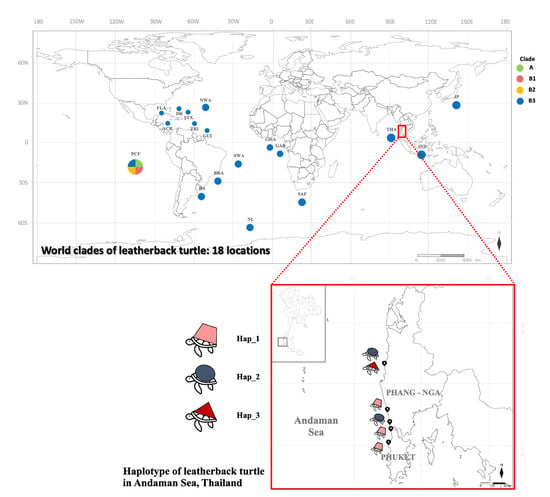
Genetic Diversity in Leatherback Turtles (Dermochelys coriacea) along the Andaman Sea of Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Microsatellite Amplification and Genotyping
2.3. Mitochondrial DNA
2.4. Data Analysis
2.4.1. Microsatellite Analysis
2.4.2. Population Structure
2.4.3. Kinship Analysis
2.4.4. Mitochondrial DNA Control Region Analysis
2.4.5. The Median Joining Network (MJN) Analysis and Genetic Diversity
2.4.6. Phylogenetic Trees Construction
2.4.7. Bayesian Skyline Plots (BSPs)
3. Results
3.1. Microsatellite Primers
3.2. Genetic Diversity
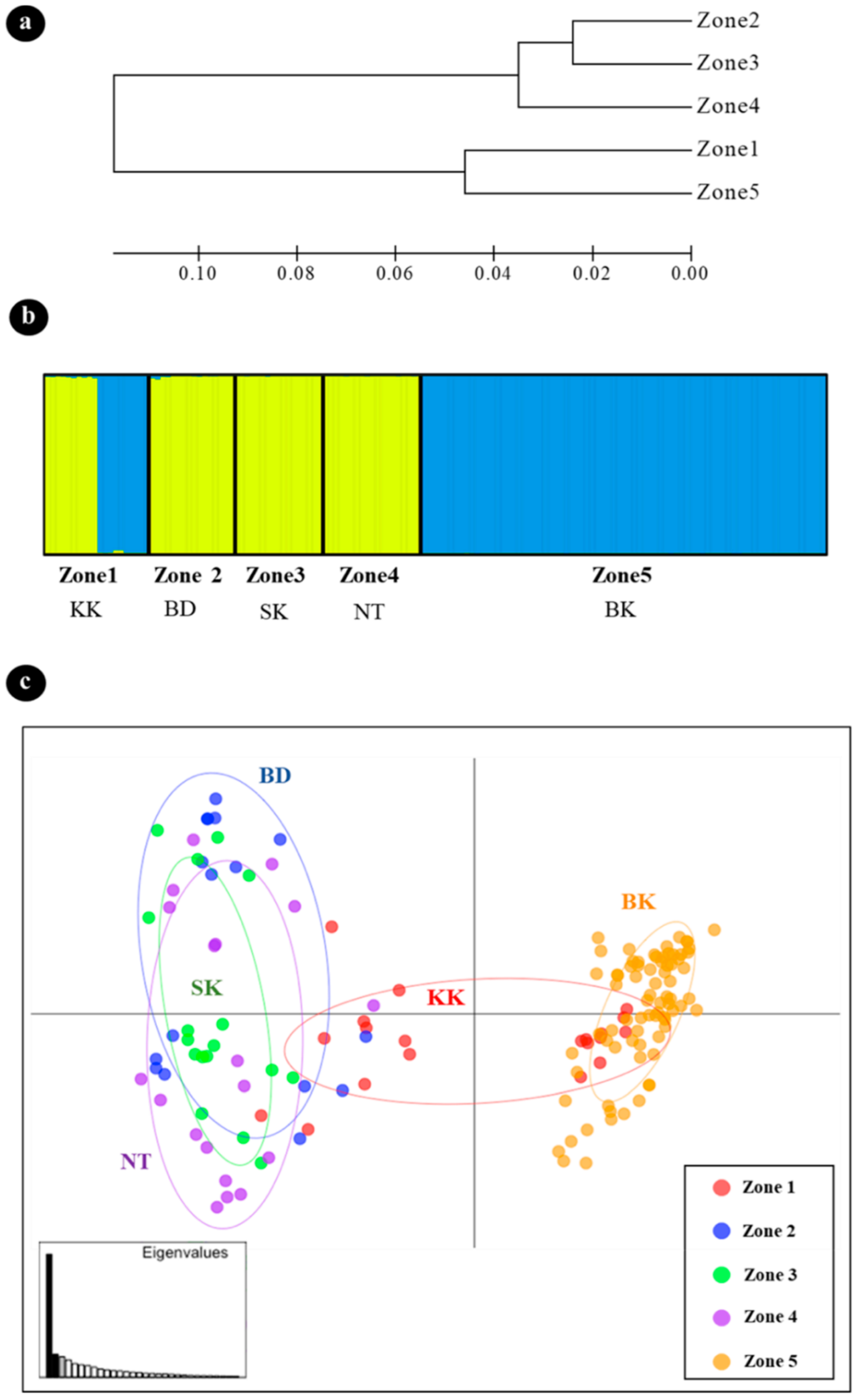
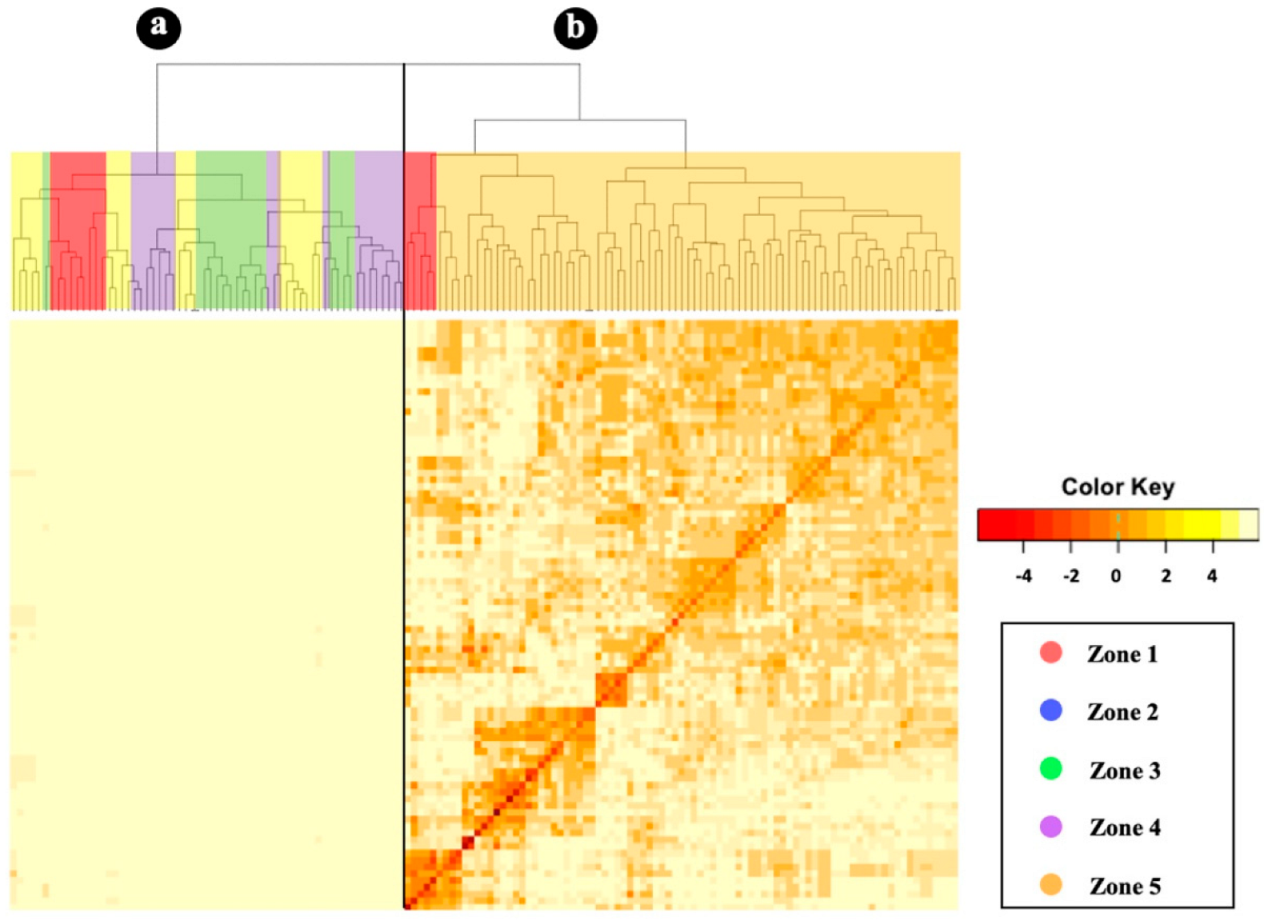
3.3. Genetic Differentiation and Population Structure
3.4. Kinship Analysis
3.5. Mitochondrial DNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Oceanic and Atmospheric Administration. Leatherback Turtle. 2022. Available online: https://www.fisheries.noaa.gov/species/leatherback-turtle (accessed on 20 February 2022).
- World Wildlife Fund. The Leatherback Turtle. Available online: https://wwf.panda.org/discover/knowledge_hub/endangered_species/marine_turtles/leatherback_turtle/ (accessed on 20 February 2022).
- Wallace, B.P.; Saba, V.S. Environmental and anthropogenic impacts on intra-specific variation in leatherback turtles: Opportunities for targeted research and conservation. Endanger. Species Res. 2009, 7, 11–21. [Google Scholar] [CrossRef]
- CITES. Dermochelys Coriacea. Available online: https://bit.ly/36i3E4m (accessed on 20 February 2022).
- Areekijseree, M.; Pumipaiboon, M.; Nuamsukon, S.; Kittiwattanawong, K.; Thongchai, C.; Sikiwat, S.; Chuen-Im, T. Leatherback turtle (Dermochelys coriacea) after incubation eggshell in Andaman Sea, Thailand study: Microanalysis on ultrastructure and elemental composition. Int. J. Bioeng. Life Sci. 2013, 7, 820–824. [Google Scholar]
- Tiwari, M.; Wallace, B.; Girondot, M. Dermochelys coriacea West Pacific Ocean subpopulation. In The IUCN Red List of Threatened Species 2013; IUCN: Gland, Switzerland, 2013; p. T46967817A46967821. [Google Scholar]
- Spotila, J.R.; Dunham, A.E.; Leslie, A.J.; Steyermark, A.C.; Plotkin, P.T.; Paladino, F.V. Worldwide population decline of Dermochelys coriacea: Are leatherback turtles going extinct? Reducción mundial de la población de Dermochelys coriaceae: Están las tortugas baula en vía de extinción. Chelonian Conserv. Biol. 1996, 2, 209–222. [Google Scholar]
- Spotila, J.R.; Reina, R.D.; Steyermark, A.C.; Plotkin, P.T.; Paladino, F.V. Pacific leatherback turtles face extinction. Nature 2000, 405, 529–530. [Google Scholar] [PubMed]
- Chan, E. Decline of the leatherback population in Terengganu, Malaysia, 1956–1995. Chelonian Conserv. Biol. 1996, 2, 196–203. [Google Scholar]
- Bolten, A.B.; Lutz, P.; Musick, J.; Wyneken, J. Variation in sea turtle life history patterns: Neritic vs. oceanic developmental stages. Biol. Sea Turt. 2003, 2, 243–257. [Google Scholar]
- Dutton, P.H.; Bowen, B.W.; Owens, D.W.; Barragan, A.; Davis, S.K. Global phylogeography of the leatherback turtle (Dermochelys coriacea). J. Zool. 1999, 248, 397–409. [Google Scholar] [CrossRef]
- Gaspar, P.; Benson, S.R.; Dutton, P.H.; Réveillère, A.; Jacob, G.; Meetoo, C.; Dehecq, A.; Fossette, S. Oceanic dispersal of juvenile leatherback turtles: Going beyond passive drift modeling. Mar. Ecol. Prog. Ser. 2012, 457, 265–284. [Google Scholar]
- Tomillo, P.S.; Oro, D.; Paladino, F.V.; Piedra, R.; Sieg, A.E.; Spotila, J.R. High beach temperatures increased female-biased primary sex ratios but reduced output of female hatchlings in the leatherback turtle. Biol. Conserv. 2014, 176, 71–79. [Google Scholar] [CrossRef]
- Santidrián Tomillo, P.; Saba, V.S.; Blanco, G.S.; Stock, C.A.; Paladino, F.V.; Spotila, J.R. Climate driven egg and hatchling mortality threatens survival of Eastern Pacific leatherback turtles. PLoS ONE 2012, 7, e37602. [Google Scholar]
- Lewison, R.L.; Freeman, S.A.; Crowder, L.B. Quantifying the effects of fisheries on threatened species: The impact of pelagic longlines on loggerhead and leatherback sea turtles. Ecol. Lett. 2004, 7, 221–231. [Google Scholar] [CrossRef]
- Plot, V.; Georges, J.-Y. Plastic debris in a nesting leatherback turtle in French Guiana. Chelonian Conserv. Biol. 2010, 9, 267–270. [Google Scholar] [CrossRef]
- Caracappa, S.; Persichetti, M.F.; Gentile, A.; Caracappa, G.; Curro, V.; Freggi, D.; Arculeo, M. New records of leatherback sea turtle, Dermochelys coriacea (Vandelli, 1761) (Testudines: Dermochelyidae) in the Strait of Sicily. Cah. Biol. Mar. 2017, 58, 353–357. [Google Scholar]
- Dutton, P.H.; Hitipeuw, C.; Zein, M.; Benson, S.R.; Petro, G.; Pita, J.; Rei, V.; Ambio, L.; Bakarbessy, J. Status and genetic structure of nesting populations of leatherback turtles (Dermochelys coriacea) in the western Pacific. Chelonian Conserv. Biol. 2007, 6, 47–53. [Google Scholar] [CrossRef]
- Dutton, P.H.; Roden, S.E.; Stewart, K.R.; LaCasella, E.; Tiwari, M.; Formia, A.; Thomé, J.C.; Livingstone, S.R.; Eckert, S.; Chacon-Chaverri, D. Population stock structure of leatherback turtles (Dermochelys coriacea) in the Atlantic revealed using mtDNA and microsatellite markers. Conserv. Genet. 2013, 14, 625–636. [Google Scholar] [CrossRef]
- Carreras, C.; Godley, B.J.; Leon, Y.M.; Hawkes, L.A.; Revuelta, O.; Raga, J.A.; Tomas, J. Contextualising the last survivors: Population structure of marine turtles in the Dominican Republic. PLoS ONE 2013, 8, e66037. [Google Scholar] [CrossRef]
- Molfetti, E.; Torres Vilaça, S.; Georges, J.-Y.; Plot, V.; Delcroix, E.; Le Scao, R.; Lavergne, A.; Barrioz, S.; Dos Santos, F.R.; De Thoisy, B. Recent demographic history and present fine-scale structure in the Northwest Atlantic leatherback (Dermochelys coriacea) turtle population. PLoS ONE 2013, 8, e58061. [Google Scholar]
- Stewart, K.R.; LaCasella, E.L.; Roden, S.E.; Jensen, M.P.; Stokes, L.W.; Epperly, S.P.; Dutton, P.H. Nesting population origins of leatherback turtles caught as bycatch in the US pelagic longline fishery. Ecosphere 2016, 7, e01272. [Google Scholar] [CrossRef] [Green Version]
- Vargas, S.M.; Araújo, F.C.; Monteiro, D.S.; Estima, S.C.; Almeida, A.P.; Soares, L.S.; Santos, F.R. Genetic diversity and origin of leatherback turtles (Dermochelys coriacea) from the Brazilian coast. J. Hered. 2008, 99, 215–220. [Google Scholar] [CrossRef]
- Prosdocimi, L.; Dutton, P.; Albareda, D.; Remis, M.I. Origin and genetic diversity of leatherbacks (Dermochelys coriacea) at Argentine foraging grounds. J. Exp. Mar. Biol. Ecol. 2014, 458, 13–19. [Google Scholar] [CrossRef]
- Kaewmong, P.; Punyapornwithaya, V.; Wongfu, C.; Prasitwiset, W.; Jongjit, P.; Boonkasemsanti, A.; Kittiwattanawong, K.; Bansiddhi, P. Nest Relocation of Leatherback Turtles (Dermochelys coriacea) Decrease the Rate of Non-Developed Eggs. Vet. Integr. Sci. 2022, 20, 279–289. [Google Scholar] [CrossRef]
- Komoroske, L.M.; Jensen, M.P.; Stewart, K.R.; Shamblin, B.M.; Dutton, P.H. Advances in the application of genetics in marine turtle biology and conservation. Front. Mar. Sci. 2017, 4, 156. [Google Scholar] [CrossRef]
- Bagda, E.; Bardakci, F.; Turkozan, O. Lower genetic structuring in mitochondrial DNA than nuclear DNA among the nesting colonies of green turtles (Chelonia mydas) in the Mediterranean. Biochem. Syst. Ecol. 2012, 43, 192–199. [Google Scholar] [CrossRef]
- Roden, S.E.; Dutton, P.H. Isolation and characterization of 14 polymorphic microsatellite loci in the leatherback turtle (Dermochelys coriacea) and cross-species amplification. Conserv. Genet. Resour. 2011, 3, 49–52. [Google Scholar] [CrossRef]
- Abreu-Grobois, F.A.; Horrocks, J.; Formia, A.; Dutton, P.; LeRoux, R.; Vélez-Zuazo, X.; Soares, L.; Meylan, P. New mtDNA Dloop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analyses. In Proceedings of the 26th Annual Symposium on Sea Turtle Biology, Island of Crete, Greece, 3–8 April 2006; ISTS: Nashville, TN, USA, 2006; p. 179. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.; Teslenko, M.; Nylander, J. MrBayes Version 3.2 Manual: Tutorials and Model Summaries. Available online: mrbayes.sourceforge.net/mb32_manual.pdf (accessed on 19 October 2018).
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2, 2–3. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Wagner, A.P.; Taper, M.L. ML-Relate: A computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 2006, 6, 576–579. [Google Scholar] [CrossRef]
- Blouin, M.S. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol. Evol. 2003, 18, 503–511. [Google Scholar] [CrossRef]
- Wagner, A.; Creel, S.; Kalinowski, S. Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity 2006, 97, 336–345. [Google Scholar] [CrossRef]
- Milligan, B.G. Maximum-likelihood estimation of relatedness. Genetics 2003, 163, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.; Rambaut, A.; Bouckaert, R.; Xie, W. Divergence Dating Tutorial with BEAST 2.0. 2012. Available online: http://pstorage-plos-3567654.s3.amazonaws.com/1460094/Text_S1.pdf (accessed on 13 February 2022).
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef]
- Vargas, S.M.; Lins, L.S.; Molfetti, É.; Ho, S.Y.; Monteiro, D.; Barreto, J.; Colman, L.; Vila-Verde, L.; Baptistotte, C.; Thomé, J.C.A. Revisiting the genetic diversity and population structure of the critically endangered leatherback turtles in the South-west Atlantic Ocean: Insights for species conservation. J. Mar. Biol. Assoc. UK 2019, 99, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Stumpf, M.P. Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur. J. Hum. Genet. 2004, 12, 469–477. [Google Scholar] [CrossRef]
- Dutton, P.H.; Squires, D. Reconciling biodiversity with fishing: A holistic strategy for Pacific sea turtle recovery. Ocean. Dev. Int. Law 2008, 39, 200–222. [Google Scholar] [CrossRef]
- Barbara, T.; Palma-Silva, C.; Paggi, G.M.; Bered, F.; Fay, M.F.; Lexer, C. Cross-species transfer of nuclear microsatellite markers: Potential and limitations. Mol. Ecol. 2007, 16, 3759–3767. [Google Scholar] [CrossRef] [PubMed]
- Torres-Pratts, H.; Schärer, M.T.; Schizas, N.V. Genetic diversity of Chelonibia caretta, commensal barnacles of the endangered hawksbill sea turtle Eretmochelys imbricata from the Caribbean (Puerto Rico). J. Mar. Biol. Assoc. UK 2009, 89, 719–725. [Google Scholar] [CrossRef]
- Daza-Criado, L.; Hernández-Fernández, J. Molecular identification and first report of mitochondrial COI gene haplotypes in the hawksbill turtle Eretmochelys imbricata (Testudines: Cheloniidae) in the Colombian Caribbean nesting colonies. Genet. Mol. Res. 2014, 13, 7123–7132. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, H.R.; Figueroa, D.F.; George, J.A. Mitochondrial genomes and genetic structure of the Kemp's ridley sea turtle (Lepidochelys kempii). Ecol. Evol. 2020, 10, 249–262. [Google Scholar] [CrossRef]
- Prakas, P.; Butkauskas, D.; Švažas, S.; Bea, A.; Yanenko, V.; Ragauskas, A.; Vaitkuvienė, D. The Genetic Diversity and Structure of the European Turtle Dove Streptopelia turtur. Animals 2021, 11, 1283. [Google Scholar] [CrossRef]
- Hamann, M.; Limpus, C.; Hughes, G.; Mortimer, J.; Pilcher, N. Assessment of the Conservation Status of the Leatherback Turtle in the Indian Ocean and South East Asia, including Consideration of the Impacts of the December 2004 Tsunami on Turtles and Turtle Habitats; IOSEA Marine Turtle MoU Secretariat: Bangkok, Thailand, 2006. [Google Scholar]
- Rius, R.; Cowley, M.J.; Riley, L.; Puttick, C.; Thorburn, D.R.; Christodoulou, J. Biparental inheritance of mitochondrial DNA in humans is not a common phenomenon. Genet. Med. 2019, 21, 2823–2826. [Google Scholar] [CrossRef]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.-C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J. Biparental inheritance of mitochondrial DNA in humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039–13044. [Google Scholar] [CrossRef]
- Tikochinski, Y.; Carreras, C.; Tikochinski, G.; Vilaça, S.T. Population-specific signatures of intra-individual mitochondrial DNA heteroplasmy and their potential evolutionary advantages. Sci. Rep. 2020, 10, 211. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Cano, D.; Mariño-Ramírez, L.; Hernández-Fernández, J. Detection of high heteroplasmy in complete loggerhead and hawksbill sea turtles mitochondrial genomes using RNAseq. Mitochondrial DNA Part A 2021, 32, 106–114. [Google Scholar] [CrossRef]
- Avens, L.; Goshe, L.R.; Zug, G.R.; Balazs, G.H.; Benson, S.R.; Harris, H. Regional comparison of leatherback sea turtle maturation attributes and reproductive longevity. Mar. Biol. 2020, 167, 4. [Google Scholar] [CrossRef]
- Domiciano, I.G.; Domit, C.; Bracarense, A.P.F.R.L. The green turtle Chelonia mydas as a marine and coastal environmental sentinels: Anthropogenic activities and diseases. Semin. Ciênc. Agrárias 2017, 38, 3417–3434. [Google Scholar] [CrossRef]
- Hernández-Fernández, J.; Pinzón, A.; Mariño-Ramírez, L. De novo transcriptome assembly of loggerhead sea turtle nesting of the Colombian Caribbean. Genom. Data 2017, 13, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Figgener, C.; Chacón-Chaverri, D.; Jensen, M.P.; Feldhaar, H. Paternity re-visited in a recovering population of Caribbean leatherback turtles (Dermochelys coriacea). J. Exp. Mar. Biol. Ecol. 2016, 475, 114–123. [Google Scholar] [CrossRef]



| Province | Beach | Nest Number | Number of Samples | Latitude and Longitude |
|---|---|---|---|---|
| Phang-nga | Khuk Khak (Zone 1) | 3 | 20 | 8.67829, 98.241 |
| Bor Dan (Zone 2) | 1 | 16 | 8.32848, 98.263 | |
| Bang Kwan (Zone 3) | 8 | 77 | 8.20880, 98.282 | |
| Phuket | Sai Keaw (Zone 4) | 1 | 17 | 8.19015, 98.287 |
| Nai Thon (Zone 5) | 1 | 19 | 8.05605, 98.277 | |
| Total | 14 | 149 |
| Primers | Sequence (5′-3′) | Repeat Motif | Ta (°C) | Length (bp) |
|---|---|---|---|---|
| LB99 | F: CACCCATTTTTTCCCATTG | (TG)11 | 55 | 113–133 |
| R: ATTTGAGCATAAGTTTTCGTGG | ||||
| LB110 | F: TAGCAACTGCAGGAGC | (CT)15 | 58 | 182–198 |
| R: CATTCCCTAAGATTATCACC | ||||
| LB123 | F: TGTAGTCaGGTGTCCAATG | (GT)12 | 51 | 170–186 |
| R: CCAAGCCAAAGAAAGAA | ||||
| LB125 | F: AACTAATGCCTTACAGAG | (AC)13 | 55 | 182–196 |
| R: CCTTAGAGGGAGAATCT | ||||
| LB128 | F: AAGCATGGAGGAGAAGG | (GT)11 | 55 | 157–165 |
| R: GGTTCTTTGCCCCAGTA | ||||
| LB133 | F: AGAGGCAGCAGAGCAAGG | (AC)13 | 60 | 156–200 |
| R: GGCTGAGGGTGGTGAGG | ||||
| LB141 | F: CATCCTCATGTTCCCATC | (TC)20(AC)9 | 55 | 166–192 |
| R: CATTGCCTCATAATAAGAGAAA | ||||
| LB142 | F: GGCCAACTTTCCTTTCTTATTA | (CA)16 | 58 | 219–237 |
| R: CTGTGTGTATCTGCACCCA | ||||
| LB143 | F: CCTATGGGCCACTGCAATGACA | (GT)10 | 58 | 181–197 |
| R: CAGCTGGAGGGATGCAAGATGT | ||||
| LB145 | F: GGCCTCCACACAAATAAATAAA | (AC)28 | 55 | 121–197 |
| R: CATTCACCTTACGCAGAAGAA | ||||
| LB157 | F: GGCATGAGTGTGAGTGA | (AC)13 | 60 | 72–102 |
| R: CCTGGTTAAAGCTGTCTC | ||||
| LB158 | F: AGGACAAGGCATTCTAGC | (CA)12 | 55 | 162–178 |
| R: ATGTACTTGCCCATCTGC |
| Locus | Fis | Fit | Fst | Nm | Genetic Variabilities | Overall HWE (p-Value) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Na | Ne | I | Ho | He | ||||||
| LB99 | −0.08 | 0.15 | 0.22 | 0.91 | 13.00 | 5.18 | 1.89 | 0.70 | 0.81 | 0.00 * |
| LB110 | −0.13 | 0.05 | 0.16 | 1.32 | 7.00 | 4.21 | 1.56 | 0.70 | 0.76 | 0.00 * |
| LB141 | 0.11 | 0.50 | 0.44 | 0.32 | 9.00 | 3.39 | 1.41 | 0.43 | 0.71 | 0.00 * |
| LB157 | −0.03 | 0.29 | 0.31 | 0.55 | 6.00 | 1.81 | 0.81 | 0.29 | 0.45 | 0.00 * |
| LB128 | −0.03 | 0.19 | 0.22 | 0.91 | 7.00 | 3.66 | 1.54 | 0.38 | 0.73 | 0.00 * |
| LB142 | −0.37 | 0.19 | 0.41 | 0.37 | 6.00 | 3.53 | 1.39 | 0.69 | 0.72 | 0.00 * |
| LB143 | 0.26 | 0.39 | 0.17 | 1.20 | 9.00 | 6.20 | 1.97 | 0.48 | 0.84 | 0.00 * |
| LB158 | 0.11 | 0.25 | 0.16 | 1.34 | 5.00 | 2.22 | 1.00 | 0.45 | 0.55 | 0.00 * |
| Mean | −0.02 | 0.25 | 0.26 | 0.86 | ||||||
| ±SE | ±0.07 | ±0.05 | ±0.04 | ±0.15 | ||||||
| Population | Forensic Parameters | Diversity Indices | ||||||
|---|---|---|---|---|---|---|---|---|
| PD | PE | MP | Na | Ne | I | Ho | He | |
| Zone 1 | 0.9999996241 | 0.9996082621 | 0.0000003759 | 5.13 ± 0.79 | 3.30 ± 0.45 | 1.26 ± 0.17 | 0.65 ± 0.10 | 0.63 ± 0.07 |
| Zone 2 | 0.9999495824 | 0.9910400379 | 0.0000504176 | 3.38 ± 0.80 | 2.40 ± 0.42 | 0.86 ± 0.20 | 0.44 ± 0.09 | 0.48 ± 0.09 |
| Zone 3 | 0.9998545164 | 0.9805346366 | 0.0001454836 | 3.25 ± 0.95 | 2.11 ± 0.26 | 0.80 ± 0.16 | 0.49 ± 0.11 | 0.46 ± 0.08 |
| Zone 4 | 0.9999254295 | 0.9853515299 | 0.0000745705 | 3.38 ± 0.46 | 2.15 ± 0.25 | 0.86 ± 0.11 | 0.50 ± 0.09 | 0.50 ± 0.05 |
| Zone 5 | 0.9999359368 | 0.9869546837 | 0.0000640632 | 4.00 ± 0.33 | 2.20 ± 0.26 | 0.88 ± 0.12 | 0.51 ± 0.10 | 0.50 ± 0.06 |
| Overall | 0.9999999512 | 0.9999021520 | 0.0000000488 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wongfu, C.; Prasitwiset, W.; Poommouang, A.; Buddhachat, K.; Brown, J.L.; Chomdej, S.; Kampuansai, J.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. Genetic Diversity in Leatherback Turtles (Dermochelys coriacea) along the Andaman Sea of Thailand. Diversity 2022, 14, 764. https://doi.org/10.3390/d14090764
Wongfu C, Prasitwiset W, Poommouang A, Buddhachat K, Brown JL, Chomdej S, Kampuansai J, Kaewmong P, Kittiwattanawong K, Nganvongpanit K. Genetic Diversity in Leatherback Turtles (Dermochelys coriacea) along the Andaman Sea of Thailand. Diversity. 2022; 14(9):764. https://doi.org/10.3390/d14090764
Chicago/Turabian StyleWongfu, Chutima, Wareerat Prasitwiset, Anocha Poommouang, Kittisak Buddhachat, Janine L. Brown, Siriwadee Chomdej, Jatupol Kampuansai, Patcharaporn Kaewmong, Kongkiat Kittiwattanawong, and Korakot Nganvongpanit. 2022. "Genetic Diversity in Leatherback Turtles (Dermochelys coriacea) along the Andaman Sea of Thailand" Diversity 14, no. 9: 764. https://doi.org/10.3390/d14090764
APA StyleWongfu, C., Prasitwiset, W., Poommouang, A., Buddhachat, K., Brown, J. L., Chomdej, S., Kampuansai, J., Kaewmong, P., Kittiwattanawong, K., & Nganvongpanit, K. (2022). Genetic Diversity in Leatherback Turtles (Dermochelys coriacea) along the Andaman Sea of Thailand. Diversity, 14(9), 764. https://doi.org/10.3390/d14090764