Phylogeny of Trachelomonas and Strombomonas (Euglenaceae) Based on Morphological and Molecular Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains Collection, Cultivation and Morphological Study
2.2. DNA Extraction, Amplification and Sequencing
2.3. Phylogenetic Analyses
3. Result
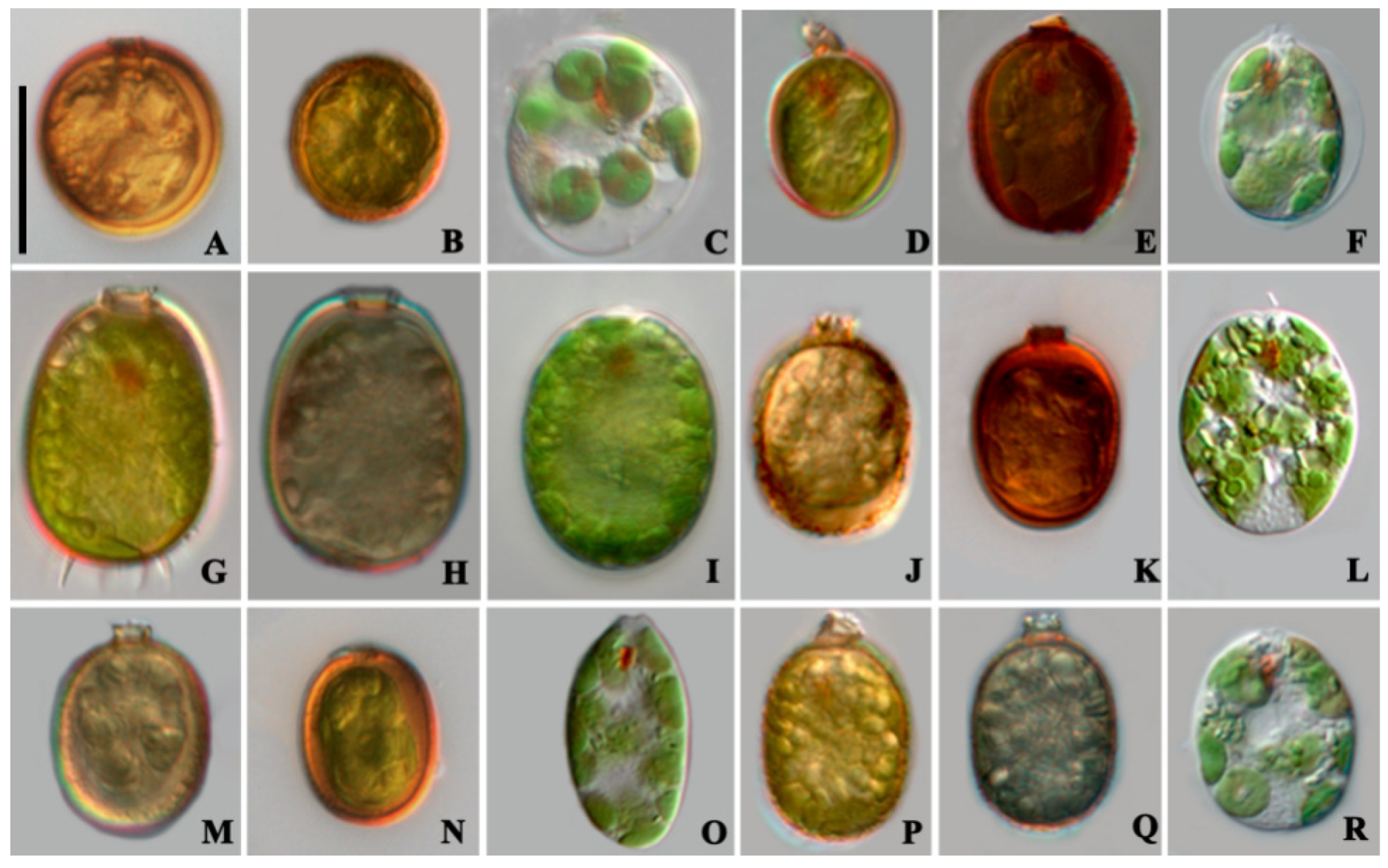
3.1. Morphological Characteristics
- Trachelomonas cervicula A.Stokes (Figure 1A–C)
3.2. Stability of Loricae Morphology
3.3. Phylogenetic Analysis and Morphological Characteristics of the Clades
4. Discussion
4.1. Stable Morphological Characteristics
4.2. The Relationship between Phylogenetic Tree and Morphological Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehrenberg, C.G. Dritter Beitrag zur Enkenntniss grosser Organisation in der Richtung des Kleinsten Raumes; Abh, K., Ed.; Akad. Wiss: Berlin, Germany, 1833; pp. 145–336. [Google Scholar]
- Deflandre, G. Monographie du Genre Trachelomonas Ehr.; Imprimerie Andre Lesot: Nemours, France, 1926; pp. 1–162. [Google Scholar]
- Dunlap, J.R.; Walne, P.L.; Kivic, P.A. Cytological and taxonomic studies of the Euglenales. II. Comparative microarchitecture and cytochemistry of envelopes of Strombomonas and Trachelomonas. Brit. Phycol. J. 1986, 21, 399–405. [Google Scholar] [CrossRef]
- Pringsheim, E.G. Observations on some species of Trachelomonas grown in culture. New Phytol. 1953, 52, 93–113. [Google Scholar] [CrossRef]
- Singh, K.P. Studies in the genus Trachelomonas. I. description of six organisms in cultivation. Amer. J. Bot. 1956, 43, 258–266. [Google Scholar] [CrossRef]
- Rosowski, J.R.; Vadas, R.L.; Kugrens, P. Surface configuration of the lorica of the euglenoid Trachelomonas as revealed with scanning electron microscopy. Amer. J. Bot. 1975, 62, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.S.D.; Walne, P.L.; Dunlap, J.R. Cytological and taxonomic studies of the Euglenales. I. Ultrastructure and envelope elemental composition in Trachelomonas. Brit. Phycol. J. 1986, 21, 387–397. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Liu, H.; Yu, J.; Sun, S.; Zhang, D.; Bao, W. Comparative studies on the fine structure and elemental composition of envelopes of Trachelomonas and Strombomonas (Euglenophyta). Acta Bot. Sin. 2003, 45, 601–607. [Google Scholar]
- Müllner, A.N.; Angler, D.G.; Samuel, R.; Linton, E.W.; Triemer, R.E. Phylogenetic analysis of phagotrophic, phototrophic, and osmotrophic euglenoids by using the nuclear 18S rDNA sequence. Int. J. Syst. Evol. Microbiol. 2001, 51, 783–791. [Google Scholar] [CrossRef]
- Marin, B.; Palm, A.; Klingberg, M.A.X.; Melkonian, M. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 2003, 154, 99–145. [Google Scholar] [CrossRef]
- Triemer, R.E.; Linton, E.; Shin, W.; Nudelman, A.; Monfils, A.; Bennett, M.; Brosnan, S. Phylogeny of the Euglenales based upon combined SSU and LSU rDNA sequence comparisons and description of Discoplastis Gen. Nov. (Euglenophyta). J. Phycol. 2006, 42, 731–740. [Google Scholar] [CrossRef]
- Ciugulea, I.; Nudelman, M.A.; Brosnan, S.; Triemer, R.E. Phylogeny of the euglenoid loricate genera Trachelomonas and Strombomonas (Euglenophyta) inferred from nuclear SSU and LSU rDNA. J. Phycol. 2008, 44, 406–418. [Google Scholar] [CrossRef]
- Linton, E.W.; Karnkowska-Ishikawa, A.; Kim, J.I.; Shin, W.; Bennett, M.S.; Kwiatowski, J.; Zakryś, B.; Triemer, R.E. Reconstructing euglenoid evolutionary relationships using three genes: Nuclear SSU and LSU, and chloroplast SSU rDNA sequences and the description of Euglenaria Gen. Nov. (Euglenophyta). Protist 2010, 161, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.S.; Triemer, R.E. Chloroplast genome evolution in the Euglenaceae. J. Eukaryot Microbiol. 2015, 62, 773–785. [Google Scholar] [CrossRef]
- Karnkowska-Ishikawa, A.; Milanowski, R.; Triemer, R.E.; Zakryś, B. Taxonomic revisions of morphologically similar species from two euglenoid genera: Euglena (E. granulata and E. velata) and Euglenaria (Eu. anabaena, Eu. caudata, and Eu. clavata). J. Phycol. 2012, 48, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Shin, W. Phylogeny of the Euglenales inferred from plastid LSU rDNA sequences. J. Phycol. 2008, 44, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Čalasan, A.Ž.; Kretschmann, J.; Gottschling, M. Contemporary integrative taxonomy for sexually deprived protists: A case study of Trachelomonas (Euglenaceae) from western Ukraine. Taxon 2020, 69, 93–113. [Google Scholar]
- Kim, J.I.; Linton, E.W.; Shin, W. Taxon-rich multigene phylogeny of the photosynthetic euglenoids (Euglenophyceae). Front. Ecol. Evol. 2015, 3, 98. [Google Scholar] [CrossRef] [Green Version]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World–Wide Electronic Publication, National University of Ireland, Galway. Available online: http://www.algaebase.org/ (accessed on 8 June 2022).
- Shi, Z.; Wang, Q.; Xie, S.; Dai, J. Flora Algarum Sinicarum Aquae Dulcis. Tomus VI. Euglenophyta; Science Press: Beijing, China, 1999; pp. 93–98. [Google Scholar]
- Jiang, X.; Chen, X.; Pang, W.; Wang, Q. A new species of Trachelomonas (Euglenaceae, Euglenophyceae) from China. Fottea 2022, 22, 30–38. [Google Scholar] [CrossRef]
- Fells, A.; Jiang, X.; Chaber, K.; Łukomska-Kowalczyk, M.; Milanowski, R.; Wang, Q.; Zakryś, B. Molecular and morphological delimitation of species in Strombomonas (Euglenida) including a protocol for DNA obtainment utilising a chelating resin. Taxon 2022, in press. [Google Scholar]
- Watanabe, M.M.; Hiroki, M. Nies-Collection List of Strains, 5th ed.; National Institute of Environmental Studies: Tsukuba, Japan, 1997; pp. 1–127.
- Collins, T.J. Introduction to ImageJ for light microscopy. Microsc. Microanal. 2007, 13, S02. [Google Scholar] [CrossRef]
- Pallant, J. Spss survival manual: A step by step guide to data analysis using spss for windows. Aust. Nz. J. Publ. Health 2013, 37, 597–598. [Google Scholar]
- Kosmala, S.; Bereza, M.; Milanowski, R.; Kwiatowski, J.; Zakryś, B. Morphological and molecular examination of relationships and epitype establishment of Phacus pleuronectes, Phacus orbicularis, and Phacus hamelii. J. Phycol. 2007, 43, 1071–1082. [Google Scholar] [CrossRef]
- Kim, J.I.; Shin, W.; Triemer, R.E. Multigene analyses of photosynthetic euglenoids and new family, Phacaceae (Euglenales). J. Phycol. 2010, 46, 1278–1287. [Google Scholar] [CrossRef]
- Łukomska-Kowalczyk, M.; Chaber, K.; Fells, A.; Milanowski, R.; Zakryś, B. Description of Flexiglena gen. nov. and new members of Discoplastis and Euglenaformis (Euglenida). J. Phycol. 2021, 57, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids. Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and eff ective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Poniewozik, M.; Ziba, E.; Sajnaga, E. Envelope development and variation in Trachelomonas hispida (Euglenophyta). Algae 2018, 33, 305–318. [Google Scholar] [CrossRef]
- West, L.K.; Walne, P.L. Trachelomonas hispida var. coronata (Euglenophyceae). II. Envelope substructure. J. Phycol. 1980, 16, 498–506. [Google Scholar] [CrossRef]
- Brosnan, S.; Brown, P.J.; Farmer, M.A.; Triemer, R.E. Morphological separation of the euglenoid genera Trachelomonas and Strombomonas based on lorica development and posterior strip reduction. J. Phycol. 2005, 41, 590–605. [Google Scholar] [CrossRef]
- Deflandre, G. Strombomonas noveau genre de’ Euglenacees. Arch. Protistenkd. 1930, 69, 551–614. [Google Scholar]
- Huber-Pestalozzi, G. Das Phytoplankton des Süsswassers; Systematik und Biologie: 4 Teil; Euglenophyceen; E. Schweizerbartsche Verlagsbuchhandlung: Stuttgart, Germany, 1955; pp. 1–606. [Google Scholar]




| Taxon | Isolate/Strain | Length (µm) | Width (µm) | Number of Chloroplasts | Pyrenoid | Loricae Shape |
|---|---|---|---|---|---|---|
| T. armata (Ehrenberg) Stein | SHNUS50B3 | 32–49 | 15–22 | 10–15 | Haplopyrenoid | Ovoid with short collar, posterior end with long cone thorns |
| T. bernardinensis Vischer | SHNUWeijian | 31–58 | 18–29 | 6–10 | Haplopyrenoid | Fusiform with collar, collar with toothed edge, a visible process at the posterior end |
| T. cervicula A.Stokes | SHNUS39D1 | 20–29 | 20–29 | 5–10 | None | Spherical without collar, surface smooth |
| T. cf. bacillifera var. minima | SHNUQ4B2 | 24–25 | 20–21 | 8–10 | Haplopyrenoid | Elliptical without collar, surface with rod spines |
| T. cf. crebea | SHNUS39B2 | 20–37 | 20–27 | 8–10 | Diplopyrenoid | Elliptical with collar, surface with granular process |
| T. cf. sydneyensis var. minima | SHNUC1D1 | 23–26 | 19–20 | 10–12 | Haplopyrenoid | Elliptical without collar, surface with short conical spines |
| T. hispida (Perty) Stein | SHNUQ1D6 | 30–33 | 22–24 | 7–10 | Diplopyrenoid | Oblong without collar, surface with short cone spines |
| T. lefevrei Deflandre | SHNUCC | 27–31 | 20–24 | 9–10 | Haplopyrenoid | Elliptical with collar, collar with toothed edge |
| T. planctonica var. oblonga Svirenko | SHNUS2C1 | 20–23 | 17–20 | 10–20 | Haplopyrenoid | Elliptical with collar, collar with toothed edge |
| T. similis Stokes | SHNUS41C3 | 20–23 | 16–17 | 5–8 | Haplopyrenoid | Wide elliptical with oblique collar, surface punctured |
| T. playfairii Deflandre | SHNUN14B1 | 19–26 | 16–20 | 8–10 | Haplopyrenoid | Elliptical with oblique collar, collar with toothed edge |
| Trachelomonas sp. | SHNUS26A3 | 16–19 | 13–16 | 7–10 | Haplopyrenoid | Elliptical without collar, anterior end with annular thickenings, surface with short spines |
| Trachelomonas sp. | SHNUC4C3 | 21–23 | 21–23 | 5–8 | Diplopyrenoid | Spherical without collar, surface with short conical spines |
| Trachelomonas sp. | SHNUS12C2 | 20–22 | 17–20 | 10–12 | Haplopyrenoid | Elliptical without collar, surface with fine spines |
| T. subplanctonica Jiang & Pang | SHNUS17C2 | 23–34 | 19–30 | 6–10 | Haplopyrenoid | Ellipsoidal with collar, collar with toothed edge, surface punctured |
| T. undulaticollum Shi | SHNUS35D5 | 28–30 | 20–22 | 8–10 | Haplopyrenoid | Elliptical with long collar |
| S. borystheniensis (Roll) Popova | SHNUQ6C2 | 22–30 | 19–20 | 6–11 | Diplopyrenoid | Wide ellipsoidal with a wide short collar, without tail |
| S. cf. borystheniensis | SHNUQ6C3 | 27–38 | 18–25 | 5–7 | Haplopyrenoid | Ellipsoidal with collar, without tail |
| S. gibberosa (Playfair) Deflandre | SHNUQ6A1 | 40–53 | 21–35 | 7–9 | Haplopyrenoid | Middle wide-rhomboidal, loricae with a wide short collar and a long tail |
| S. fluviatilis (Lemmermann) Deflandre | SHNUQ6B2 | 53–71 | 21–28 | 9–13 | Haplopyrenoid | Fusiform with straight collar and a long tail |
| S. maxima (Skvortsov) Deflandre | SHNUS6A3 | 70–109 | 28–43 | 8–17 | Haplopyrenoid | Broadly ovate with straight collar and a long tail |
| S. triquetra (Playfair) Deflandre | SHNUQ6A4 | 34–43 | 20–21 | 6–10 | Haplopyrenoid | Inverted triangle with short collar and a short caudate process |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Chen, X.; Pang, W.; Wang, Q. Phylogeny of Trachelomonas and Strombomonas (Euglenaceae) Based on Morphological and Molecular Data. Diversity 2022, 14, 623. https://doi.org/10.3390/d14080623
Jiang X, Chen X, Pang W, Wang Q. Phylogeny of Trachelomonas and Strombomonas (Euglenaceae) Based on Morphological and Molecular Data. Diversity. 2022; 14(8):623. https://doi.org/10.3390/d14080623
Chicago/Turabian StyleJiang, Xiaodie, Xi Chen, Wanting Pang, and Quanxi Wang. 2022. "Phylogeny of Trachelomonas and Strombomonas (Euglenaceae) Based on Morphological and Molecular Data" Diversity 14, no. 8: 623. https://doi.org/10.3390/d14080623
APA StyleJiang, X., Chen, X., Pang, W., & Wang, Q. (2022). Phylogeny of Trachelomonas and Strombomonas (Euglenaceae) Based on Morphological and Molecular Data. Diversity, 14(8), 623. https://doi.org/10.3390/d14080623






