Abstract
Early blight (EB) is a destructive disease affecting potato and tomato plants in Russia, caused by a heterogeneous group of plant pathogenic Alternaria fungi. The current species delimitation in Alternaria sect. Porri with medium to large conidia and a long (filamentous) beak is based on molecular data. In this study, the ITS, GAPDH, RPB2, TEF1, and Alt a 1 gene regions were analyzed in 41 large-spored Alternaria isolates obtained from diseased potato and tomato plants collected from 13 regions in Russia. Our data revealed five pathogenic species (A. alternariacida, A. grandis, A. linariae, A. protenta, and A. solani). Two species (A. solani and A. linariae) were found to be associated with early blight of tomato. Alternaria linariae and A. protenta were confirmed as the major causal agents of tomato and potato early blight, respectively. There were no phylogenetic groupings among tested Russian Alternaria isolates associated with their locality.
1. Introduction
In Russia, early blight (EB) is considered to be one of the most destructive diseases of potato (Solanum tuberosum L.) and tomato (S. lycopersicum L.) plants, the leading vegetable crops in the country. The volume of potato production in Russia is about 30 million tons and financial losses from the development of potato diseases can be very significant. A wide variety of plant pathogenic Alternaria fungi cause early blight. It is characterized by necrotic lesions in the aerial parts of plants. Species delimitation among Alternaria spp. pathogens was always challenging, and all large-spored species were generally considered to be Alternaria solani Sorauer. Based on conidial morphology, Simmons described 21 species occurring on Solanaceae plants [1]. The main EB-inducing agents for potato were A. solani and A. grandis E.G. Simmons. A. tomatophila, A. cretica, and A. subcylindrica were identified to cause EB on tomatoes [2]. In 2014, a large-scale phylogenetic reconstruction of large-spored Alternaria species was performed [3]. Multilocus analyses using concatenated phylogeny of internal transcribed spacers 1 and 2 and 5.8S gene (ITS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), DNA-dependent RNA polymerase II (RPB2) gene, transcription elongation factor 1α (TEF1) and Alternaria major allergen gene (Alt a 1) have separated Alternaria species into different groups according to their original hosts [3]. At present, this is the most extensive phylogenetic study of large-spored Alternaria. There were only A. solani, A. protenta, and A. grandis strains isolated from potato plants in the study; A. alternariacida and A. linaria from tomato plants; and A. nitrimali and A. solani-nigri from other Solanaceae plants.
There have only been a few studies conducted on species affecting potato and tomato plants according to the revised Porri classification proposed by Woudenberg et al. [3]. As far as we know, such work has not been conducted in Russia, the former Soviet Union, or Eastern Europe. Thus, our work aimed to revise large-spored Alternaria strains that infect potato and tomato plants by DNA sequencing species-specific regions in accordance with the research of Woudenberg et al. [3].
2. Materials and Methods
2.1. Isolates
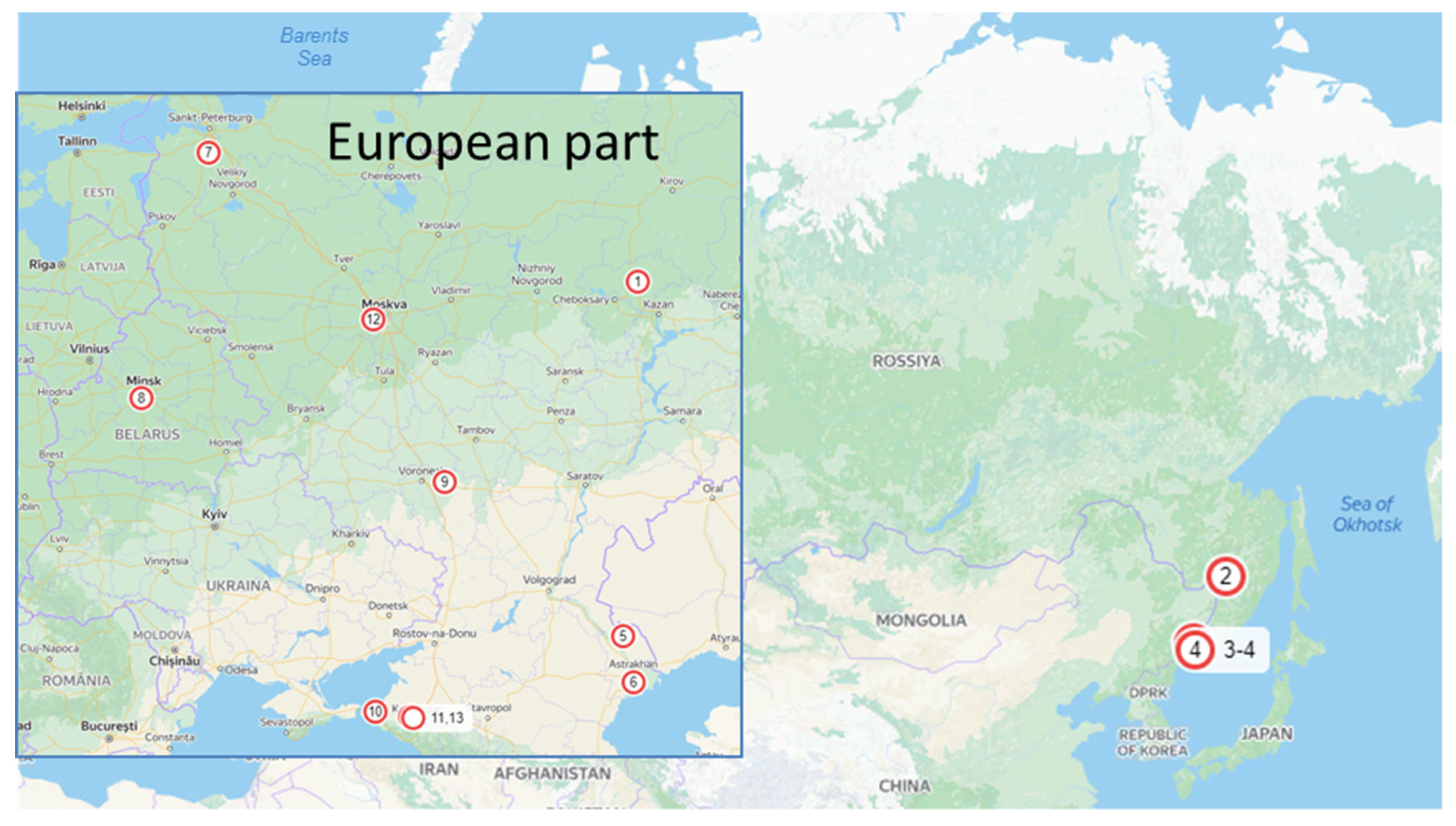
Alternaria isolates were collected from commercial potato and tomato fields and small private gardens in different regions of Russia (Figure 1, Table 1): Voronezh (site 9), Leningrad (7), Astrakhan (5,6), Krasnodar (10,11,13), Moscow (12), Primorsky (3,4), Khabarovsk (2) regions, and Tatarstan republic (1) as well as in Belarus (8). For direct isolation, plant material was incubated for 24 h in moist chambers. Under a binocular microscope (MBS 10, Russia), conidia were transferred with a preparation needle to potato dextrose agar (PDA) medium with an antibiotic solution (1000 U/mL benzylpenicillin sodium). After that, the hyphal tips were transferred under a binocular microscope onto another Petri dish with PDA medium.
Figure 1.
Collection sites of Alternaria isolates. 1—Mari El republic; 2—Khabarovsky krai; 3—Primorsky krai, Ussurijsk district; 4—Primorsky krai, Vladivostok district, 5—Astrakhan region, Habarlin district; 6—Astrakhan region, Kamyzyak district; 7—Leningrad region; 8—Republic of Belarus; 9—Voronezh region; 10—Krasnodar region, Strelka village; 11—Krasnodar region, Temryuk district; 12—Moscow region; 13—Krasnodar region, Anapa district.

Table 1.
Isolates used in this study.
2.2. PCR and Sequencing
In order to isolate the DNA, the mycelium of fungi was grown on liquid pea medium [4]. DNA was extracted according to the standard CTAB protocol [5]. The internal transcribed spacer 1 (ITS1) and ITS2 regions and the 5.8S ribosomal DNA (rDNA) region of the fungi were amplified with ITS1 and ITS4 primers [6], parts of the GAPDH gene—with gpd1 and gpd2 [7], the RPB2 gene—with RPB2-5F2 [8] and fRPB2-7cR [9], and the TEF1 gene—with the primers EF1-728F and EF1-986R [10]. PCR was performed with GenPak® PCR Core kit (Isogene Lab., Moscow, Russia). The PCR program consisted of an initial denaturing step at 94 °C for 5 min, 35 amplification cycles, and an additional extending step at 72 °C for 3 min. For the primer pairs ITS1/ITS4, RPB2-5F2/fRPB2-7cR, EF1-728F/EF1-986R, and Alt-for/Alt-rev, the amplification cycles were 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. For the primer pair gpd1/gpd2, the amplification cycles were 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 1 min. After the reaction, the length and purity of the PCR products were monitored by electrophoresis in 1% agarose gel. Ethidium bromide was used to visualize the PCR product. A piece of gel containing the amplicon of the desired size was cut with a sterile scalpel and placed in a microtube. Then, the instructions specified in the description of the CleanUp Standard kit for DNA isolation from the gel (Evrogen Co Ltd., Moscow, Russia) were followed. DNA sequencing was carried out according to the Sanger method at the Evrogen company with both forward and reverse primers. The raw sequence reads were assembled into the consensus sequence in Geneious v. 7.13 (Biomatters Limited, Auckland, New Zealand) using default settings.
2.3. Phylogenetic Analysis
The sequences of each gene were aligned and cut at the ends. Specifically, for the ALT-A1 gene, a fragment of about 475 nucleotides (nt) was considered; for the ITS—538 nt, for the GAPDH gene—580 nt, for the RPB2—772 nt, and for the TEF1—355 nt. Multiple sequence alignments were generated with MAFTT algorithm plugin in Geneious ver. 7.13 (Biomatters Ltd., Auckland, New Zealand). Sequences of fragments of ITS-5,8S-ITS2, GAPDH, RPB2, TEF1, and Alt a1 genes were used for phylogenetic study (Table 2). Additionally, gene sequences, including the outgroup, were retrieved from Woudenberg et al. [3] (Table 2).

Table 2.
Reference isolates from Solanaceus plants from Woudenberg et al., 2014 [3].
2.4. Bioinformatic Methods
Sequences were aligned with the MAFFT version 7 web tool (http://mafft.cbrc.jp/alignment/server/ accessed on 1 December 2021) with subsequent manual processing. Phylogenetic reconstructions were performed with maximum likelihood (ML) and Bayesian (BI) analyses. Nucleotide substitution models for BI were chosen with TOPALI v. 2.5 (The Apache Software Foundation, Maryland, CA, USA) based on the Bayesian information criterion (BIC). Bayesian analyses were performed with Geneious v. 7.13. In these analyses, three parallel runs with four chains each and other default parameters were run for one million generations. A burn-in of 25% was used in the final analyses, ensuring the average standard deviation of split frequencies had reached <0.01 for all data sets. Support at nodes was indicated when posterior probabilities were ≥0.6. For ML analyses, the best-fit substitution model for the alignment was estimated based on the Akaike information criterion (AIC) using the IQ-TREE Web Service (http://iqtree.cibiv.univie.ac.at/ accessed 12 December 2021). The Tamura–Nei (TN) [11] model plus empirical base frequencies allowing for a proportion of invariable sites was chosen for the “potato” dataset. For the “tomato” dataset, TN plus empirical base frequencies and a freeRate model with 2 categories were used. The RAxML program ver. 7.0.3 (The Exelixis Lab, Heidelberg, Germany) was used for the heuristic search.
3. Results
3.1. Bioinformatic Analysis
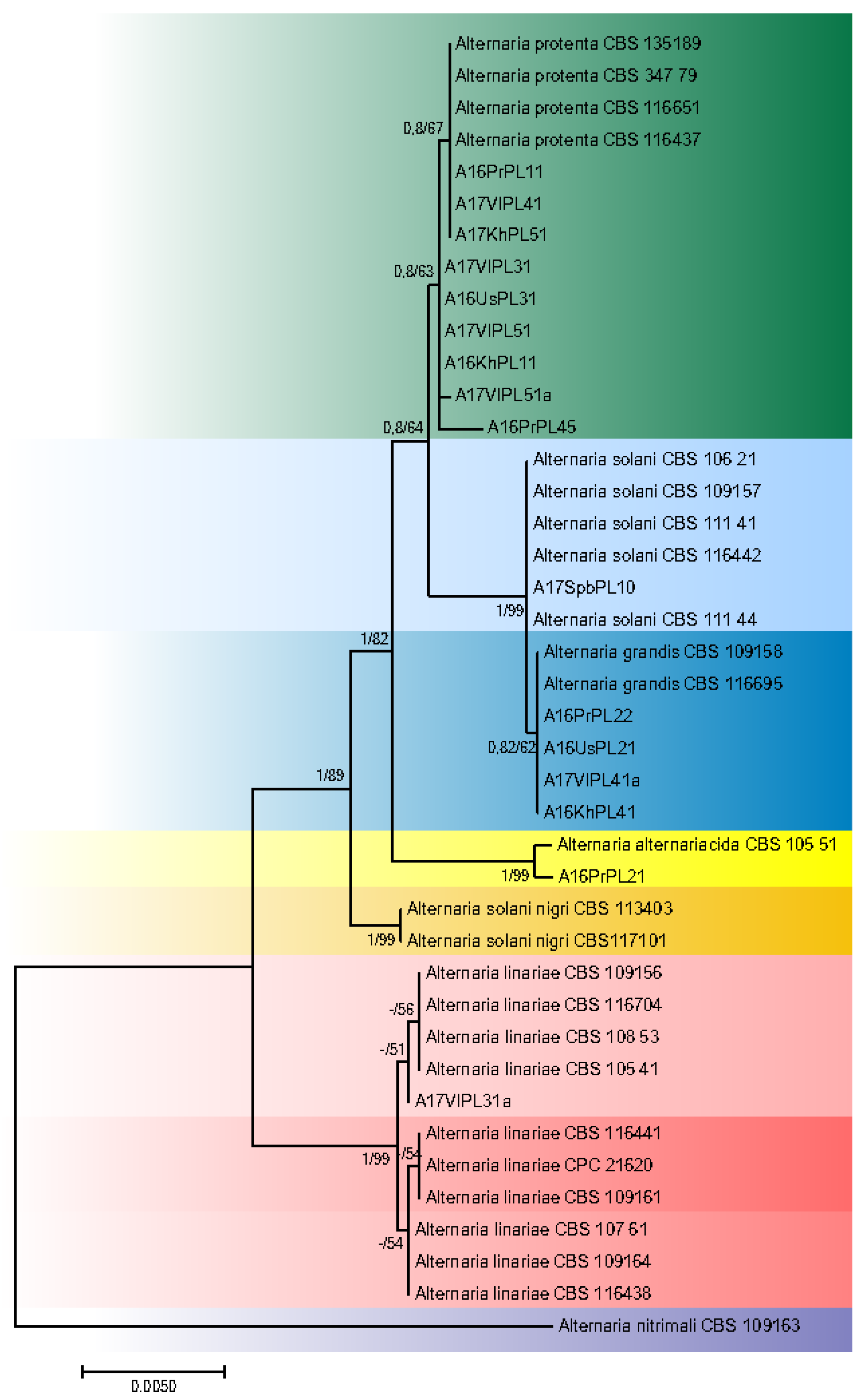
The aligned sequences of ITS, GAPDH, RPB2, TEF1, and Alt a1 regions had a total length of 2690 characters for the alignments of both potato- and tomato-related strains, with 2, 6, 28, 6, and 8 unique site patterns, respectively. The aligned “potato” data set for seven species included 2572 constant sites and 54 parsimony informative sites. Alignment of the “tomato” data set sequences resulted in 2574 constant sites and 47 parsimony informative sites. The phylogenetic analyses based upon Maximum Likelihood inference of ITS, GAPDH, RPB2, TEF1, and Alt a1 regions of 41 Alternaria isolates are shown in Figure 2 and Figure 3. Bayesian Inference and ML returned similar topologies and relevant support values. Two species, A. solani-nigri (R. Dubey, S.K. Singh and Kamal) and A. nitrimali (E.G. Simmons and M.E. Palm), which also occur on Solanaceae plants, were used in tree A. nitrimali and found to be a proper out-group as indicated by its clear segregation from the other strains used in the study.
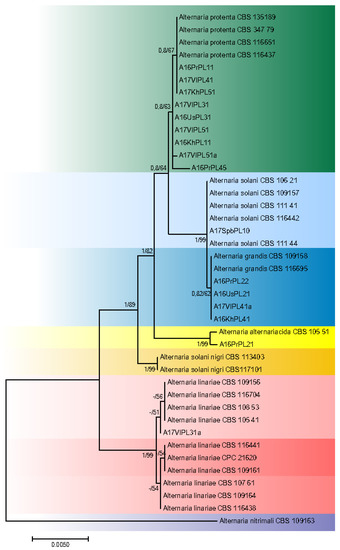
Figure 2.
Phylogenetic tree based on the combined gene sequences of ITS, GAPDH, Alt a1, TEF1, and RPB2 of strains isolated from S. tuberosum plants. Bayesian posterior probabilities followed by ML bootstrap values are shown at nodes.
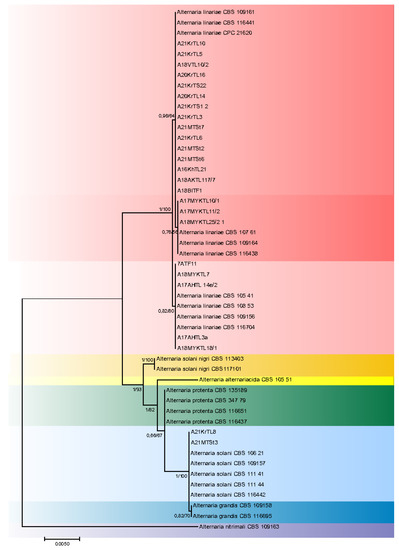
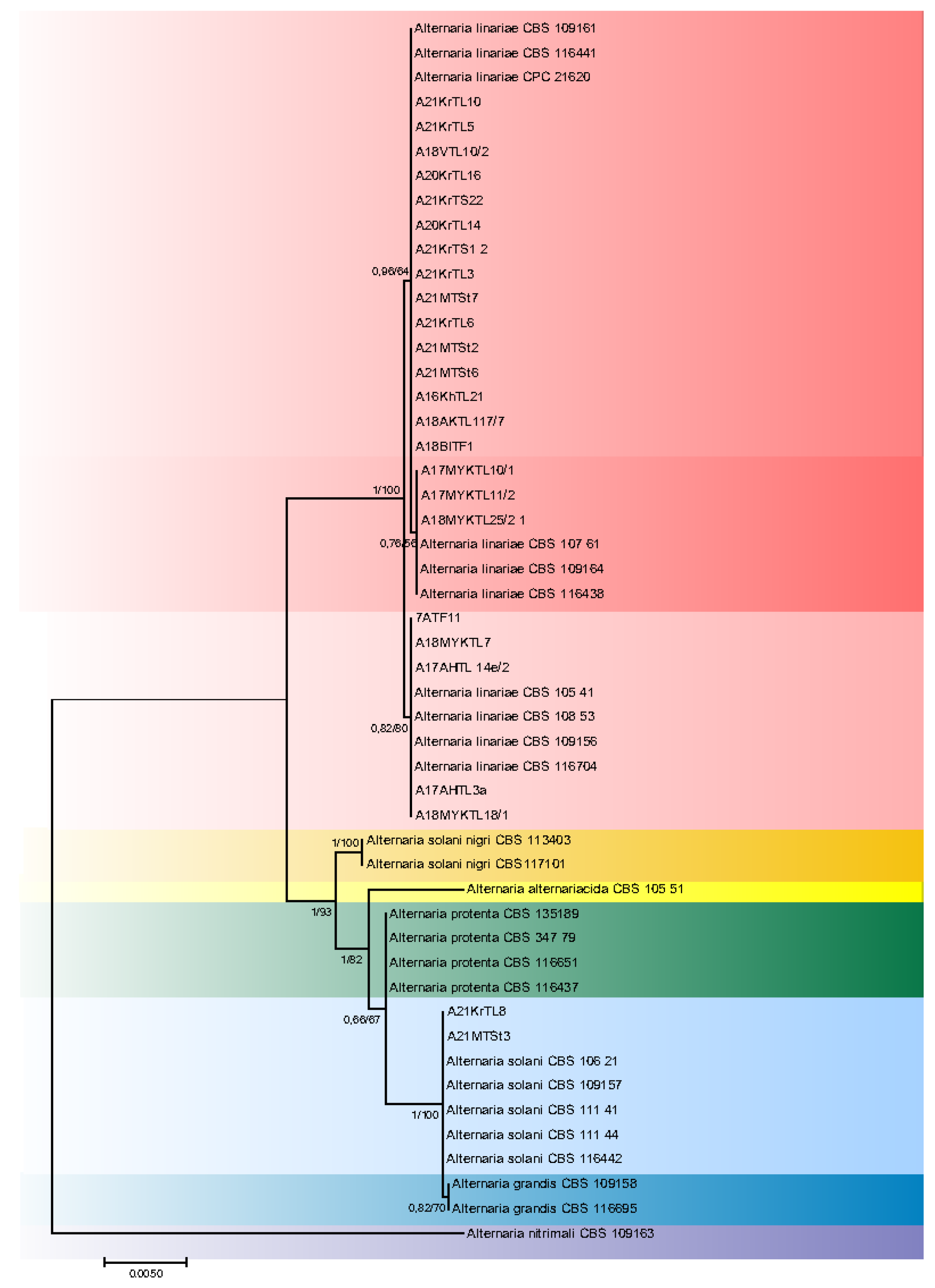
Figure 3.
Combined phylogenetic ITS, GAPDH, Alt a1, TEF1 and RPB2 topology from maximum likelihood analysis of strains isolated from S. lycopersicum plants. Bayesian posterior probabilities followed by ML bootstrap values are shown at nodes.
3.2. Phylogeny
Isolates from affected potato leaves included species of Alternaria alternariacida, A. grandis, A. linariae, A. protenta, A. solani (Figure 2, Table 1). Most of the potato strains (9) were grouped with A. protenta. Among them are isolates from the eastern and western parts of Russia. Seven strains were completely identical to the reference isolate CBS 116651; the sequence of the GADPH gene of strain A17VoPL51a differed from the reference only by one nucleotide. A16PrPL45 strain from the Far East belongs to the A. protenta clade as well but differed from CBS 116651 by one nucleotide in each of four gene regions (ITS, GADPH, TEF1, Alt). Among the strains isolated from the Far East, four were A. grandis. Their DNA sequences were completely identical to that of the A. grandis CBS 109158 and CBS 116695 reference strains. From all tested potato isolates, the only strain isolated in northern Europe in 2017 belonged to A. solani and was identical to CBS 109157. Although A. solani clustered with A. grandis and differs by only one nt in its GAPDH sequence and one in the ITS sequence from A. grandis, although they were retained as a distinct species [3]. The A16PrPL21 strain from the Far East clustered with A. alternariacida CBS 105.51, although it differed from the reference strain by one nucleotide deletion in the ITS region. Among the strains studied was one that belonged to the clade A. linariae. This isolate from the Far East, A17VlPL31a, was closest to the first subclade of A. linariae.
The tomato strains studied belong to the A. linariae and A. solani (Figure 3) species. Twenty-three out of twenty-five tomato strains studied belong to the A. linariae species. The isolates came from leaves, fruits, and stems of tomato plants cultivated in different Russian regions and in different years. According to the reference strains CPC 21620, CBS 109164, and CBS 108.53, A. linariae strains were grouped into three subclades. There were two strains found in the European part of Russia in 2021 that were identified as A. solani and were analogous to CBS reference strains.
4. Discussion
In our study, five species of large-spore Alternaria were identified on potato leaves: A. grandis, A. solani, A. alternariacida, A. protenta, A. linariae. The A. alternariacida description (Woudenberg) is based on the strain isolated from the fruit of Solanum lycopersicum. In the present work, we first discovered a strain of this species on potato. We confirmed disease caused by A. alternaricida on potato plants [12]. Moreover, two species, A. solani and A. linariae, were found to be associated with tomato. There have been similar observations elsewhere in the world: in Algeria, A. protenta, A. linariae, A. solani and A. grandis have been found on potato leaves [13,14,15,16]; A. solani was found on a potato tuber in Egypt [17]. We have also found a large-spore Alternaria strain on potatoes grown in Uganda. The DNA sequence analysis indicated that it was similar to the reference A. linariae strain, differing by one nucleotide in the GADPH gene and two nucleotides in the Alt a 1 gene (OL450058and OL450057). In Wisconsin, USA, [18] strains isolated from potato leaves were found to be A. protenta or A. solani. Researchers did not analyze the sequence of the rpb2 gene that differentiates the two species, so a more precise identification was not possible. Einspanier et al.’s [19] genome-wide study involved 43 large-spore Alternaria isolates collected from potato plants in Europe and the United States. By analyzing the sequences of species-specific markers, it was observed that eight of the isolates studied were identical to strain CBS 116651, which belongs, according to Woudenberg et al. [3], to the A. protenta species. Whole-genome analysis of strains revealed that large-spore species have high levels of single nucleotide substitution rates. This corresponds well with our results. Only one-point substitutions separated the strains that had no complete similarity with the reference strains. This may be caused by the absence of sexual reproduction in large-spore Alternaria populations.
Most of the isolates from the leaves, stems, and fruits of affected tomato plants belonged to the A. linariae species. This is not surprising, since the revised A. linariae species include A. tomatophila, A. cretica and A. subcylindrica [3], which were previously considered the main species of Alternaria infecting tomato [2]. Additionally, a potato A17VPL31a isolate was included in the A. linariae clade. In the Moscow and Krasnodar regions (European part of Russia), two strains of A. solani were isolated from affected leaves and stems of tomato plants. Tomato plants in Algeria have also been found affected by both A. linariae and A. solani [15].
There are relatively few molecular genetic studies of the species structure of large-spore Alternaria in the world. As a result, it is difficult to compare the species and intraspecific composition of Alternaria that infect different Solanaceae plants. Yet, if we look at our and literature-based data, we can find some patterns. Our study found that almost all tomato isolates belonged to the A. linariae species, and only two isolates belonged to the A. solani species. Furthermore, of the tested potato isolates, only one was A. linariae and one was A. solani, and the rest were distributed among the species A. grandis, A. alternariacida, and A. protenta. The CBS A. protenta reference strains were isolated from both S. lycopersicum and S. tuberosum plants. An isolate of A. solani CBS 111.41 was isolated from S. aviculare. It was identical to the A. solani strains we observed causing early blight on potatoes and tomatoes. The A. linariae strains that cause early blight on tomatoes and potatoes were also similar. This indicates that there is no evidence to support the assumption of species-specificity. This corresponds well with the results of studies in North Carolina and Wisconsin involving the Alternaria species from tomato and potato plants [20]. However, the hypothesis of a lack of host specialization needs to be confirmed by cross-inoculation of Alternaria isolates on tomato and potato plants. Our previous studies of the virulence of Alternaria alternata detected intraspecific differences in the virulence and aggressiveness of strains towards potato and tomato cultivars. Some isolates successfully infected cultivars that were highly resistant to other isolates, suggesting that potato and tomato cultivars have genes of specific resistance to A. alternata [21].
The phylogenies of the single-gene trees were not congruent with the consensus tree. Only RPB2 gene trees had the same topology as the consensus tree. We found that the sequences of the ITS region, Alt A1 and GAPDH genes alone could not resolve the phylogeny of closely related Alternaria pathogens of Solanaceae. These results agree with Lourencßo et al. [22] and Peixoto et al. [23] which also found a relatively low number of polymorphisms in the Alt a1 gene sequence among EB-inducing isolates from potato and tomato plants. Therefore, the RPB2 gene is the most relevant for this species complex.
We hypothesized that the genetic diversity of species can vary between different locations, at least in the European region and the Far East. Despite this, we found no relationship between the variable characters and the geographical spread between species. The low number of differences corresponds well with whole-genome results [19], suggesting the existence of true clones that have been transported by seed tubers. In North Carolina and Wisconsin, Adhakiri et al. [20] analyzed field populations of three Alternaria species, finding that A. solani had much lower diversity than A. alternata and A. linariae. Indeed, we found three different haplotypes in A. linariae species. However, it has been shown that A. solani in China has relatively high levels of genetic variation, suggesting parasexual reproduction [24].
Thus, we found five pathogenic Alternaria species on potato plants and two species on tomato plants in Russia. These findings allow us to study the host range and possible options for disease control. We found no phylogenetic groupings among Russian Alternaria isolates associated with their locality. Yet, the sister relationship between the potato and tomato plants makes these species excellent subjects for studying the model of genetic divergence and speciation. A better understanding of their virulence and fungicide resistance can help in the elaboration of the most effective methods of plant protection.
Author Contributions
Conceptualization, L.Y.K. and S.N.E.; Methodology, M.M.Y. and E.M.C.; Investigation, Z.G.K.; Writing—original draft preparation, L.Y.K.; Writing—review and editing, P.N.B.; Project administration, S.N.E.; Funding acquisition, S.N.E. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the RUDN University Scientific Grant System, project № 202193-2-174.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Simmons, E.G. Alternaria themes and variation (244–286) Species on Solanaceae. Mycotaxon 2000, 75, 1–115. [Google Scholar]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Elansky, S.N.; Chudinova, E.M.; Elansky, A.S.; Kah, M.O.; Sandzhieva, D.A.; Mukabenova, B.A.; Dedov, A.G. Microorganisms in spent water-miscible metalworking fluids as a resource of strains for their disposal. J. Clean. Prod. 2022, 350, 131438. [Google Scholar] [CrossRef]
- Kutuzova, I.A.; Kokaeva, L.Y.; Pobedinskaya, M.A.; Krutyakov, Y.A.; Scolotneva, E.S.; Chudinova, E.M.; Elansky, S.N. Resistance of Helminthosporium solani strains to the fungicides applied for tuber treatment. J. Plant Pathol. 2017, 99, 635–642. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Kokaeva, L.Y.; Elansky, S.N. First report of Alternaria alternariacida causing potato leaf blight in the Far East, Russia. Plant Dis. 2022; in press. [Google Scholar]
- Ayad, D.; Aribi, D.; Hamon, B.; Kedad, A.; Simoneau, P.; Bouznad, Z. Distribution of large-spored Alternaria species associated with early blight of potato and tomato in Algeria. Phytopathol. Mediterr. 2019, 58, 139–149. [Google Scholar] [CrossRef]
- Ayad, D.; Hamon, B.; Kedad, A.; Bouznad, Z.; Simoneau, P. First Report of Early Blight Caused by Alternaria linariae on Potato in Algeria. Plant Dis. 2018, 102, 2651. [Google Scholar] [CrossRef]
- Ayad, D.; Leclerc, S.; Hamon, B.; Kedad, A.; Bouznad, Z.; Simoneau, P. First Report of Early Blight Caused by Alternaria protenta on Potato in Algeria. Plant Dis. 2017, 101, 836. [Google Scholar] [CrossRef]
- Bessadat, N.; Berruyer, R.; Hamon, B.; Bataille-Simoneau, N.; Benichou, S.; Kihal, M.; Henni, D.E.; Simoneau, P. Alternaria species associated with early blight epidemics on tomato and other Solanaceae crops in northwestern Algeria. Eur. J. Plant Pathol. 2017, 148, 181–197. [Google Scholar] [CrossRef]
- El-Ganainy, S.M.; El-Abeid, S.E.; Ahmed, Y.; Iqbal, Z. Morphological and molecular characterization of large-spored Alternaria species associated with potato and tomato early blight in Egypt. Int. J. Agric. Biol. 2021, 25, 1101–1110. [Google Scholar] [CrossRef]
- Ding, S.; Meinholz, K.; Cleveland, K.; Jordan, S.A.; Gevens, A.J. Diversity and Virulence of Alternaria spp. Causing Potato Early Blight and Brown Spot in Wisconsin. Phytopathology 2019, 109, 436–445. [Google Scholar] [CrossRef]
- Einspanier, S.; Susanto, T.; Metz, N.; Wolters, P.J.; Vleeshouwers, V.G.; Lankinen, Å.; Liljeroth, E.; Landschoot, S.; Ivanović, Ž.; Hückelhoven, R.; et al. Whole genome sequencing elucidates the species-wide diversity and evolution of fungicide resistance in the early blight pathogen Alternaria solani. Evol. Appl. 2022, 01, 1–16. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Muzhinji, N.; Halterman, D.; Louws, F.J. Genetic diversity and population structure of Alternaria species from tomato and potato in North Carolina and Wisconsin. Sci. Rep. 2021, 11, 17024. [Google Scholar] [CrossRef]
- Kudryavtseva, N.N.; Pobedinskaya, M.A.; Balabko, P.N.; Kokaeva, L.Y.; Zaichik, B.T.; Kutuzova, N.A.; Azarkovich, M.I.; Elansky, S.N.; Valueva, T.A. The proteolytic activity and virulence of Alternaria alternata strains, isolated from tomato. Mikol. I Fitopatol. 2017, 51, 110–116. [Google Scholar]
- Lourenço, V.; Moya, A.; González-Candelas, F.; Carbone, I.; Maffia, L.A.; Mizubuti, E.S.G. Population Biology Molecular Diversity and Evolutionary Processes of Alternaria solani in Brazil Inferred Using Genealogical and Coalescent Approaches. Phytopathology 2009, 99, 765–774. [Google Scholar] [CrossRef]
- Peixoto, C.C.; Cabral, C.S.; Fonseca, M.E.N.; Boiteux, L.S.; Reis, A. Species diversity, novel interactions and absence of well-supported host-guided phylogenetic groupings of Neotropical Alternaria isolates causing foliar lesions in Solanaceae. J. Appl. Microbiol. 2021, 131, 2466–2487. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Fan, S.; Zhang, D.; Pan, Y.; Gu, Q.; Wang, J.; Yang, Z.; Zhu, J. Parasexual reproduction in Alternaria solani: Simple sequence repeat molecular evidence for haploidization. Mycologia 2021, 113, 949–955. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).