Chhotaray, S.; Panigrahi, M.; Pal, D.; Ahmad, S.F.; Bhanuprakash, V.; Kumar, H.; Parida, S.; Bhushan, B.; Gaur, G.K.; Mishra, B.P.; et al. Genome-wide estimation of inbreeding coefficient, effective population size and haplotype blocks in Vrindavani crossbred cattle strain of India. Biol. Rhythm Res. 2019, 52, 666–679. [Google Scholar] [CrossRef]
- Olšanská, B.; Kasarda, R.; Lehocká, K.; Moravčíková, N. Genome-wide characterisation of regions under intense selection based on runs of homozygosity in Charolais cattle. Acta Fytotech. Zootech. 2020, 23, 350–355. [Google Scholar] [CrossRef]
- Zhan, H.; Zhang, S.; Zhang, K.; Peng, X.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Genome-wide patterns of homozygosity and relevant characterizations on the population structure in Piétrain pigs. Genes 2020, 11, 577. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Tolone, M.; Sardina, M.T.; Sottile, G.; Sutera, A.M.; Di Gerlando, R.; Portolano, B. Genome-wide scan for runs of homozygosity identifies potential candidate genes associated with local adaptation in Valle del Belice sheep. Genet. Sel. Evol. 2017, 49, 84. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.S.; Gao, L.; Xie, X.L.; Ren, Y.L.; Shen, Z.Q.; Wang, F.; Shen, M.; Eypórsdóttir, E.; Hallsson, J.H.; Kiseleva, T.; et al. Genome-wide association analyses highlight the potential for different genetic mechanisms for litter size among sheep breeds. Front. Genet. 2018, 9, 118. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Ben Jemaa, S.; Sottile, G.; Casu, S.; Portolano, B.; Ciani, E.; Pilla, F. Combined approaches to identify genomic regions involved in phenotypic differentiation between low divergent breeds: Application in Sardinian sheep populations. J. Anim. Breed. Genet. 2019, 136, 526–534. [Google Scholar] [CrossRef]
- Magi, A.; Giangregorio, T.; Semeraro, R.; Carangelo, G.; Palombo, F.; Romeo, G.; Seri, M.; Pippucci, T. AUDACITY: A comprehensive approach for the detection and classification of Runs of Homozygosity in medical and population genomics. Comput. Struct. Biotechnol. J. 2020, 18, 1956–1967. [Google Scholar] [CrossRef]
- Jiang, R.; Cheng, J.; Cao, X.K.; Ma, Y.L.; Chaogetu, B.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Hu, L.Y.; Chen, H. Copy number variation of the SHE gene in sheep and its association with economic traits. Animals 2019, 9, 531. [Google Scholar] [CrossRef] [Green Version]
- Escaramís, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief. Funct. Genom. 2015, 14, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Tobar, K.M.C.; Álvarez, D.C.L.; Franco, L.Á.Á. Genome-wide association studies in sheep from Latin America. Review. Rev. Mex. Cienc. Pecu. 2020, 11, 659–683. [Google Scholar] [CrossRef]
- Xiong, H.; Xiong, H.; He, X.; Li, J.; Li, J.; Liu, X.; Peng, C.; Xi, D.; Deng, W. Genetic diversity and genetic origin of Lanping black-boned sheep investigated by genome-wide single-nucleotide polymorphisms (SNPs). Arch. Anim. Breed. 2020, 63, 193–201. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Di Gerlando, R.; Sardina, M.T.; Sutera, A.M.; Moscarelli, A.; Tolone, M.; Cortellari, M.; Marletta, D.; Crepaldi, P.; Portolano, B. Genome-wide patterns of homozygosity reveal the conservation status in five italian goat populations. Animals 2021, 11, 1510. [Google Scholar] [CrossRef]
- Fang, Y.; Hao, X.; Xu, Z.; Sun, H.; Zhao, Q.; Cao, R.; Zhang, Z.; Ma, P.; Sun, Y.; Qi, Z.; et al. Genome-Wide Detection of Runs of Homozygosity in Laiwu Pigs Revealed by Sequencing Data. Front. Genet. 2021, 12, 463. [Google Scholar] [CrossRef]
- He, S.; Di, J.; Han, B.; Chen, L.; Liu, M.; Li, W. Genome-wide scan for runs of homozygosity identifies candidate genes related to economically important traits in Chinese merino. Animals 2020, 10, 524. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Montiel, W.; Martínez-Núñez, M.A.; Ramón-Ugalde, J.P.; Román-Ponce, S.I.; Calderón-Chagoya, R.; Zamora-Bustillos, R. Genome-Wide Association Study Reveals Candidate Genes for Litter Size Traits in Pelibuey Sheep. Animals 2020, 10, 434. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 1–16. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Barbato, M.; Orozco-terWengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [Green Version]
- Ceballos, F.C.; Hazelhurst, S.; Ramsay, M. Runs of Homozygosity in sub-Saharan African populations provide insights into a complex demographic and health history Francisco. bioRxiv 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Ballan, M.; Bovo, S.; Schiavo, G.; Schiavitto, M.; Negrini, R.; Fontanesi, L. Genomic diversity and signatures of selection in meat and fancy rabbit breeds based on high-density marker data. Genet. Sel. Evol. 2022, 54, 3. [Google Scholar] [CrossRef] [PubMed]
- Meyermans, R.; Gorssen, W.; Buys, N.; Janssens, S. How to study runs of homozygosity using plink? a guide for analyzing medium density snp data in livestock and pet species. BMC Genom. 2020, 21, 94. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes. Available online: https://www.semanticscholar.org/paper/detectRUNS%3A-Detect-runs-of-homozygosity-and-runs-of-Biscarini-Cozzi/ccc3091c370ae99f7c1b56aa5521b80cd47a4258 (accessed on 20 August 2021).
- Zhao, G.; Zhang, T.; Liu, Y.; Wang, Z.; Xu, L.; Zhu, B.; Gao, X.; Zhang, L.; Gao, H.; Liu, G.E.; et al. Genome-wide assessment of runs of homozygosity in chinese wagyu beef cattle. Animals 2020, 10, 1425. [Google Scholar] [CrossRef]
- Woodruff, D.S. Populations, Species, and Conservation Genetics. In Encyclopedia of Biodiversity; Levin, S.A., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 811–829. [Google Scholar]
- Daetwyler, H.D.; Villanueva, B.; Bijma, P.; Woolliams, J.A. Inbreeding in genome-wide selection. J. Anim. Breed. Genet. 2007, 124, 369–376. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Ramilo, S.T.; Elsen, J.M.; Legarra, A. Inbreeding and effective population size in French dairy sheep: Comparison between genomic and pedigree estimates. J. Dairy Sci. 2019, 102, 4227–4237. [Google Scholar] [CrossRef]
- Ortiz Sanchéz, Y.; Martínez Guzmán, M.; Kübler, I.; Ariza, M.F.; Castro Molina, S.; Infante-González, J. Diversidad genética del Ovino Criollo de Pelo Colombiano mediante el uso del marcador molecular de tipo polimorfismos de nucleótido simple (SNP). Rev. Investig. Vet. Perú 2021, 32, e19487. [Google Scholar] [CrossRef]
- Prieur, V.; Clarke, S.M.; Brito, L.F.; McEwan, J.C.; Lee, M.A.; Brauning, R.; Dodds, K.G.; Auvray, B. Estimation of linkage disequilibrium and effective population size in New Zealand sheep using three different methods to create genetic maps. BMC Genet. 2017, 18, 68. [Google Scholar] [CrossRef]
- Tao, L.; He, X.; Wang, X.; Di, R.; Chu, M. Litter size of sheep (Ovis aries): Inbreeding depression and homozygous regions. Genes 2021, 12, 109. [Google Scholar] [CrossRef]
- McHugo, G.P.; Browett, S.; Randhawa, I.A.S.; Howard, D.J.; Mullen, M.P.; Richardson, I.W.; Park, S.D.E.; Magee, D.A.; Scraggs, E.; Dover, M.J.; et al. A Population Genomics Analysis of the Native Irish Galway Sheep Breed. Front. Genet. 2019, 10, 927. [Google Scholar] [CrossRef] [Green Version]
- Weeks, A.R.; Stoklosa, J.; Hoffmann, A.A. Conservation of genetic uniqueness of populations may increase extinction likelihood of endangered species: The case of Australian mammals. Front. Zool. 2016, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Sigdel, A.; Bisinotto, R.S.; Peñagaricano, F. Genes and pathways associated with pregnancy loss in dairy cattle. Sci. Rep. 2021, 11, 13329. [Google Scholar] [CrossRef]
- Williams, S.M.; An, J.Y.; Edson, J.; Watts, M.; Murigneux, V.; Whitehouse, A.J.O.; Jackson, C.J.; Bellgrove, M.A.; Cristino, A.S.; Claudianos, C. An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Mol. Psychiatry 2019, 24, 1707–1719. [Google Scholar] [CrossRef] [Green Version]
- Killeen, A.P.; Morris, D.G.; Kenny, D.A.; Mullen, M.P.; Diskin, M.G.; Waters, S.M. Global gene expression in endometrium of high and low fertility heifers during the mid-luteal phase of the estrous cycle. BMC Genom. 2014, 15, 234. [Google Scholar] [CrossRef] [Green Version]
- Müller, P.S.; Schulz, R.; Maretto, S.; Costello, I.; Srinivas, S.; Bikoff, E.; Robertson, E. The fibronectin leucine-rich repeat transmembrane protein Flrt2 is required in the epicardium to promote heart morphogenesis. Development 2011, 138, 1297–1308. [Google Scholar] [CrossRef] [Green Version]
- Gonda, X.; Eszlari, N.; Torok, D.; Gal, Z.; Bokor, J.; Millinghoffer, A.; Baksa, D.; Petschner, P.; Antal, P.; Breen, G.; et al. Genetic underpinnings of affective temperaments: A pilot GWAS investigation identifies a new genome-wide significant SNP for anxious temperament in ADGRB3 gene. Transl. Psychiatry 2021, 11, 337. [Google Scholar] [CrossRef]
- Lu, X.; Abdalla, I.M.; Nazar, M.; Fan, Y.; Zhang, Z.; Wu, X.; Xu, T.; Yang, Z. Genome-wide association study on reproduction-related body-shape traits of chinese holstein cows. Animals 2021, 11, 1927. [Google Scholar] [CrossRef]
- Sinha, N.; Roy, S.; Huang, B.; Wang, J.; Padmanabhan, V.; Sen, A. Developmental programming: Prenatal testosterone-induced epigenetic modulation and its effect on gene expression in sheep ovary. Biol. Reprod. 2020, 102, 1045–1054. [Google Scholar] [CrossRef]
- Cicvaric, A.; Yang, J.; Bulat, T.; Zambon, A.; Dominguez-Rodriguez, M.; Kühn, R.; Sadowicz, M.G.; Siwert, A.; Egea, J.; Pollak, D.D.; et al. Enhanced synaptic plasticity and spatial memory in female but not male FLRT2-haplodeficient mice. Sci. Rep. 2018, 8, 3703. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Yu, Q.; Liu, H.; Huang, T.; Zhao, S.; Ma, J.; Zhao, H. Growth Hormone Promotes in vitro Maturation of Human Oocytes. Front. Endocrinol. 2019, 10, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abedel-Majed, M.A.; Romereim, S.M.; Davis, J.S.; Cupp, A.S. Perturbations in Lineage Specification of Granulosa and Theca Cells May Alter Corpus Luteum Formation and Function. Front. Endocrinol. 2019, 10, 832. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.C.; Bøtkjær, J.A.; Østrup, O.; Petersen, K.B.; Yding Andersen, C.; Grøndahl, M.L.; Englund, A.L.M. Two waves of transcriptomic changes in periovulatory human granulosa cells. Hum. Reprod. 2020, 35, 1230–1245. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Sasaki, S.; Gotoh, Y.; Nakamura, Y.; Aoyagi, Y.; Kawahara, T.; Sugimoto, Y. Genetic variants related to gap junctions and hormone secretion influence conception rates in cows. Proc. Natl. Acad. Sci. USA 2013, 110, 19495–19500. [Google Scholar] [CrossRef] [Green Version]
- Selva, D.M.; Hirsch-Reinshagen, V.; Burgess, B.; Zhou, S.; Chan, J.; McIsaac, S.; Hayden, M.R.; Hammond, G.L.; Vogl, A.W.; Wellington, C.L. The ATP-binding cassette transporter 1 mediates lipid efflux from Sertoli cells and influences male fertility. J. Lipid Res. 2004, 45, 1040–1050. [Google Scholar] [CrossRef] [Green Version]
- Qian, H.; Zhao, X.; Cao, P.; Lei, J.; Yan, N.; Gong, X. Structure of the Human Lipid Exporter ABCA1. Cell 2017, 169, 1228–1239.e10. [Google Scholar] [CrossRef] [Green Version]
- Bibikova, M.; Le, J.; Barnes, B.; Saedinia-Melnyk, S.; Zhou, L.; Shen, R.; Gunderson, K.L. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics 2009, 1, 177–200. [Google Scholar] [CrossRef]
- Hendricks, D.T.; Taylor, R.; Reed, M.; Birrer, M.J. FHIT Gene Expression in Human Ovarian, Endometrial, and Cervical Cancer Cell Lines. Cancer Res. 1997, 57, 2112–2115. [Google Scholar]
- Fan, T.; Hu, Y.; Xin, J.; Zhao, M.; Wang, J. Analyzing the genes and pathways related to major depressive disorder via a systems biology approach. Brain Behav. 2020, 10, e01502. [Google Scholar] [CrossRef] [Green Version]
- La, Y.; Liu, Q.; Zhang, L.; Chu, M. Single nucleotide polymorphisms in SLC5A1, CCNA1, and ABCC1 and the association with litter size in small-tail han sheep. Animals 2019, 9, 432. [Google Scholar] [CrossRef] [Green Version]
- Brubaker, D.; Liu, Y.; Wang, J.; Tan, H.; Zhang, G.; Jacobsson, B.; Muglia, L.; Mesiano, S.; Chance, M.R. Finding lost genes in GWAS via integrative-omics analysis reveals novel sub-networks associated with preterm birth. Hum. Mol. Genet. 2016, 25, 5254–5264. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, L.; Cao, J.; Wu, M.; Ma, X.; Liu, Z.; Liu, R.; Zhao, F.; Wei, C.; Du, L. Genome-wide specific selection in three domestic sheep breeds. PLoS ONE 2015, 10, e0128688. [Google Scholar] [CrossRef] [Green Version]
- Eydivandi, S.; Roudbar, M.A.; Karimi, M.O.; Sahana, G. Genomic scans for selective sweeps through haplotype homozygosity and allelic fixation in 14 indigenous sheep breeds from Middle East and South Asia. Sci. Rep. 2021, 11, 2834. [Google Scholar] [CrossRef]
- An, X.; Ma, H.; Han, P.; Zhu, C.; Cao, B.; Bai, Y. Genome-wide differences in DNA methylation changes in caprine ovaries between oestrous and dioestrous phases. J. Anim. Sci. Biotechnol. 2018, 9, 85. [Google Scholar] [CrossRef]


| Farm | n | |
|---|---|---|
| Prolific | No Prolific | |
| Las Potrancas | 7 | 13 |
| El Rodeo | 8 | 6 |
| San Alberto | 5 | 5 |
| El Cortijo | 4 | - |
| Total | 24 | 24 |
| ID | Farm | Animals | Ho | He ± SD | F | Fis |
|---|---|---|---|---|---|---|
| Prolific | El Cortijo | 4 | 0.280 | 0.216 × 104 | −0.044 | 0.027 |
| Las Potrancas | 7 | 0.289 | 0.288 × 104 | 0.006 | −0.004 | |
| El Rodeo | 8 | 0.299 | 0.288 × 104 | 0.064 | −0.040 | |
| San Alberto | 5 | 0.288 | 0.288 × 104 | 0.001 | 0.001 | |
| No-prolific | San Alberto | 5 | 0.283 | 0.288 × 104 | −0.026 | 1.56 × 102 |
| Las Potrancas | 13 | 0.200 | 0.284 × 104 | −0.029 | 2.936 × 101 | |
| El Rodeo | 6 | 0.289 | 0.288 × 104 | 0.010 | −0.006 |
| Generation Ago | Ne | Dist | r2 | r2 ± SD |
|---|---|---|---|---|
| 13 | 192 | 3,749,483 | 0.0335209 | 0.045845 |
| 15 | 212 | 3,273,237 | 0.0346997 | 0.0475146 |
| 17 | 235 | 2,844,328 | 0.0360788 | 0.0494267 |
| 20 | 260 | 2,459,811 | 0.0376715 | 0.0514669 |
| 23 | 290 | 2,116,245 | 0.0391051 | 0.0537381 |
| 27 | 327 | 1,811,153 | 0.0405171 | 0.0552067 |
| 32 | 364 | 1,541,204 | 0.0426174 | 0.057825 |
| 38 | 411 | 1,303,353 | 0.0446124 | 0.0606928 |
| 45 | 468 | 1,095,152 | 0.0464675 | 0.0632997 |
| 54 | 535 | 914,214 | 0.0486655 | 0.065538 |
| Category | 0–6 Mb | 6–12 Mb | 12–24 Mb | 24–48 Mb | >48 Mb |
|---|---|---|---|---|---|
| Prolific | 2010 | 114 | 44 | 17 | 8 |
| No-prolific | 2055 | 99 | 28 | 3 | 0 |
| Chr | SNP1 | Gen 1 | SNP2 | Gen 2 | Pos1 pb | Pos2 pb | SNP | Kb | Density |
|---|---|---|---|---|---|---|---|---|---|
| 1 | OAR1_215101913.1 | DGKG | OAR1_221977733.1 | PEX5L | 19,915,7665 | 20,534,2835 | 107 | 6185.1 | 57.8 |
| 2 | OAR2_103911840.1 | LINGO2 | DU300339_104.1 | -- | 96,505,293 | 11,001,2279 | 283 | 13,506.9 | 47.7 |
| 3 | OAR3_195973885.1 | PKP2 | OAR3_209916841.1 | -- | 18,173,2438 | 19,472,4802 | 261 | 12,992.3 | 49.7 |
| 7 | s07208.1 | FLRT2 | OAR7_108923470.1 | TRIM69 | 94,069,579 | 99,977,231 | 125 | 5907.6 | 47.2 |
| 9 | OAR9_5634331.1 | ADGRB3 | OAR9_18493683.1 | -- | 5,696,218 | 17,666,651 | 143 | 11,970.4 | 51.1 |
| 13 | OAR13_35436626.1 | CACNB2 | s11802.1 | DIP2C | 32,041,727 | 45,925,125 | 265 | 13,883.3 | 52.3 |
| 16 | OAR16_55041078.1 | CDH12 | OAR16_62246248.1 | -- | 50,543,439 | 57,002,028 | 132 | 6458.5 | 48.9 |
| 25 | s62494.1 | NRG3 | OAR25_48288071_X.1 | -- | 36,775,576 | 45,193,605 | 177 | 8418.0 | 47.5 |
| Terms | Genes | List of Genes | p-Value |
|---|---|---|---|
| GO Biological Process | |||
| (GO:0051965) positive regulation of synapse assembly | 7.97 × 105 | LINGO2, NLGN1, FLRT2, ADGRB3, ADGRB1, IL1RAP, EPHB1 | 2.88 |
| (GO:0035556) intracellular signal transduction | 9.05 × 105 | DGKG, SRPK2, DGKE, PRKCH, DGKB, STAC, PRKCE, DCDC2, STK38, ARHGEF3, ARHGEF4, DGKI, RGS6 | 5.34 |
| (GO:0007205) protein kinase C-activating G-protein coupled receptor signaling pathway | 1.646090535 | DGKG, DGKE, DGKB, DGKI | 1.64 |
| (GO:0007155) cell adhesion | 0.004436121 | DSCAM, CNTN5, NINJ2, PRKCE, NCAM1, CTNNA3, NCAM2 | 2.880658436 |
| (GO:0032926) negative regulation of activin receptor signaling pathway | 0.099286868 | ACVR1, MAGI2 | 0.823045267 |
| GO Molecular Function | |||
| (GO:0005096) GTPase activator activity | 0.026660316 | ARHGAP10, ADGRB3, NPRL3, DAB2IP, SMAP1, RGS6 | 2.469135802 |
| (GO:0005432) calcium: sodium antiporter activity | 0.054904043 | SLC8A3, SLC8A1 | 0.823045267 |
| (GO:0005548) phospholipid transporter activity | 0.06815558 | ABCA1, ABCG1 | 0.823045267 |
| GO Cellular Component | |||
| (GO:0005887) integral component of plasma membrane | 0.029326849 | ABCA1, SLC8A3, ALK, NLGN1, DSCAM, FLRT2, EPHB1, HCN1, DDR2 | 3.703703704 |
| (GO:0048471) perinuclear region of cytoplasm | 0.048576171 | ABCA1, SLC8A3, PKHD1, ATP7B, DAB1, PRKCE, PTPRM, TMEM192, DGKI | 3.703703704 |
| (GO:1990454) L-type voltage-gated calcium channel complex | 0.052429865 | CACNB2, CACNA2D1 | 0.823045267 |
| KEGG pathway | |||
| oas04921: Oxytocin signaling pathway | CACNB2, PPP3R2, CACNB4, CACNA2D1, CACNA2D3, ITPR1 | 0.026681693 | |
| oas04020: Calcium signaling pathway | SLC8A3, ITPKB, PPP3R2, ITPR1, SLC8A1, GRM1 | 0.053913811 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Montiel, W.; Cob-Calan, N.N.; Cahuich-Tzuc, L.E.; Rueda, J.A.; Quiroz-Valiente, J.; Meza-Villalvazo, V.; Zamora-Bustillos, R. Runs of Homozygosity and Gene Identification in Pelibuey Sheep Using Genomic Data. Diversity 2022, 14, 522. https://doi.org/10.3390/d14070522
Hernández-Montiel W, Cob-Calan NN, Cahuich-Tzuc LE, Rueda JA, Quiroz-Valiente J, Meza-Villalvazo V, Zamora-Bustillos R. Runs of Homozygosity and Gene Identification in Pelibuey Sheep Using Genomic Data. Diversity. 2022; 14(7):522. https://doi.org/10.3390/d14070522
Chicago/Turabian StyleHernández-Montiel, Wilber, Nubia Noemi Cob-Calan, Lilia E. Cahuich-Tzuc, José A. Rueda, Jorge Quiroz-Valiente, Víctor Meza-Villalvazo, and Roberto Zamora-Bustillos. 2022. "Runs of Homozygosity and Gene Identification in Pelibuey Sheep Using Genomic Data" Diversity 14, no. 7: 522. https://doi.org/10.3390/d14070522
APA StyleHernández-Montiel, W., Cob-Calan, N. N., Cahuich-Tzuc, L. E., Rueda, J. A., Quiroz-Valiente, J., Meza-Villalvazo, V., & Zamora-Bustillos, R. (2022). Runs of Homozygosity and Gene Identification in Pelibuey Sheep Using Genomic Data. Diversity, 14(7), 522. https://doi.org/10.3390/d14070522






