Phylogenetic Reconstruction of the Rainforest Lineage Fontainea Heckel (Euphorbiaceae) Based on Chloroplast DNA Sequences and Reduced-Representation SNP Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study System and Sampling Strategy
2.2. Chloroplast Sequence Data
2.3. Reduced-Representation Genome Sequencing
2.4. Phylogenetic Reconstruction
2.5. Estimates of Datasets Diverging to Different Topology
2.6. Genetic Structure
3. Results
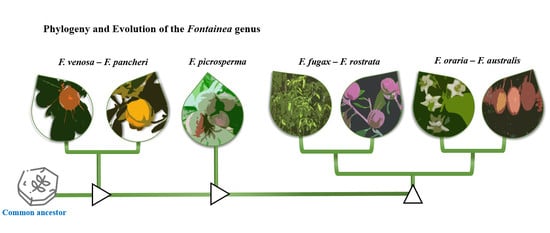
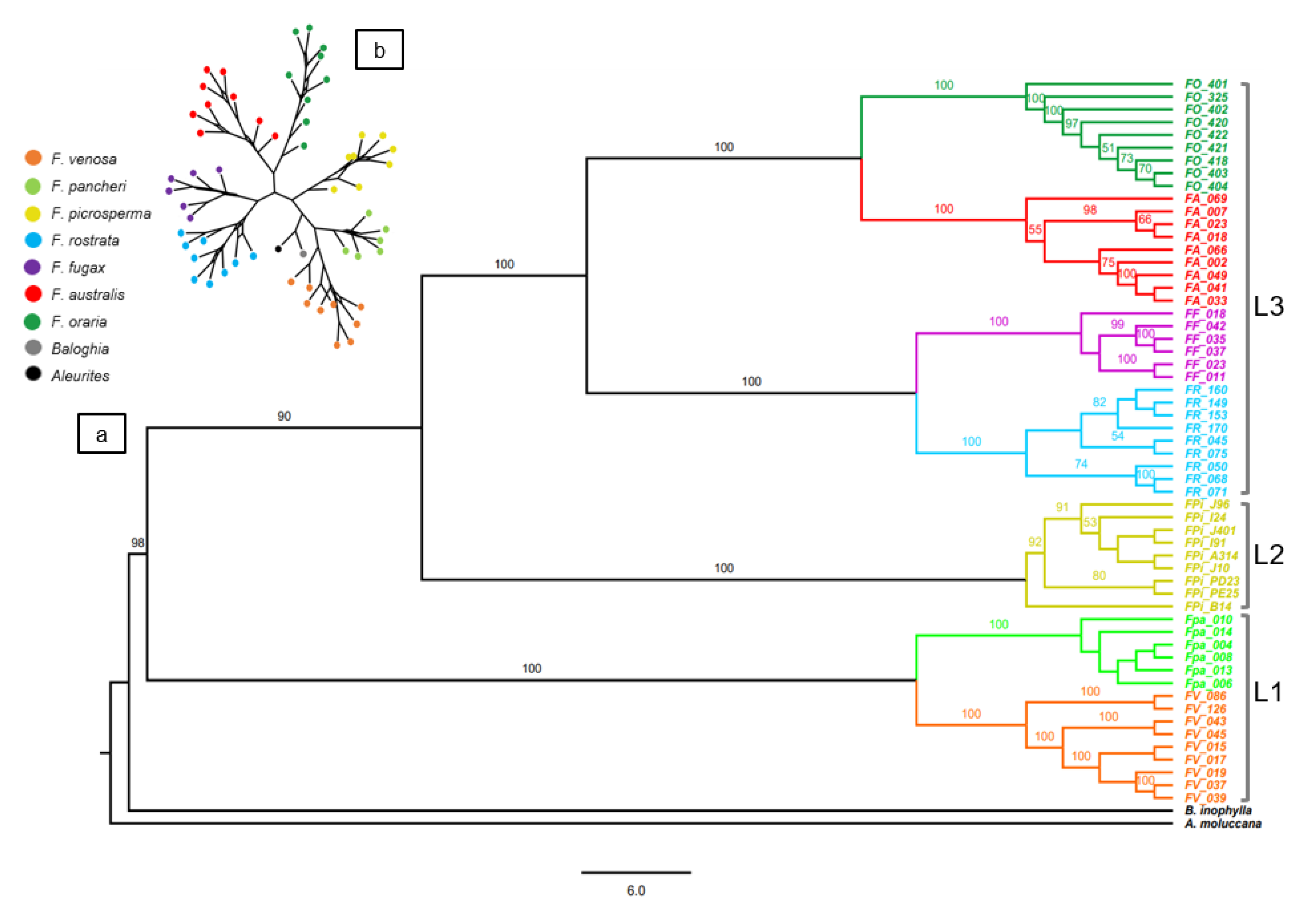
3.1. CpDNA Sequence Phylogeny
3.2. DArTseq SNP Phylogeny
3.3. Fontainea Population-Level Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webster, G.L. Synopsis of the Genera and Suprageneric Taxa of Euphorbiaceae. Ann. Mo. Bot. Gard. 1994, 81, 33. [Google Scholar] [CrossRef]
- Rozefelds, A.C.; Milroy, A.K.; Dettmann, M.E.; Clifford, H.T.; Maksimenko, A. Synchrotron computer tomographic (CT) scans complement traditional techniques in understanding the internal anatomy of permineralised Fontainocarpa (Crotonoideae, Euphorbiaceae) fruits from the Oligocene of eastern Australia. Rev. Palaeobot. Palynol. 2017, 242, 43–57. [Google Scholar] [CrossRef]
- Jessup, L.W.; Guymer, G.P. A revision of Fontainea heckel (euphorbiaceae—cluytieae). Austrobaileya 1985, 2, 112–125. [Google Scholar]
- Forster, P.I.J.A. Three new species of Fontainea Heckel (Euphorbiaceae) from Australia and Papua New Guinea. Austrobaileya 1997, 5, 29–37. [Google Scholar]
- Forster, P.I.; Bostock, P.D.; Bird, L.H.; Bean, A.R. Vineforest Plant Atlas for South-East Queensland; Queensland Herbarium: Brisbane, Australia, 1991. [Google Scholar]
- Boyle, G.M.; D'Souza, M.M.A.; Pierce, C.J.; Adams, R.A.; Cantor, A.S.; Johns, J.P.; Maslovskaya, L.; Gordon, V.A.; Reddell, P.; Parsons, P. Intra-Lesional Injection of the Novel PKC Activator EBC-46 Rapidly Ablates Tumors in Mouse Models. PLoS ONE 2014, 9, e108887. [Google Scholar] [CrossRef]
- De Ridder, T.R.; Campbell, J.E.; Burke-Schwarz, C.; Clegg, D.; Elliot, E.L.; Geller, S.; Kozak, W.; Pittenger, S.T.; Pruitt, J.B.; Riehl, J.; et al. Randomized controlled clinical study evaluating the efficacy and safety of intratumoral treatment of canine mast cell tumors with tigilanol tiglate (EBC-46). J. Veter-Intern. Med. 2020, 35, 415–429. [Google Scholar] [CrossRef]
- Wurdack, K.J.; Hoffmann, P.; Chase, M.W. Molecular phylogenetic analysis of uniovulate Euphorbiaceae (Euphorbiaceae sensu stricto) using plastid RBCL and TRNL-F DNA sequences. Am. J. Bot. 2005, 92, 1397–1420. [Google Scholar] [CrossRef]
- Matamoro-Vidal, A.; Furness, C.A.; Gouyon, P.-H.; Wurdack, K.J.; Albert, B. Evolutionary stasis in Euphorbiaceae pollen: Selection and constraints. J. Evol. Biol. 2012, 25, 1077–1096. [Google Scholar] [CrossRef]
- van Welzen, P.C.; Guerrero, S.A.; Arifiani, D.; Bangun, T.J.F.; Bouman, R.W.; Eurlings, M.C.M.; Gushilman, I.; Phillipson, P.B.; Tabak, I.; Winkel, E.; et al. Weda, a new genus with two new species of Euphorbiaceae-Crotonoideae from Halmahera (North Maluku, Indonesia) and phylogenetic relationships of the Australasian tribe Ricinocarpeae. J. Syst. Evol. 2021, 59, 1000–1017. [Google Scholar] [CrossRef]
- Rozefelds, A.C.; Dettmann, M.E.; Clifford, H.T.; Ekins, M. An Australian origin for the candle nut (Aleurites, Crotonoideae, Euphorbiaceae) and the fossil record of the Euphorbiaceae and related families in Australia and New Zealand. Rev. Palaeobot. Palynol. 2017, 241, 39–48. [Google Scholar] [CrossRef]
- Downie, S.R.; Palmer, J.D. Use of Chloroplast DNA Rearrangements in Reconstructing Plant Phylogeny. In Molecular Systematics of Plants; Soltis, P.S., Soltis, D.E., Doyle, J.J., Eds.; Springer: Boston, MA, USA, 1992; pp. 14–35. [Google Scholar]
- Steane, D.A.; Nicolle, D.; Sansaloni, C.P.; Petroli, C.D.; Carling, J.; Kilian, A.; Myburg, A.A.; Grattapaglia, D.; Vaillancourt, R.E. Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Mol. Phylogenet. Evol. 2011, 59, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Crisp, M.D.; Cook, L.G.; Meusemann, K.; Edwards, R.D.; Toon, A.; Külheim, C. Identifying genetic markers for a range of phylogenetic utility–From species to family level. PLoS ONE 2019, 14, e0218995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamont, R.W.; Conroy, G.C.; Reddell, P.; Ogbourne, S.M. Population genetic analysis of a medicinally significant Australian rainforest tree, Fontainea picrosperma C.T. White (Euphorbiaceae): Biogeographic patterns and implications for species domestication and plantation establishment. BMC Plant Biol. 2016, 16, 57. [Google Scholar] [CrossRef]
- Conroy, G.C.; Shimizu-Kimura, Y.; Lamont, R.W.; Ogbourne, S.M. A multidisciplinary approach to inform assisted migration of the restricted rainforest tree, Fontainea rostrata. PLoS ONE 2019, 14, e0210560. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, M.; McNally, J.; Henry, R.J.; Hunter, J.; Matthes, M. Conservation genetics of an endangered rainforest tree (Fontainea oraria—Euphorbiaceae) and implications for closely related species. Conserv. Genet. 2000, 1, 217–229. [Google Scholar] [CrossRef]
- Grant, E.L.; Wallace, H.M.; Trueman, S.J.; Reddell, P.W.; Ogbourne, S.M. Floral and reproductive biology of the medicinally significant rainforest tree, Fontainea picrosperma (Euphorbiaceae). Ind. Crop. Prod. 2017, 108, 416–422. [Google Scholar] [CrossRef]
- Grant, E.L.; Conroy, G.C.; Lamont, R.W.; Reddell, P.; Wallace, H.M.; Ogbourne, S.M. Short distance pollen dispersal and low genetic diversity in a subcanopy tropical rainforest tree, Fontainea picrosperma (Euphorbiaceae). Heredity 2019, 123, 503–516. [Google Scholar] [CrossRef]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, 54. [Google Scholar] [CrossRef]
- Obeng-Bio, E.; Badu-Apraku, B.; Ifie, B.E.; Danquah, A.; Blay, E.T.; Abu Dadzie, M.; Noudifoulè, G.T.; Talabi, A.O. Genetic diversity among early provitamin A quality protein maize inbred lines and the performance of derived hybrids under contrasting nitrogen environments. BMC Genet. 2020, 21, 78. [Google Scholar] [CrossRef]
- Wilf, P.; Escapa, I.H. Green Web or megabiased clock? Plant fossils from Gondwanan Patagonia speak on evolutionary radiations. New Phytol. 2014, 207, 283–290. [Google Scholar] [CrossRef]
- Crisp, M.D.; Cook, L.G. How Was the Australian Flora Assembled Over the Last 65 Million Years? A Molecular Phylogenetic Perspective. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 303–324. [Google Scholar] [CrossRef]
- Rossetto, M.; Bragg, J.; Kilian, A.; McPherson, H.; Van Der Merwe, M.; Wilson, P. Restore and Renew: A genomics-era framework for species provenance delimitation. Restor. Ecol. 2018, 27, 538–548. [Google Scholar] [CrossRef] [Green Version]
- Tokuoka, T. Molecular phylogenetic analysis of Euphorbiaceae sensu stricto based on plastid and nuclear DNA sequences and ovule and seed character evolution. J. Plant Res. 2007, 120, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Naeem, R.; Su, J.-X.; Cao, Z.-Y.; Burleigh, J.G.; Soltis, P.S.; Soltis, D.E.; Chen, Z.-D. Phylogeny of the Rosidae: A dense taxon sampling analysis. J. Syst. Evol. 2016, 54, 363–391. [Google Scholar] [CrossRef]
- Byrne, M.; Hankinson, M. Testing the variability of chloroplast sequences for plant phylogeography. Aust. J. Bot. 2012, 60, 569–574. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Tate, J.A.; Simpson, B.B. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 2003, 28, 723–737. [Google Scholar]
- Silva, O.L.M.; Riina, R.; Cordeiro, I. Phylogeny and biogeography of Astraea with new insights into the evolutionary history of Crotoneae (Euphorbiaceae). Mol. Phylogenetics Evol. 2020, 145, 106738. [Google Scholar] [CrossRef]
- Maya-Lastra, C.A.; Steinmann, V.W. Evolution of the untouchables: Phylogenetics and classification of Cnidoscolus (Euphorbiaceae). TAXON 2019, 68, 692–713. [Google Scholar] [CrossRef]
- Watts, C.; Fisher, A.E.; Shrum, C.D.; Newbold, W.L.; Hansen, S.; Liu, C.; Kelchner, S.A. The D4 set: Primers that target highly variable intron loops in plant chloroplast genomes. Mol. Ecol. Resour. 2008, 8, 1344–1347. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, S.; Rossetto, M.; Bragg, J.; McPherson, H.; Benson, D.; Bonser, S.P.; Wilson, P.G. Speciation in the presence of gene flow: Population genomics of closely related and diverging Eucalyptus species. Heredity 2018, 121, 126–141. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; FigTree. Tree Figure Drawing Tool. 2009. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 March 2022).
- Holland, B.; Huber, K.T.; Moulton, V.; Lockhart, P.J. Using Consensus Networks to Visualize Contradictory Evidence for Species Phylogeny. Mol. Biol. Evol. 2004, 21, 1459–1461. [Google Scholar] [CrossRef]
- Jombart, T.J.B. Adegenet: An R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. DiveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in ecology and evolution. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Hill, R.S.; Woinarski, J.C.Z.; Williams, J. History of the Australian Vegetation: Cretaceous to Recent; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Kooyman, R.; Rossetto, M.; Allen, C.; Cornwell, W. Australian Tropical and Subtropical Rain Forest Community Assembly: Phylogeny, Functional Biogeography, and Environmental Gradients. Biotropica 2012, 44, 668–679. [Google Scholar] [CrossRef]
- Ladiges, P.Y.; Cantrill, D. New Caledonia–Australian connections: Biogeographic patterns and geology. Aust. Syst. Bot. 2007, 20, 383–389. [Google Scholar] [CrossRef]
- Grandicolas, P.; Murienne, J.M.; Robillard, T.; Desutter-Grandcolas, L.; Jourdan, H.; Guilbert, E.; Deharveng, L. New Caledonia: A very old Darwinian island? Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3309–3317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranitz, M.L.; Biffin, E.; Clark, A.; Hollingsworth, M.L.; Ruhsam, M.; Gardner, M.F.; Thomas, P.; Mill, R.R.; Ennos, R.A.; Gaudeul, M. Evolutionary diversification of new Caledonian Araucaria. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Nattier, R.; Pellens, R.; Robillard, T.; Jourdan, H.; Legendre, F.; Caesar, M.; Nel, A.; Grandcolas, P. Updating the phylogenetic dating of New Caledonian biodiversity with a meta-analysis of the available evidence. Sci. Rep. 2017, 7, 1–9. [Google Scholar]
- Harbaugh, D.T. A taxonomic revision of Australian northern sandalwood (Santalum lanceolatum, Santalaceae). Aust. Syst. Bot. 2007, 20, 409–416. [Google Scholar] [CrossRef]
- Barrabe, L.; Maggia, L.; Pillon, Y.; Rigault, F.; Mouly, A.; Davis, A.P.; Buerki, S. New Caledonian lineages of Psychotria (Rubiaceae) reveal different evolutionary histories and the largest documented plant radiation for the archipelago. Mol. Phylogenet. Evol. 2014, 71, 15–35. [Google Scholar] [CrossRef]
- Costion, M.; Edwards, W.; Ford, A.J.; Metcalfe, D.J.; Cross, H.B.; Harrington, M.G.; Richardson, J.-E.; Hilbert, D.W.; Lowe, A.; Crayn, D. Using phylogenetic diversity to identify ancient rain forest refugia and diversification zones in a biodiversity hotspot. Divers. Distrib. 2014, 21, 279–289. [Google Scholar] [CrossRef]
- Crayn, D.M.; Costion, C.; Harrington, M.G. The Sahul-Sunda floristic exchange: Dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics. J. Biogeogr. 2014, 42, 11–24. [Google Scholar] [CrossRef]
- Kooyman, R.M.; Wilf, P.; Barreda, V.D.; Carpenter, R.J.; Jordan, G.J.; Sniderman, J.K.; Allen, A.; Brodribb, T.J.; Crayn, D.; Feild, T.S. Paleo-Antarctic rainforest into the modern Old World tropics: The rich past and threatened future of the “southern wet forest survivors”. Am. J. Bot. 2014, 101, 2121–2135. [Google Scholar] [CrossRef]
- Kooyman, R.M.; Rossetto, M.; Sauquet, H.; Laffan, S.W. Landscape patterns in rainforest phylogenetic signal: Isolated islands of refugia or structured continental distributions? PLoS ONE 2013, 8, e80685. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, A.P.; Bretherton, S.C.; Van Der Kaars, S. A complete pollen record of the last 230 ka from Lynch's Crater, north-eastern Australia. Palaeogeography, Palaeoclimatology, Palaeoecology 2007, 251, 23–45. [Google Scholar] [CrossRef]
- Wagner, V.; Treiber, J.; Danihelka, J.; Ruprecht, E.; Wesche, K.; Hensen, I. Declining genetic diversity and increasing genetic isolation toward the range periphery of Stipa pennata, a Eurasian feather grass. Int. J. Plant Sci. 2012, 173, 802–811. [Google Scholar] [CrossRef]
- Rutherford, S.; Wan, J.S.; Cohen, J.M.; Benson, D.; Rossetto, M. Looks can be deceiving: Speciation dynamics of co-distributed Angophora (Myrtaceae) species in a varying landscape. Evolution 2020, 75, 310–329. [Google Scholar] [CrossRef]
- Rossetto, M.; Allen, C.B.; Thurlby, K.A.; Weston, P.H.; Milner, M.L. Genetic structure and bio-climatic modeling support allopatric over parapatric speciation along a latitudinal gradient. BMC Evol. Biol. 2012, 12, 149. [Google Scholar] [CrossRef]
- Environment Protection and Biodiversity Conservation Act 1999; Australian Government: Canberra, Australia, 1999.
- Border Ranges Rainforest Biodiversity Management Plan-NSW and Queensland; Department of Environment: Sydney, Australia, 2010.
- Ikabanga, D.U.; Stévart, T.; Koffi, K.G.; Monthe, F.K.; Doubindou, E.C.N.; Dauby, G.; Souza, A.; M’Batchi, B.; Hardy, O.J. Combining morphology and population genetic analysis uncover species delimitation in the widespread African tree genus Santiria (Burseraceae). Phytotaxa 2017, 321, 166–180. [Google Scholar] [CrossRef]
- Kooyman, R.; Rossetto, M.; Cornwell, W.; Westoby, M. Phylogenetic tests of community assembly across regional to continental scales in tropical and subtropical rain forests. Glob. Ecol. Biogeogr. 2011, 20, 707–716. [Google Scholar]
- Sutherland, F.L. Diversity within geodiversity, underpinning habitats in New South Wales volcanic areas. Proc. Linn. Soc. New South Wales 2011, 132, 37–54. [Google Scholar]
- Parkes, T.; Delaney, M.; Dunphy, M.; Woodford, R.; Bower, H.; Bower, S.; Bailey, D.; Joseph, R.; Nagle, J.; Roberts, T.; et al. Big Scrub: A cleared landscape in transition back to forest? Ecol. Manag. Restor. 2012, 13, 212–223. [Google Scholar] [CrossRef]
- Hocknull, S.A.; Zhao, J.-X.; Feng, Y.-X.; Webb, G.E. Responses of Quaternary rainforest vertebrates to climate change in Australia. Earth Planet. Sci. Lett. 2007, 264, 317–331. [Google Scholar] [CrossRef]
- Dennis, A.J.; Westcott, D.A. Reducing complexity when studying seed dispersal at community scales: A functional classification of vertebrate seed dispersers in tropical forests. Oecologia 2006, 149, 620–634. [Google Scholar] [CrossRef] [PubMed]
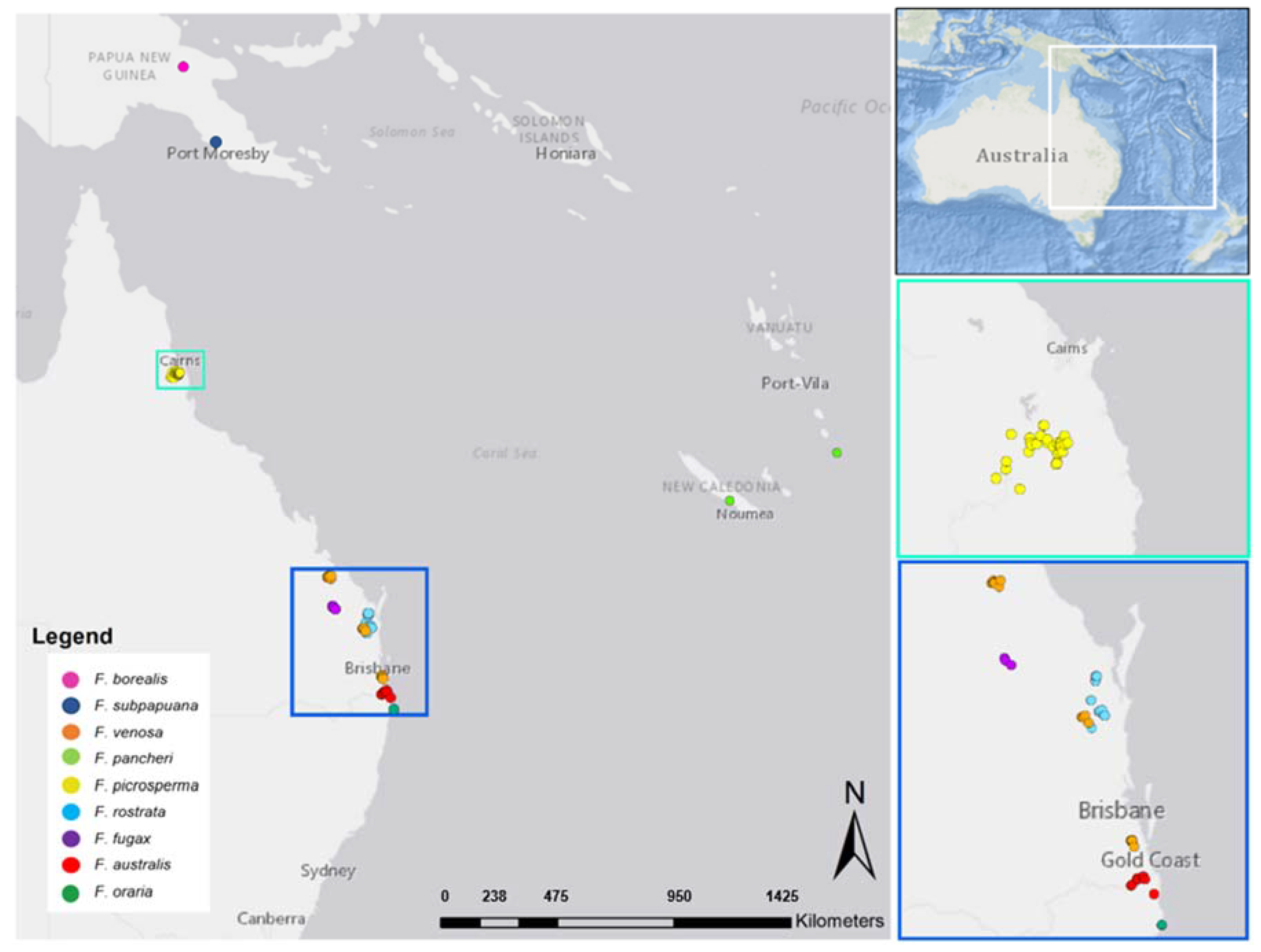


| Species | Conservation Status | Accession Distribution | N | Biome | Forest Type | Elevation (Above Sea Level) |
|---|---|---|---|---|---|---|
| F. pancheri | LC–1 | New Caledonia, | 6 | Subtropical | Seasonally dry | ~100–400 m |
| F. venosa | V–2 | Gladstone, and Gympie, QLD, Australia | 9 | Subtropical | Seasonally dry | ~150–350 m |
| F. picrosperma | LC–2 | Atherton-Evelyn Tablelands, QLD, Australia | 9 | Tropical | Upland wet tropical | ~700–1100 m |
| F. rostrata | V–2, 3 | Gympie and Maryborough, QLD, Australia | 9 | Subtropical | Seasonally dry | ~150–350 m |
| F. fugax | E–2 | Gayndah, QLD, Australia | 6 | Subtropical | Seasonally dry | ~350–400 m |
| F. australis | E–2, 3, 4 | Tweed Caldera, QLD and NSW, Australia | 9 | Subtropical | Aseasonally wet subtropical | ~50–900 m |
| F. oraria | CE–3, 4 | Lennox Head, NSW, Australia | 9 | Subtropical | Littoral (Coastal) rainforest | ~60 m |
| F. venosa | F. pancheri | F. picrosperma | F. fugax | F. rostrata | F. oraria | F. australis | |
|---|---|---|---|---|---|---|---|
| F. venosa | - | 0.667 | 0.97 | 0.965 | 0.963 | 0.962 | 0.963 |
| F. pancheri | 0.667 | - | 0.981 | 0.976 | 0.975 | 0.971 | 0.972 |
| F. picrosperma | 0.97 | 0.981 | - | 0.965 | 0.961 | 0.953 | 0.954 |
| F. fugax | 0.965 | 0.976 | 0.965 | - | 0.539 | 0.863 | 0.865 |
| F. rostrata | 0.963 | 0.975 | 0.961 | 0.539 | - | 0.861 | 0.861 |
| F. oraria | 0.962 | 0.971 | 0.953 | 0.863 | 0.861 | - | 0.294 |
| F. australis | 0.963 | 0.972 | 0.954 | 0.865 | 0.861 | 0.294 | - |

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunton, A.J.; Lamont, R.W.; Conroy, G.C.; Yap, S.; Rossetto, M.; Taylor-Brown, A.; Maggia, L.; Reddell, P.W.; Ogbourne, S.M. Phylogenetic Reconstruction of the Rainforest Lineage Fontainea Heckel (Euphorbiaceae) Based on Chloroplast DNA Sequences and Reduced-Representation SNP Markers. Diversity 2022, 14, 725. https://doi.org/10.3390/d14090725
Brunton AJ, Lamont RW, Conroy GC, Yap S, Rossetto M, Taylor-Brown A, Maggia L, Reddell PW, Ogbourne SM. Phylogenetic Reconstruction of the Rainforest Lineage Fontainea Heckel (Euphorbiaceae) Based on Chloroplast DNA Sequences and Reduced-Representation SNP Markers. Diversity. 2022; 14(9):725. https://doi.org/10.3390/d14090725
Chicago/Turabian StyleBrunton, Aaron J., Robert W. Lamont, Gabriel C. Conroy, Samantha Yap, Maurizio Rossetto, Alyce Taylor-Brown, Laurent Maggia, Paul W. Reddell, and Steven M. Ogbourne. 2022. "Phylogenetic Reconstruction of the Rainforest Lineage Fontainea Heckel (Euphorbiaceae) Based on Chloroplast DNA Sequences and Reduced-Representation SNP Markers" Diversity 14, no. 9: 725. https://doi.org/10.3390/d14090725
APA StyleBrunton, A. J., Lamont, R. W., Conroy, G. C., Yap, S., Rossetto, M., Taylor-Brown, A., Maggia, L., Reddell, P. W., & Ogbourne, S. M. (2022). Phylogenetic Reconstruction of the Rainforest Lineage Fontainea Heckel (Euphorbiaceae) Based on Chloroplast DNA Sequences and Reduced-Representation SNP Markers. Diversity, 14(9), 725. https://doi.org/10.3390/d14090725