Bacterial Products and Their Effect on the Shrubby Legume Calicotome villosa (Poir.) Link
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material Collection
2.2. Authentication of the Symbiotic Interactions
2.3. Phenotypic Characterization of the Bacterial Isolates
2.4. HCN Production
2.5. Ammonia (NH3) Production
2.6. Assay for Indoleacetic Acid (IAA) Production
2.7. Siderophore Production
2.8. Effects of the Rhizobacterial Strains on C. villosa and Accompanying Understory Species
2.9. Data Analysis
3. Results
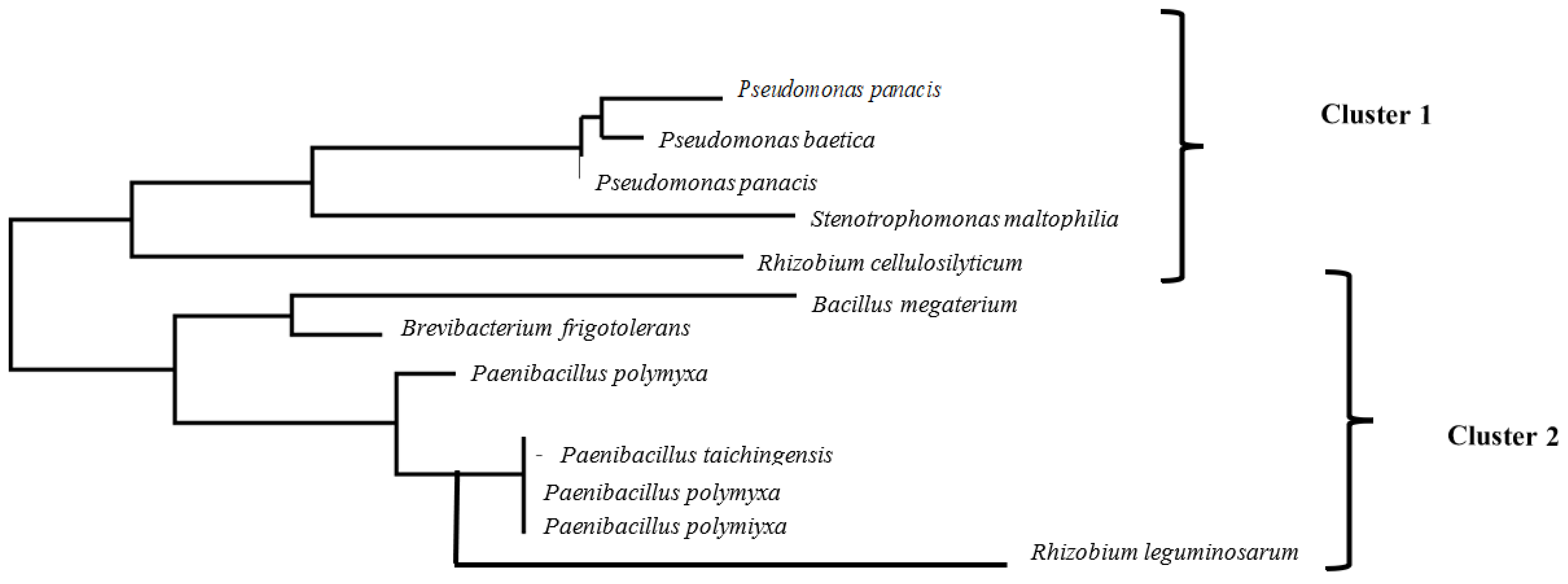
3.1. Bacterial Identification
3.2. Phenotypic Characterization of the Bacterial Isolates
3.3. HCN and NH3 Production
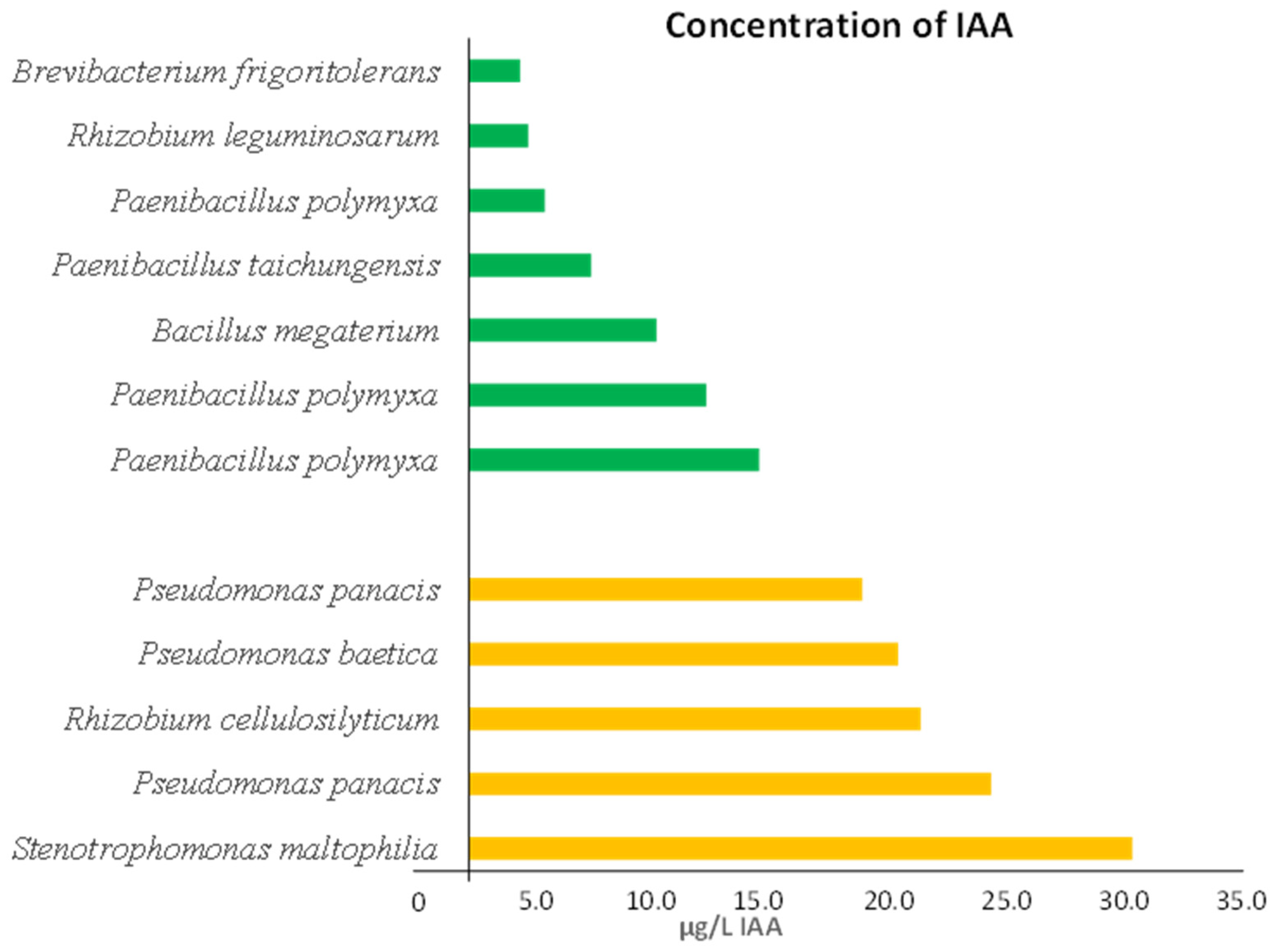
3.4. Assay for Indoleacetic Acid (IAA) Production
3.5. Siderophore Production
3.6. Effects of the Rhizobacterial Strains on C. villosa and Accompanying Understory Species
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Segarra-Moragues, J.G.; Carrión-Marco, Y.; Castellanos, M.C.; Molina, M.J.; García-Fayos, P. Ecological and historical determinants of population genetic structure and diversity in the Mediterranean shrub Rosmarinus officinalis (Lamiaceae). Bot. J. Linn. Soc. 2016, 180, 50–63. [Google Scholar] [CrossRef]
- Turco, M.; Von Hardenberg, J.; AghaKouchak, A.; Llasat, M.C.; Provenzale, A.; Trigo, R.M. On the Key Role of Droughts in the Dynamics of Summer Fires in Mediterranean Europe. Sci. Rep. 2017, 7, 81. [Google Scholar] [CrossRef]
- García, D.; Martínez, D. Species Richness Matters for the Quality of Ecosystem Services: A Test Using Seed Dispersal by Frugivorous Birds. Proc. R. Soc. B 2012, 279, 3106–3113. [Google Scholar] [CrossRef]
- Haase, P.; Pugnaire, F.I.; Fernández, E.M.; Puigdefábregas, J.; Clark, S.C.; Incoll, L.D. An Investigation of Rooting Depth of the Semiarid Shrub Retama Sphaerocarpa (L.) Boiss. by Labelling of Ground Water with a Chemical Tracer. J. Hydrol. 1996, 177, 23–31. [Google Scholar] [CrossRef]
- Thamer, S.; Schädler, M.; Bonte, D.; Ballhorn, D.J. Dual Benefit from a Belowground Symbiosis: Nitrogen Fixing Rhizobia Promote Growth and Defense against a Specialist Herbivore in a Cyanogenic Plant. Plant Soil 2011, 341, 209–219. [Google Scholar] [CrossRef]
- De Lara-Del Rey, I.A.; Pérez-Fernández, M.A. Regulatory Effect of Light and Rhizobial Inoculation on the Root Architecture and Plant Performance of Pasture Legumes. Agronomy 2023, 13, 2058. [Google Scholar] [CrossRef]
- Pérez-Fernández, M.A.; Calvo-Magro, E.; Valentine, A. Benefits of the Symbiotic Association of Shrubby Legumes for the Rehabilitation of Degraded Soils under Mediterranean Climatic Conditions: Inoculated Legumes Recover Contaminated Soil. Land Degrad. Dev. 2016, 27, 395–405. [Google Scholar] [CrossRef]
- Thrall, P.H.; Millsom, D.A.; Jeavons, A.C.; Waayers, M.; Harvey, G.R.; Bagnall, D.J.; Brockwell, J. Seed Inoculation with Effective Root-Nodule Bacteria Enhances Revegetation Success: Seed Inoculation with Root-Nodule Bacteria. J. Appl. Ecol. 2005, 42, 740–751. [Google Scholar] [CrossRef]
- Berger, J.D.; Shrestha, D.; Ludwig, C. Reproductive Strategies in Mediterranean Legumes: Trade-Offs between Phenology, Seed Size and Vigor within and between Wild and Domesticated Lupinus Species Collected along Aridity Gradients. Front. Plant Sci. 2017, 8, 548. [Google Scholar] [CrossRef]
- Alonso-Vargas, M.A.; Crespo-Villalva, M.B.; Martínez-Azorín, M.; Pérez-Botella, J. Calicotome villosa (Poir.) Link (Fabaceae), novedad para la flora de la Comunidad Valenciana. Flora Montiberica 2020, 77, 104–107. [Google Scholar]
- Lattanzi, E. The distribution of three species of the genus Calicotome in Italy. Flora Mediterr. 2008, 18, 123–125. [Google Scholar]
- Zahran, H.H. Rhizobia from wild legumes: Diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J. Biotechnol. 2001, 91, 143–153. [Google Scholar] [CrossRef]
- Zakhia, F. Diversity of Bacteria Hosts of Mediterranean Legumes in Tunisia and Lebanon. Ph.D. Thesis, Université de Montpellier II, Montpellier, France, 2004. [Google Scholar]
- Gómez-Hernández, A.; Ferrera-Cerrato, R.; López-Piñeiro, A.; Velázquez, E. Rhizobial diversity associated with Calicotome villosa (Poir.) Link, a N2-fixing shrub legume from Spain. FEMS Microbiol. Ecol. 2007, 61, 434–444. [Google Scholar]
- Marulanda, Á.; Barea, J.M.; Azcón-Aguilar, C.; Ferrol, N. Mycorrhizal arbuscular fungi and Rhizobium bacteria can improve plant growth and soil aggregation in a Calicotome villosa revegetation trial. Soil Biol. Biochem. 2009, 41, 1915–1920. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and algorithms. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Vincent, J.M. A Manual for the Practical Study of Root-Nodule Bacteria; IBP handbook no. 15; International Biological Programme; Blackwell Scientific: Oxford, UK, 1970. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The Barcode of Life Data System (BOLD). Mol. Ecol. Resour. 2013, 13, 286–293. [Google Scholar]
- Pérez-Fernández, M.A.; Hill, Y.J.; Calvo-Magro, E.; Valentine, A. Competing Bradyrhizobia Strains Determine Niche Occupancy by Two Native Legumes in the Iberian Peninsula. Plant Ecol. 2015, 216, 1537–1549. [Google Scholar] [CrossRef]
- Lorck, H. Production of Hydrocyanic Acid by Bacteria. Physiol. Plant. 1948, 1, 142–146. [Google Scholar] [CrossRef]
- Cappuccino, J.C.; Sherman, N. A Laboratory Manual. In Microbiology, 3rd ed.; Benjamin/Cummings Pub. Co.: New York, NY, USA, 1992; pp. 125–179. [Google Scholar]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid In Situ Assay for Indoleacetic Acid Production by Bacteria Immobilized on a Nitrocellulose Membrane. Appl. Env. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef]
- Loper, J.E.; Scroth, M.N. Influence of bacterial sources on indole-3 acetic acid on root elongation of sugarbeet. Phytopathology 1986, 76, 386–389. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Meyer, J.M.; Abdallah, M.A. The Fluorescent Pigment of Pseudomonas fluorescens: Biosynthesis, Purification and Physicochemical Properties. J. Gen. Microbiol. 1978, 107, 319–328. [Google Scholar] [CrossRef]
- Payne, S.M. [25] Detection, Isolation, and Characterization of Siderophores. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1994; Volume 235, pp. 329–344. [Google Scholar] [CrossRef]
- IBM SPSS Software. Available online: https://www.ibm.com/analytics/spss-statistics-software (accessed on 12 September 2023).
- Cochran, W.G. The distribution of the largest of a set of estimated variances as a fraction of their total. Ann. Eugen. 1941, 11, 47–61. [Google Scholar] [CrossRef]
- Bever, J.D.; Platt, T.G.; Morton, E.R. Microbial Population and Community Dynamics on Plant Roots and Their Feedbacks on Plant Communities. Annu. Rev. Microbiol. 2012, 66, 265–283. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant Growth-Promoting Rhizobacteria (PGPR): Emergence in Agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Lawton, J.H. A Direct Test of the Theory of Habitat Selection. Ecology 1970, 51, 704–707. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The Importance of the Microbiome of the Plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Prudent, M.; Dequiedt, S.; Sorin, C.; Girodet, S.; Nowak, V.; Duc, G.; Salon, C.; Maron, P. The Diversity of Soil Microbial Communities Matters When Legumes Face Drought. Plant Cell Env. 2020, 43, 1023–1035. [Google Scholar] [CrossRef]
- Amine-Khodja, I.R.; Boscari, A.; Riah, N.; Kechid, M.; Maougal, R.T.; Belbekri, N.; Djekoun, A. Impact of Two Strains of Rhizobium leguminosarum on the Adaptation to Terminal Water Deficit of Two Cultivars Vicia Faba. Plants 2022, 11, 515. [Google Scholar] [CrossRef]
- Antolín, M.C.; Yoller, J.; Sánchez-Díaz, M. Effects of Temporary Drought on Nitrate-Fed and Nitrogen-Fixing Alfalfa Plants. Plant Sci. 1995, 107, 159–165. [Google Scholar] [CrossRef]
- Lodeiro, A.R.; González, P.; Hernández, A.; Balagué, L.J.; Favelukes, G. Comparison of Drought Tolerance in Nitrogen-Fixing and Inorganic Nitrogen-Grown Common Beans. Plant Sci. 2000, 154, 31–41. [Google Scholar] [CrossRef]
- Staudinger, C.; Mehmeti-Tershani, V.; Gil-Quintana, E.; Gonzalez, E.M.; Hofhansl, F.; Bachmann, G.; Wienkoop, S. Evidence for a Rhizobia-Induced Drought Stress Response Strategy in Medicago truncatula. J. Proteom. 2016, 136, 202–213. [Google Scholar] [CrossRef]
- Turetschek, R.; Staudinger, C.; Wienkoop, S. Rhizobial Symbiosis Influences Response to Early Salt and Drought Stress of the Medicago truncatula Root Proteome. In The Model Legume Medicago truncatula; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 253–260. [Google Scholar] [CrossRef]
- Frindte, K.; Pape, R.; Werner, K.; Löffler, J.; Knief, C. Temperature and Soil Moisture Control Microbial Community Composition in an Arctic–Alpine Ecosystem along Elevational and Micro-Topographic Gradients. ISME J. 2019, 13, 2031–2043. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Ritz, K.; Wheatley, R.; Kuan, H.L.; Boag, B.; Christensen, S.; Ekelund, F.; Sørensen, S.J.; Muller, S.; Bloem, J. An Examination of the Biodiversity–Ecosystem Function Relationship in Arable Soil Microbial Communities. Soil Biol. Biochem. 2001, 33, 1713–1722. [Google Scholar] [CrossRef]
- Maire, V.; Gross, N.; Da Silveira Pontes, L.; Picon-Cochard, C.; Soussana, J.-F. Trade-off between Root Nitrogen Acquisition and Shoot Nitrogen Utilization across 13 Co-Occurring Pasture Grass Species. Funct. Ecol. 2009, 23, 668–679. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant Species and Soil Type Cooperatively Shape the Structure and Function of Microbial Communities in the Rhizosphere: Plant Species, Soil Type and Rhizosphere Communities. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina Del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic Acid Modulates Colonization of the Root Microbiome by Specific Bacterial Taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef]
- Panke-Buisse, K.; Poole, A.C.; Goodrich, J.K.; Ley, R.E.; Kao-Kniffin, J. Selection on Soil Microbiomes Reveals Reproducible Impacts on Plant Function. ISME J. 2015, 9, 980–989. [Google Scholar] [CrossRef]

| pH | Tolerance to Salinity | Resistance to Drought | HCN | NH3 | ||||
|---|---|---|---|---|---|---|---|---|
| Bacterial Strain | 1% | 3% | 7% | 10% | ||||
| Pseudomonas panacis | Yellow | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Pseudomonas baetica | Yellow | Yes | Yes | Yes | Yes | Yes | Yes | |
| Pseudomonas panacis | Yellow | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Stenotrophomonas maltophilia | Green | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Rhizobium cellulosilyticum | Green | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Bacillus megaterium | Yellow | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Brevibacterium frigoritolerans | Yellow | Yes | Yes | Yes | Yes | No | No | Yes |
| Paenibacillus polymyxa | Green | Yes | Yes | Yes | Yes | No | No | Yes |
| Paenibacillus taichungensis | Yellow | Yes | Yes | Yes | No | No | No | Yes |
| Paenibacillus polymyxa | Green | Yes | Yes | No | No | No | No | Yes |
| Paenibacillus polymyxa | Green | Yes | Yes | No | No | Yes | No | Yes |
| Rhizobium leguminosarum | Green | Yes | Yes | Yes | Yes | No | Yes | Yes |
| C. villosa | O. compressus | V. sativa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shoot Length (cm) | Shoot Length (cm) | Shoot Mass (mg) | Root Mass (mg) | Shoot Length (cm) | Root Length (cm) | Shoot Mass (mg) | Root Mass (mg) | Shoot Length (cm) | Root Length (cm) | Shoot Mass (mg) | Root Mass (mg) | |
| Pseudomonas panacis | 2.95 | 2.93 | 54.09 | 64.09 | 3.07 | 5.90 | 110.17 | −227.45 | 2.65 | 2.38 | 156.45 | −75.81 |
| Pseudomonas baetica | −1.40 | 0.40 | 80.09 | −87.40 | −2.05 | −1.80 | 148.90 | 80.00 | −6.22 | −2.39 | 211.45 | 26.67 |
| Pseudomonas panacis | 0.02 | −2.08 | 132.54 | −89.42 | −4.20 | −1.20 | 226.93 | 67.25 | −3.86 | 0.31 | 322.25 | 22.42 |
| Rhizobium cellulosilyticum | 7.60 | 4.90 | 1977.23 | 331.75 | 8.35 | 14.06 | 1483.67 | 835.23 | 7.02 | 13.43 | 2106.82 | 278.41 |
| Stenotrophomonas maltophilia | 8.72 | 4.29 | 911.82 | 439.61 | 10.11 | 21.89 | 493.67 | 251.23 | 20.60 | 14.53 | 701.02 | 83.75 |
| Bacillus megaterium | 6.98 | 3.27 | 613.85 | 199.73 | 2.28 | 11.44 | 943.03 | 632.26 | 15.80 | 10.13 | 1339.11 | 210.76 |
| Brevibacterium frigoritolerans | 0.30 | 0.51 | 280.24 | −52.77 | 0.48 | 0.53 | 446.70 | 299.49 | 0.45 | 0.45 | 634.32 | 99.83 |
| Paenibacillus polymyxa | 0.53 | 0.55 | 20.33 | −102.96 | 0.30 | 0.89 | 59.99 | −18.57 | −0.34 | 0.19 | 85.19 | −6.19 |
| Paenibacillus taichungensis | 0.02 | −0.20 | 42.53 | −94.28 | −0.22 | 0.09 | 93.01 | 36.44 | −0.20 | −0.61 | 132.08 | 12.15 |
| Paenibacillus polymyxa | −0.14 | 0.58 | 11.52 | −101.79 | −0.50 | −0.31 | 46.88 | −11.17 | −0.42 | 1.72 | 66.58 | −3.71 |
| Paenibacillus polymyxa | −0.97 | −0.29 | 33.04 | −103.73 | −0.82 | 0.33 | 78.90 | −23.45 | −0.28 | 0.14 | 112.04 | −7.81 |
| Rhizobium leguminosarum | 6.70 | 3.80 | 1597.17 | 797.10 | 9.42 | 6.28 | 918.21 | 615.62 | 15.65 | 12.34 | 1303.88 | 205.21 |
| C. juncea | T. repens | |||||||||||
| Shoot Length (cm) | Shoot Length (cm) | Shoot Mass (mg) | Root Mass (mg) | Shoot Length (cm) | Root Length (cm) | Shoot Mass (mg) | Root Mass (mg) | |||||
| Pseudomonas panacis | 2.45 | 2.18 | 151.27 | −33.06 | 4.65 | 2.83 | 10.28 | 10.12 | ||||
| Pseudomonas baetica | −6.42 | −2.59 | 185.64 | 30.99 | 3.07 | 5.90 | −2.91 | −2.03 | ||||
| Pseudomonas panacis | −4.06 | 0.11 | 254.89 | 28.34 | −2.05 | −1.80 | 350.60 | 184.72 | ||||
| Rhizobium cellulosilyticum | 6.82 | 13.23 | 1870.25 | 1188.33 | −4.20 | −1.20 | 406.33 | 175.61 | ||||
| Stenotrophomonas maltophilia | 20.40 | 14.33 | 491.63 | 66.67 | 8.35 | 14.06 | 1404.01 | 824.17 | ||||
| 10.11 | 21.89 | 696.86 | 407.02 | |||||||||
| Bacillus megaterium | 15.60 | 9.93 | 1290.43 | 146.05 | −4.65 | −2.83 | −10.28 | −10.12 | ||||
| Brevibacterium frigoritolerans | 0.25 | 0.25 | 481.94 | 76.72 | 2.28 | 11.44 | 1017.83 | 679.19 | ||||
| Paenibacillus polymyxa | −0.54 | −0.01 | 106.73 | 10.46 | 0.48 | 0.53 | 663.31 | 441.50 | ||||
| Paenibacillus taichungensis | −0.40 | −0.81 | 136.04 | 21.92 | 0.30 | 0.89 | 387.09 | 214.31 | ||||
| Paenibacillus polymyxa | −0.62 | 1.52 | 95.10 | 12.01 | −0.22 | 0.09 | 410.68 | 253.60 | ||||
| Paenibacillus polymyxa | −0.48 | −0.06 | 123.51 | 9.44 | −0.50 | −0.31 | 377.73 | 219.59 | ||||
| Rhizobium leguminosarum | 15.45 | 12.14 | 2668.41 | 1142.58 | −0.82 | 0.33 | 400.60 | 210.82 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Fernández, M.A.; De-Lara-Del-Rey, I.A.; Magadlela, A. Bacterial Products and Their Effect on the Shrubby Legume Calicotome villosa (Poir.) Link. Diversity 2023, 15, 1101. https://doi.org/10.3390/d15101101
Pérez-Fernández MA, De-Lara-Del-Rey IA, Magadlela A. Bacterial Products and Their Effect on the Shrubby Legume Calicotome villosa (Poir.) Link. Diversity. 2023; 15(10):1101. https://doi.org/10.3390/d15101101
Chicago/Turabian StylePérez-Fernández, María A., Irene Ariadna De-Lara-Del-Rey, and Anathi Magadlela. 2023. "Bacterial Products and Their Effect on the Shrubby Legume Calicotome villosa (Poir.) Link" Diversity 15, no. 10: 1101. https://doi.org/10.3390/d15101101
APA StylePérez-Fernández, M. A., De-Lara-Del-Rey, I. A., & Magadlela, A. (2023). Bacterial Products and Their Effect on the Shrubby Legume Calicotome villosa (Poir.) Link. Diversity, 15(10), 1101. https://doi.org/10.3390/d15101101







