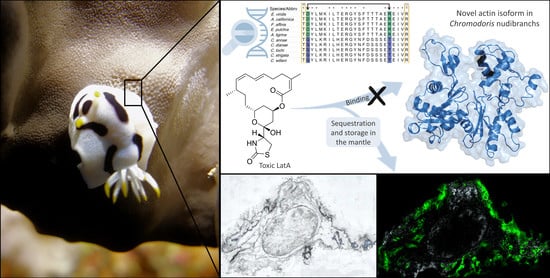
Is a Modified Actin the Key to Toxin Resistance in the Nudibranch Chromodoris? A Biochemical and Molecular Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Chemical Analysis
2.3. MALDI-MS Imaging
2.4. Fluorescence Microscopy
2.5. In Vivo Toxicity Assay
2.6. Comparative Analysis of Heterobranchia Actin Genes
3. Results
3.1. Chemical Investigation of Chromodoris Nudibranchs and Cacospongia mycofijiensis
3.2. MALDI-MS Imaging
3.3. Fluorescence Microscopy
3.4. In Vivo Toxicity Assay
3.5. Comparative Analysis of Heterobranchia Actin Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wägele, H.; Klussmann-Kolb, A. Opisthobranchia (Mollusca, Gastropoda)—More than Just Slimy Slugs. Shell Reduction and Its Implications on Defence and Foraging. Front. Zool. 2005, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Faulkner, D.; Ghiselin, M. Chemical Defense and Evoluti—Onary Ecology of Dorid Nudibranchs and Some Other Opisthobranch Gastropods. Mar. Ecol. Prog. Ser. 1983, 13, 295–301. [Google Scholar] [CrossRef]
- Avila, C.; Núñez-Pons, L.; Moles, J. From the Tropics to the Poles: Chemical Defense Strategies in Sea Slugs (Mollusca: Heterobranchia). In Chemical Ecology; CRC Press: Boca Raton, FL, USA, 2018; ISBN 978-0-429-45346-5. [Google Scholar]
- Dietz, L.; Dömel, J.S.; Leese, F.; Lehmann, T.; Melzer, R.R. Feeding Ecology in Sea Spiders (Arthropoda: Pycnogonida): What Do We Know? Front. Zool. 2018, 15, 7. [Google Scholar] [CrossRef] [Green Version]
- Mehrotra, R.; Monchanin, C.; Scott, C.M.; Phongsuwan, N.; Gutierrez, M.C.; Chavanich, S.; Hoeksema, B.W. Selective Consumption of Sacoglossan Sea Slugs (Mollusca: Gastropoda) by Scleractinian Corals (Cnidaria: Anthozoa). PLoS ONE 2019, 14, e0215063. [Google Scholar] [CrossRef] [Green Version]
- Anker, A.; Ivanov, Y. First Record of the Predation upon Sea Slugs (Cephalaspidea and Nudibranchia) by the Peculiar Elbow Crab Lambrachaeus Ramifer Alcock, 1895 (Decapoda: Parthenopidae). Mar. Biodivers. 2020, 50, 24. [Google Scholar] [CrossRef]
- Battini, N.; Bravo, G. Unexpected Meal: First Record of Predation upon a Potentially Neurotoxic Sea Slug by the European Green Crab Carcinus Maenas. N. Z. J. Zool. 2020, 48, 166–173. [Google Scholar] [CrossRef]
- Battini, N.; Giachetti, C.; Castro, K.; Bortolus, A.; Schwindt, E. Predator–Prey Interactions as Key Drivers for the Invasion Success of a Potentially Neurotoxic Sea Slug. Biol. Invasions 2021, 23, 1–23. [Google Scholar] [CrossRef]
- Avila, C. Natural Products of Opisthobranch Molluscs: A Biological Review. In Oceanography and Marine Biology—An Annual Review, Vol 33; Ansell, A.D., Gibson, R.N., Barnes, M., Eds.; U C L Press Ltd: London, UK, 1995; Volume 33, pp. 487–559. ISBN 978-1-85728-363-1. [Google Scholar]
- Cimino, G.; Ghiselin, M.T. Chemical Defense and Evolutionary Trends in Biosynthetic Capacity among Dorid Nudibranchs (Mollusca: Gastropoda: Opisthobranchia). Chemoecology 1999, 9, 187–207. [Google Scholar] [CrossRef]
- Cheney, K.L.; White, A.; Mudianta, I.W.; Winters, A.E.; Quezada, M.; Capon, R.J.; Mollo, E.; Garson, M.J. Choose Your Weaponry: Selective Storage of a Single Toxic Compound, Latrunculin A, by Closely Related Nudibranch Molluscs. PLoS ONE 2016, 11, e0145134. [Google Scholar] [CrossRef] [Green Version]
- Dean, L.J.; Prinsep, M.R. The Chemistry and Chemical Ecology of Nudibranchs. Nat. Prod. Rep. 2017, 34, 1359–1390. [Google Scholar] [CrossRef] [Green Version]
- Fisch, K.M.; Hertzer, C.; Böhringer, N.; Wuisan, Z.G.; Schillo, D.; Bara, R.; Kaligis, F.; Wägele, H.; König, G.M.; Schäberle, T.F. The Potential of Indonesian Heterobranchs Found around Bunaken Island for the Production of Bioactive Compounds. Mar. Drugs 2017, 15, 384. [Google Scholar] [CrossRef] [Green Version]
- Avila, C.; Angulo-Preckler, C. Bioactive Compounds from Marine Heterobranchs. Mar. Drugs 2020, 2020, 657. [Google Scholar] [CrossRef]
- Winters, A.E.; Chan, W.; White, A.M.; van den Berg, C.P.; Garson, M.J.; Cheney, K.L. Weapons or Deterrents? Nudibranch Molluscs Use Distinct Ecological Modes of Chemical Defence against Predators. J. Anim. Ecol. 2022, 91, 831–844. [Google Scholar] [CrossRef]
- Johnson, R.F.; Gosliner, T.M. Traditional Taxonomic Groupings Mask Evolutionary History: A Molecular Phylogeny and New Classification of the Chromodorid Nudibranchs. PLoS ONE 2012, 7, e33479. [Google Scholar] [CrossRef] [Green Version]
- Layton, K.K.S.; Gosliner, T.M.; Wilson, N.G. Flexible Colour Patterns Obscure Identification and Mimicry in Indo-Pacific Chromodoris Nudibranchs (Gastropoda: Chromodorididae). Mol. Phylogenet. Evol. 2018, 124, 27–36. [Google Scholar] [CrossRef]
- Undap, N.; Papu, A.; Schillo, D.; Ijong, F.G.; Kaligis, F.; Lepar, M.; Hertzer, C.; Böhringer, N.; König, G.M.; Schäberle, T.F.; et al. First Survey of Heterobranch Sea Slugs (Mollusca, Gastropoda) from the Island Sangihe, North Sulawesi, Indonesia. Diversity 2019, 11, 170. [Google Scholar] [CrossRef] [Green Version]
- Layton, K.; Carvajal, J.; Wilson, N. Mimicry and Mitonuclear Discordance in Nudibranchs: New Insights from Exon Capture Phylogenomics. Ecol. Evol. 2020, 10, 11966–11982. [Google Scholar] [CrossRef]
- Padula, V.; Bahia, J.; Stöger, I.; Camacho-García, Y.; Malaquias, M.A.E.; Cervera, J.L.; Schrödl, M. A Test of Color-Based Taxonomy in Nudibranchs: Molecular Phylogeny and Species Delimitation of the Felimida Clenchi (Mollusca: Chromodorididae) Species Complex. Mol. Phylogenet. Evol. 2016, 103, 215–229. [Google Scholar] [CrossRef]
- Nakano, R.; Hirose, E. Field Experiments on the Feeding of the Nudibranch Gymnodoris Spp. (Nudibranchia: Doridina: Gymnodorididae) in Japan. Veliger 2011, 51, 66–75. [Google Scholar]
- Avila, C. Terpenoids in Marine Heterobranch Molluscs. Mar. Drugs 2020, 18, 162. [Google Scholar] [CrossRef] [Green Version]
- Pawlik, J.R.; Kernan, M.R.; Molinski, T.F.; Harper, M.K.; Faulkner, D.J. Defensive Chemicals of the Spanisch Dancer Nudibranch Hexabranchus Sanguineus and Its Egg Ribbons: Macrolides Derived from a Sponge Diet. J. Exp. Mar. Biol. Ecol. 1988, 119, 99–109. [Google Scholar] [CrossRef]
- Proksch, P. Defensive Roles for Secondary Metabolites from Marine Sponges and Sponge-Feeding Nudibranchs. Toxicon 1994, 32, 639–655. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.W.; Hill, R.T.; Piel, J.; Thacker, R.W.; Hentschel, U. Soaking It up: The Complex Lives of Marine Sponges and Their Microbial Associates. ISME J. 2007, 1, 187–190. [Google Scholar] [CrossRef]
- Putz, A.; Proksch, P. Chemical Defence in Marine Ecosystems. In Annual Plant Reviews Volume 39: Functions and Biotechnology of Plant Secondary Metabolites; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010; pp. 162–213. ISBN 978-1-4443-1887-6. [Google Scholar]
- Ng, T.P.T.; Saltin, S.H.; Davies, M.S.; Johannesson, K.; Stafford, R.; Williams, G.A. Snails and Their Trails: The Multiple Functions of Trail-Following in Gastropods. Biol. Rev. 2013, 88, 683–700. [Google Scholar] [CrossRef]
- Puglisi, M.P.; Sneed, J.M.; Sharp, K.H.; Ritson-Williams, R.; Paul, V.J. Marine Chemical Ecology in Benthic Environments. Nat. Prod. Rep. 2014, 31, 1510–1553. [Google Scholar] [CrossRef]
- Mollo, E.; Fontana, A.; Roussis, V.; Polese, G.; Amodeo, P.; Ghiselin, M.T. Sensing Marine Biomolecules: Smell, Taste, and the Evolutionary Transition from Aquatic to Terrestrial Life. Chem. Biol. 2014, 2, 92. [Google Scholar] [CrossRef] [Green Version]
- Rohde, S.; Nietzer, S.; Schupp, P.J. Prevalence and Mechanisms of Dynamic Chemical Defenses in Tropical Sponges. PLoS ONE 2015, 10, e0132236. [Google Scholar] [CrossRef]
- Venuleo, M.; Raven, J.A.; Giordano, M. Intraspecific Chemical Communication in Microalgae. New Phytol. 2017, 215, 516–530. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.; Caspers, B.A.; Gadau, J.; Kaiser, S. The Power of Infochemicals in Mediating Individualized Niches. Trends Ecol. Evol. 2020, 35, 981–989. [Google Scholar] [CrossRef]
- Okuda, R.K. Chemical Ecology of Some Opisthobranch Mollusks. Ph.D. Thesis, University of Hawai’i at Manoa, Honolulu, HI, USA, 1983. [Google Scholar]
- Okuda, R.K.; Scheuer, P.J. Latrunculin-A, Ichthyotoxic Constituent of the NudibranchChromodoris Elisabethina. Experientia 1985, 41, 1355–1356. [Google Scholar] [CrossRef]
- Pika, J.; John Faulkner, D. Unusual Chlorinated Homo-Diterpenes from the South African Nudibranch Chromodoris Hamiltoni. Tetrahedron 1995, 51, 8189–8198. [Google Scholar] [CrossRef]
- Kakou, Y.; Crews, P.; Bakus, G.J. Dendrolasin and Latrunculin A from the Fijian Sponge Spongia Mycofijiensis and an Associated Nudibranch Chromodoris Lochi. J. Nat. Prod. 1987, 50, 482–484. [Google Scholar] [CrossRef]
- Houssen, W.E.; Jaspars, M.; Wease, K.N.; Scott, R.H. Acute Actions of Marine Toxin Latrunculin A on the Electrophysiological Properties of Cultured Dorsal Root Ganglion Neurones. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2006, 142, 19–29. [Google Scholar] [CrossRef]
- Hamilton, B.R.; Chan, W.; Cheney, K.L.; Sullivan, R.K.P.; Floetenmeyer, M.; Garson, M.J.; Wepf, R. Cryo-Ultramicrotomy and Mass Spectrometry Imaging Analysis of Nudibranch Microstructures. J. Am. Soc. Mass Spectrom. 2022, 33, 592–597. [Google Scholar] [CrossRef]
- McPhail, K.; Davies-Coleman, M.T. New Spongiane Diterpenes from the East African Nudibranch Chromodoris Hamiltoni. Tetrahedron 1997, 53, 4655–4660. [Google Scholar] [CrossRef]
- Mebs, D. Chemical Defense of a Dorid Nudibranch, Glossodoris Quadricolor, from the Red Sea. J. Chem. Ecol. 1985, 11, 713–716. [Google Scholar] [CrossRef]
- Ilan, M. Reproductive Biology, Taxonomy, and Aspects of Chemical Ecology of Latrunculiidae (Porifera). Biol. Bull. 1995, 188, 306–312. [Google Scholar] [CrossRef]
- Guo, Y. Chemical Studies of the Novel Bioactive Secondary Metabolites from the Benthic Invertebrates: Isolation and Structure Characterization. Ph.D. Thesis, University of Naples, Naples, Italy, 1997. [Google Scholar]
- Amagata, T.; Johnson, T.A.; Cichewicz, R.H.; Tenney, K.; Mooberry, S.L.; Media, J.; Edelstein, M.; Valeriote, F.A.; Crews, P. Interrogating the Bioactive Pharmacophore of the Latrunculin Chemotype by Investigating the Metabolites of Two Taxonomically Unrelated Sponges. J. Med. Chem. 2008, 51, 7234–7242. [Google Scholar] [CrossRef] [Green Version]
- Sonnenschein, R.N.; Johnson, T.A.; Tenney, K.; Valeriote, F.A.; Crews, P. A Reassignment of (–)-Mycothiazole and the Isolation of a Related Diol. J. Nat. Prod. 2006, 69, 145–147. [Google Scholar] [CrossRef] [Green Version]
- Kashman, Y.; Groweiss, A.; Shmueli, U. Latrunculin, a New 2-Thiazolidinone Macrolide from the Marine Sponge Latrunculia Magnifica. Tetrahedron Lett. 1980, 21, 3629–3632. [Google Scholar] [CrossRef]
- Spector, I.; Shochet, N.R.; Kashman, Y.; Groweiss, A. Latrunculins: Novel Marine Toxins That Disrupt Microfilament Organization in Cultured Cells. Science 1983, 219, 493–495. [Google Scholar] [CrossRef]
- Smith, A.B.; Leahy, J.W.; Noda, I.; Remiszewski, S.W.; Liverton, N.J.; Zibuck, R. Total Synthesis of the Latrunculins. J. Am. Chem. Soc. 1992, 114, 2995–3007. [Google Scholar] [CrossRef]
- Oliveira, C.A.; Kashman, Y.; Mantovani, B. Effects of Latrunculin A on Immunological Phagocytosis and Macrophage Spreading-Associated Changes in the F-Actin/G-Actin Content of the Cells. Chem. Biol. Interact. 1996, 100, 141–153. [Google Scholar] [CrossRef]
- Tanaka, J.; Higa, T.; Bernardinelli, G.; Jefford, C.W. New Cytotoxic Macrolides from the Sponge Fasciospongia Rimosa. Chem. Lett. 1996, 25, 255–256. [Google Scholar] [CrossRef]
- Ayscough, K.R.; Stryker, J.; Pokala, N.; Sanders, M.; Crews, P.; Drubin, D.G. High Rates of Actin Filament Turnover in Budding Yeast and Roles for Actin in Establishment and Maintenance of Cell Polarity Revealed Using the Actin Inhibitor Latrunculin-A. J. Cell Biol. 1997, 137, 399–416. [Google Scholar] [CrossRef]
- Belmont, L.D.; Patterson, G.M.; Drubin, D.G. New Actin Mutants Allow Further Characterization of the Nucleotide Binding Cleft and Drug Binding Sites. J. Cell Sci. 1999, 112 Pt 9, 1325–1336. [Google Scholar] [CrossRef]
- Cai, S.; Liu, X.; Glasser, A.; Volberg, T.; Filla, M.; Geiger, B.; Polansky, J.R.; Kaufman, P.L. Effect of Latrunculin-A on Morphology and Actin-Associated Adhesions of Cultured Human Trabecular Meshwork Cells. Mol. Vis. 2000, 6, 132–143. [Google Scholar]
- Gillor, O.; Carmeli, S.; Rahamim, Y.; Fishelson, Z.; Ilan, M. Immunolocalization of the Toxin Latrunculin B within the Red Sea Sponge Negombata Magnifica (Demospongiae, Latrunculiidae). Mar. Biotechnol. 2000, 2, 213–223. [Google Scholar] [CrossRef]
- Morton, W.M.; Ayscough, K.R.; McLaughlin, P.J. Latrunculin Alters the Actin-Monomer Subunit Interface to Prevent Polymerization. Nat. Cell Biol. 2000, 2, 376–378. [Google Scholar] [CrossRef] [Green Version]
- Yarmola, E.G.; Somasundaram, T.; Boring, T.A.; Spector, I.; Bubb, M.R. Actin-Latrunculin A Structure and Function. Differential Modulation of Actin-Binding Protein Function by Latrunculin, A.J. Biol. Chem. 2000, 275, 28120–28127. [Google Scholar] [CrossRef] [Green Version]
- Baluška, F.; Jasik, J.; Edelmann, H.G.; Salajová, T.; Volkmann, D. Latrunculin B-Induced Plant Dwarfism: Plant Cell Elongation Is F-Actin-Dependent. Dev. Biol. 2001, 231, 113–124. [Google Scholar] [CrossRef] [Green Version]
- Wakatsuki, T.; Schwab, B.; Thompson, N.C.; Elson, E.L. Effects of Cytochalasin D and Latrunculin B on Mechanical Properties of Cells. J. Cell Sci. 2001, 114, 1025–1036. [Google Scholar] [CrossRef]
- Pring, M.; Cassimeris, L.; Zigmond, S.H. An Unexplained Sequestration of Latrunculin A Is Required in Neutrophils for Inhibition of Actin Polymerization. Cell Motil. Cytoskeleton 2002, 52, 122–130. [Google Scholar] [CrossRef]
- Fujita, M.; Ichinose, S.; Kiyono, T.; Tsurumi, T.; Omori, A. Establishment of Latrunculin-A Resistance in HeLa Cells by Expression of R183A D184A Mutant Beta-Actin. Oncogene 2003, 22, 627–631. [Google Scholar] [CrossRef] [Green Version]
- Vilozny, B.; Amagata, T.; Mooberry, S.L.; Crews, P. A New Dimension to the Biosynthetic Products Isolated from the Sponge Negombata Magnifica. J. Nat. Prod. 2004, 67, 1055–1057. [Google Scholar] [CrossRef]
- Ethier, C.R.; Read, A.T.; Chan, D.W.-H. Effects of Latrunculin-B on Outflow Facility and Trabecular Meshwork Structure in Human Eyes. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1991–1998. [Google Scholar] [CrossRef] [Green Version]
- El Sayed, K.A.; Youssef, D.T.A.; Marchetti, D. Bioactive Natural and Semisynthetic Latrunculins. J. Nat. Prod. 2006, 69, 219–223. [Google Scholar] [CrossRef]
- Khalifa, S.; Ahmed, S.; Mesbah, M.; Youssef, D.; Hamann, M. Quantitative Determination of Latrunculins A and B in the Red Sea Sponge Negombata Magnifica by High Performance Liquid Chromatography. J. Chromatogr. B 2006, 832, 47–51. [Google Scholar] [CrossRef]
- Sierra-Paredes, G.; Oreiro-García, T.; Núñez-Rodriguez, A.; Vázquez-López, A.; Sierra-Marcuño, G. Seizures Induced by in Vivo Latrunculin A and Jasplakinolide Microperfusion in the Rat Hippocampus. J. Mol. Neurosci. 2006, 28, 151–160. [Google Scholar] [CrossRef]
- Foissner, I.; Wasteneys, G.O. Wide-Ranging Effects of Eight Cytochalasins and Latrunculin A and B on Intracellular Motility and Actin Filament Reorganization in Characean Internodal Cells. Plant Cell Physiol. 2007, 48, 585–597. [Google Scholar] [CrossRef]
- Fürstner, A.; Kirk, D.; Fenster, M.D.B.; Aïssa, C.; De Souza, D.; Nevado, C.; Tuttle, T.; Thiel, W.; Müller, O. Latrunculin Analogues with Improved Biological Profiles by “Diverted Total Synthesis”: Preparation, Evaluation, and Computational Analysis. Chem.—Eur. J. 2007, 13, 135–149. [Google Scholar] [CrossRef] [Green Version]
- Meadows, J.C.; Millar, J.; Solomon, M. Latrunculin A Delays Anaphase Onset in Fission Yeast by Disrupting an Ase1-Independent Pathway Controlling Mitotic Spindle Stability. Mol. Biol. Cell 2008, 19, 3713–3723. [Google Scholar] [CrossRef] [Green Version]
- Konishi, H.; Kikuchi, S.; Ochiai, T.; Ikoma, H.; Kubota, T.; Ichikawa, D.; Fujiwara, H.; Okamoto, K.; Sakakura, C.; Sonoyama, T.; et al. Latrunculin A Has a Strong Anticancer Effect in a Peritoneal Dissemination Model of Human Gastric Cancer in Mice. Anticancer Res. 2009, 29, 2091–2097. [Google Scholar]
- Kudrimoti, S.; Ahmed, S.A.; Daga, P.R.; Wahba, A.E.; Khalifa, S.I.; Doerksen, R.J.; Hamann, M.T. Semisynthetic Latrunculin B Analogs: Studies of Actin Docking Support a Proposed Mechanism for Latrunculin Bioactivity. Bioorg. Med. Chem. 2009, 17, 7517–7522. [Google Scholar] [CrossRef] [Green Version]
- Ketelaar, T.; Meijer, H.J.G.; Spiekerman, M.; Weide, R.; Govers, F. Effects of Latrunculin B on the Actin Cytoskeleton and Hyphal Growth in Phytophthora Infestans. Fungal Genet. Biol. 2012, 49, 1014–1022. [Google Scholar] [CrossRef]
- Saito, S. Toxins Affecting Actin Filaments and Microtubules. In Marine Toxins as Research Tools; Fusetani, N., Kem, W., Eds.; Progress in Molecular and Subcellular Biology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 46, pp. 187–219. ISBN 978-3-540-87892-6. [Google Scholar]
- Moscatelli, A.; Idilli, A.I.; Rodighiero, S.; Caccianiga, M. Inhibition of Actin Polymerisation by Low Concentration Latrunculin B Affects Endocytosis and Alters Exocytosis in Shank and Tip of Tobacco Pollen Tubes. Plant Biol. 2012, 14, 770–782. [Google Scholar] [CrossRef]
- Terashita, Y.; Yamagata, K.; Tokoro, M.; Itoi, F.; Wakayama, S.; Li, C.; Sato, E.; Tanemura, K.; Wakayama, T. Latrunculin A Treatment Prevents Abnormal Chromosome Segregation for Successful Development of Cloned Embryos. PLoS ONE 2013, 8, e78380. [Google Scholar] [CrossRef] [Green Version]
- Kopecká, M.; Yamaguchi, M.; Kawamoto, S. Effects of the F-Actin Inhibitor Latrunculin A on the Budding Yeast Saccharomyces Cerevisiae. Microbiol. Read. Engl. 2015, 161, 1348–1355. [Google Scholar] [CrossRef] [Green Version]
- Ebrahim, H.Y.; El Sayed, K.A. Discovery of Novel Antiangiogenic Marine Natural Product Scaffolds. Mar. Drugs 2016, 14, 57. [Google Scholar] [CrossRef] [Green Version]
- Varghese, S.; Rahmani, R.; Drew, D.; Williams, M.; Huang, J.; Wilkinson, M.; Tan, Y.-H.; Tonkin, C.; Beeson, J.; Baum, J.; et al. Truncated Latrunculins as Actin Inhibitors Targeting Plasmodium Falciparum Motility and Host-Cell Invasion. J. Med. Chem. 2016, 59, 10994–11005. [Google Scholar] [CrossRef]
- da Silva, S.C.G.P. Multi-Approach Analysis of the Metagenome of a Marine Sponge Containing Latrunculin A. Master’s Thesis, Ciências Farmacêuticas, Universidade de Lisboa, Faculdade de Farmácia, Lisbon, Portugal, 2017. [Google Scholar]
- Fujiwara, I.; Zweifel, M.E.; Courtemanche, N.; Pollard, T.D. Latrunculin A Accelerates Actin Filament Depolymerization in Addition to Sequestering Actin Monomers. Curr. Biol. CB 2018, 28, 3183–3192. [Google Scholar] [CrossRef] [Green Version]
- Würtemberger, J.; Tchessalova, D.; Regina, C.; Bauer, C.; Schneider, M.; Wagers, A.J.; Hettmer, S. Growth Inhibition Associated with Disruption of the Actin Cytoskeleton by Latrunculin A in Rhabdomyosarcoma Cells. PLoS ONE 2020, 15, e0238572. [Google Scholar] [CrossRef]
- Al-Tarabeen, M.; El-Neketi, M.; Albohy, A.; Müller, W.E.G.; Rasheed, M.; Ebrahim, W.; Proksch, P.; Ebada, S.S. Isolation and Molecular Docking of Cytotoxic Secondary Metabolites from Two Red Sea Sponges of the Genus Diacarnus. ChemistrySelect 2021, 6, 217–220. [Google Scholar] [CrossRef]
- Lenz, K.D.; Klosterman, K.E.; Mukundan, H.; Kubicek-Sutherland, J.Z. Macrolides: From Toxins to Therapeutics. Toxins 2021, 13, 347. [Google Scholar] [CrossRef]
- Varghese, S.; Rahmani, R.; Drew, D.R.; Beeson, J.G.; Baum, J.; Smith, B.J.; Baell, J.B. Structure-Activity Studies of Truncated Latrunculin Analogues with Antimalarial Activity. ChemMedChem 2021, 16, 679–693. [Google Scholar] [CrossRef]
- Risinger, A.L.; Du, L. Targeting and Extending the Eukaryotic Druggable Genome with Natural Products: Cytoskeletal Targets of Natural Products. Nat. Prod. Rep. 2019, 634–652. [Google Scholar] [CrossRef]
- Rubenstein, P.A. The Functional Importance of Multiple Actin Isoforms. BioEssays 1990, 12, 309–315. [Google Scholar] [CrossRef]
- Doolittle, R.F.; Gerhart, J.; Hunt, R.T.; Kirschner, M.W.; Wolpert, L. The Origins and Evolution of Eukaryotic Proteins. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1995, 349, 235–240. [Google Scholar] [CrossRef]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Mol. Cell Biol., 4th ed.; W. H. Freeman: New York, NY, USA, 2000; ISBN 978-0-7167-3136-8. [Google Scholar]
- Chhabra, E.S.; Higgs, H.N. The Many Faces of Actin: Matching Assembly Factors with Cellular Structures. Nat. Cell Biol. 2007, 9, 1110–1121. [Google Scholar] [CrossRef]
- Perrin, B.J.; Ervasti, J.M. The Actin Gene Family: Function Follows Isoform. Cytoskelet. Hoboken Nj 2010, 67, 630–634. [Google Scholar] [CrossRef]
- Dominguez, R.; Holmes, K.C. Actin Structure and Function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef] [Green Version]
- Gunning, P.W.; Ghoshdastider, U.; Whitaker, S.; Popp, D.; Robinson, R.C. The Evolution of Compositionally and Functionally Distinct Actin Filaments. J. Cell Sci. 2015, 128, 2009–2019. [Google Scholar] [CrossRef] [Green Version]
- Skau, C.T.; Waterman, C.M. Specification of Architecture and Function of Actin Structures by Actin Nucleation Factors. Annu. Rev. Biophys. 2015, 44, 285–310. [Google Scholar] [CrossRef]
- Pollard, T.D. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef] [Green Version]
- Parisis, N.; Krasinska, L.; Harker, B.; Urbach, S.; Rossignol, M.; Camasses, A.; Dewar, J.; Morin, N.; Fisher, D. Initiation of DNA Replication Requires Actin Dynamics and Formin Activity. EMBO J. 2017, 36, 3212–3231. [Google Scholar] [CrossRef]
- Pollard, T.D. What We Know and Do Not Know About Actin. Handb. Exp. Pharmacol. 2017, 235, 331–347. [Google Scholar] [CrossRef]
- Stoddard, P.R.; Williams, T.A.; Garner, E.; Baum, B. Evolution of Polymer Formation within the Actin Superfamily. Mol. Biol. Cell 2017, 28, 2461–2469. [Google Scholar] [CrossRef]
- Vedula, P.; Kashina, A. The Makings of the ‘actin Code’: Regulation of Actin’s Biological Function at the Amino Acid and Nucleotide Level. J. Cell Sci. 2018, 131, jcs215509. [Google Scholar] [CrossRef] [Green Version]
- Wada, S.; Matsunaga, S.; Saito, S.; Fusetani, N.; Watabe, S. Actin-Binding Specificity of Marine Macrolide Toxins, Mycalolide B and Kabiramide D. J. Biochem. 1998, 123, 946–952. [Google Scholar] [CrossRef]
- Belmont, L.D.; Drubin, D.G. The Yeast V159N Actin Mutant Reveals Roles for Actin Dynamics In Vivo. J. Cell Biol. 1998, 142, 1289–1299. [Google Scholar] [CrossRef] [Green Version]
- Procaccio, V.; Salazar, G.; Ono, S.; Styers, M.L.; Gearing, M.; Davila, A.; Jimenez, R.; Juncos, J.; Gutekunst, C.-A.; Meroni, G.; et al. A Mutation of β-Actin That Alters Depolymerization Dynamics Is Associated with Autosomal Dominant Developmental Malformations, Deafness, and Dystonia. Am. J. Hum. Genet. 2006, 78, 947–960. [Google Scholar] [CrossRef] [Green Version]
- Schwarzerová, K.; Vondráková, Z.; Fischer, L.; Boříková, P.; Bellinvia, E.; Eliášová, K.; Havelková, L.; Fišerová, J.; Vágner, M.; Opatrný, Z. The Role of Actin Isoforms in Somatic Embryogenesis in Norway Spruce. BMC Plant Biol. 2010, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Rivière, J.-B.; van Bon, B.W.M.; Hoischen, A.; Kholmanskikh, S.S.; O’Roak, B.J.; Gilissen, C.; Gijsen, S.; Sullivan, C.T.; Christian, S.L.; Abdul-Rahman, O.A.; et al. De Novo Mutations in the Actin Genes ACTB and ACTG1 Cause Baraitser-Winter Syndrome. Nat. Genet. 2012, 44, 440-S2. [Google Scholar] [CrossRef] [Green Version]
- Johnston, J.J.; Wen, K.-K.; Keppler-Noreuil, K.; McKane, M.; Maiers, J.L.; Greiner, A.; Sapp, J.C.; NIH Intramural Sequencing Center; Demali, K.A.; Rubenstein, P.A.; et al. Functional Analysis of a de Novo ACTB Mutation in a Patient with Atypical Baraitser-Winter Syndrome. Hum. Mutat. 2013, 34, 1242–1249. [Google Scholar] [CrossRef] [Green Version]
- Roopa, L.; Pravin, K.R.; Sudheer Mohammed, M.M. Molecular Dynamics Simulation Reveal the Mechanism of Resistance of Mutant Actins to Latrunculin A—Insight into Specific Modifications to Design Novel Drugs to Overcome Resistance. Curr. Comput. Aided Drug Des. 2016, 12, 107–118. [Google Scholar]
- Filipuzzi, I.; Thomas, J.R.; Pries, V.; Estoppey, D.; Salcius, M.; Studer, C.; Schirle, M.; Hoepfner, D. Direct Interaction of Chivosazole F with Actin Elicits Cell Responses Similar to Latrunculin A but Distinct from Chondramide. ACS Chem. Biol. 2017, 12, 2264–2269. [Google Scholar] [CrossRef]
- Singh, H.P.; Batish, D.R.; Kohli, R.K. Autotoxicity: Concept, Organisms, and Ecological Significance. Crit. Rev. Plant Sci. 1999, 18, 757–772. [Google Scholar] [CrossRef]
- Arbuckle, K.; Rodríguez de la Vega, R.C.; Casewell, N.R. Coevolution Takes the Sting out of It: Evolutionary Biology and Mechanisms of Toxin Resistance in Animals. Toxicon 2017, 140, 118–131. [Google Scholar] [CrossRef]
- Almabruk, K.H.; Dinh, L.K.; Philmus, B. Self-Resistance of Natural Product Producers: Past, Present, and Future Focusing on Self-Resistant Protein Variants. ACS Chem. Biol. 2018, 13, 1426–1437. [Google Scholar] [CrossRef]
- Ogawara, H. Comparison of Strategies to Overcome Drug Resistance: Learning from Various Kingdoms. Mol. J. Synth. Chem. Nat. Prod. Chem. 2018, 23, 1476. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Liu, N.; Tang, Y. Recent Developments in Self-Resistance Gene Directed Natural Product Discovery. Nat. Prod. Rep. 2020, 37, 879–892. [Google Scholar] [CrossRef]
- Crozier, W.J.; Crozier, W.J. The Nature of the Conical Bodies on the Mantle of Certain Nudibranchs. Nautilus 1917, 30, 103–106. [Google Scholar]
- Groweiss, A.; Shmueli, U.; Kashman, Y. Marine Toxins of Latrunculia Magnifica. J. Org. Chem. 1983, 48, 3512–3516. [Google Scholar] [CrossRef]
- Garson, M.J.; Thompson, J.E.; Larsen, R.M.; Battershill, C.N.; Murphy, P.T.; Bergquist, P.R. Terpenes in Sponge Cell Membranes: Cell Separation and Membrane Fractionation Studies with the Tropical Marine SpongeAmphimedon Sp. Lipids 1992, 27, 378–388. [Google Scholar] [CrossRef]
- Uriz, M.J.; Becerro, M.A.; Tur, J.M.; Turon, X. Location of Toxicity within the Mediterranean Sponge Crambe Crambe (Demospongiae: Poecilosclerida). Mar. Biol. 1996, 124, 583–590. [Google Scholar] [CrossRef]
- Uriz, M.J.; Turon, X.; Galera, J.; Tur, J.M. New Light on the Cell Location of Avarol within the Sponge Dysidea Avara (Dendroceratida). Cell Tissue Res. 1996, 285, 519–527. [Google Scholar] [CrossRef]
- Garson, M.J.; Flowers, A.E.; Webb, R.I.; Charan, R.D.; McCaffrey, E.J. A Sponge/Dinoflagellate Association in the Haplosclerid Sponge Haliclona Sp.: Cellular Origin of Cytotoxic Alkaloids by Percoll Density Gradient Fractionation. Cell Tissue Res. 1998, 293, 365–373. [Google Scholar] [CrossRef]
- Turon, X.; Becerro, M.A.; Uriz, M.J. Distribution of Brominated Compounds within the Sponge Aplysina Aerophoba: Coupling of X-Ray Microanalysis with Cryofixation Techniques. Cell Tissue Res. 2000, 301, 311–322. [Google Scholar] [CrossRef]
- Gerçe, B.; Schwartz, T.; Voigt, M.; Rühle, S.; Kirchen, S.; Putz, A.; Proksch, P.; Obst, U.; Syldatk, C.; Hausmann, R. Morphological, Bacterial, and Secondary Metabolite Changes of Aplysina Aerophoba upon Long-Term Maintenance Under Artificial Conditions. Microb. Ecol. 2009, 58, 865–878. [Google Scholar] [CrossRef]
- Tianero, M.D.; Balaich, J.N.; Donia, M.S. Localized Production of Defence Chemicals by Intracellular Symbionts of Haliclona Sponges. Nat. Microbiol. 2019, 4, 1149–1159. [Google Scholar] [CrossRef]
- Avila, C.; Cimino, G.; Fontana, A.; Gavagnin, M.; Ortea, J.; Trivellone, E. Defensive Strategy of Two Hypselodoris Nudibranchs from Italian and Spanish Coasts. J. Chem. Ecol. 1991, 17, 625–636. [Google Scholar] [CrossRef]
- García-Gómez, J.C.; Cimino, G.; Medina, A. Studies on the Defensive Behaviour of Hypselodoris Species (Gastropoda: Nudibranchia): Ultrastructure and Chemical Analysis of Mantle Dermal Formations (MDFs). Mar. Biol. 1990, 106, 245–250. [Google Scholar] [CrossRef]
- Fontana, A.; Giménez, F.; Marin, A.; Mollo, E.; Cimino, G. Transfer of Secondary Metabolites from the Sponges Dysidea Fragilis and Pleraplysilla Spinifera to the Mantle Dermal Formations (MDFs) of the Mudibranch Hypserlodoris Webbi. Experientia 1994, 50, 510–516. [Google Scholar] [CrossRef]
- Avila, C.; Paul, V.J. Chemical Ecology of the Nudibranch Glossodoris Pallida: Is the Location of Diet-Derived Metabolites Important for Defense? Mar. Ecol. Prog. Ser. 1997, 150, 171–180. [Google Scholar] [CrossRef]
- Wägele, H.; Willan, R.C. Phylogeny of the Nudibranchia. Zool. J. Linn. Soc. 2000, 130, 83–181. [Google Scholar] [CrossRef]
- Winters, A.E.; White, A.M.; Dewi, A.S.; Mudianta, I.W.; Wilson, N.G.; Forster, L.C.; Garson, M.J.; Cheney, K.L. Distribution of Defensive Metabolites in Nudibranch Molluscs. J. Chem. Ecol. 2018, 44, 384–396. [Google Scholar] [CrossRef]
- Carbone, M.; Gavagnin, M.; Haber, M.; Guo, Y.-W.; Fontana, A.; Manzo, E.; Genta-Jouve, G.; Tsoukatou, M.; Rudman, W.B.; Cimino, G.; et al. Packaging and Delivery of Chemical Weapons: A Defensive Trojan Horse Stratagem in Chromodorid Nudibranchs. PLoS ONE 2013, 8, e62075. [Google Scholar] [CrossRef] [Green Version]
- Wägele, H. Potential Key Characters in Opisthobranchia (Gastropoda, Mollusca) Enhancing Adaptive Radiation. Org. Divers. Evol. 2004, 4, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Winters, A.E.; Green, N.F.; Wilson, N.G.; How, M.J.; Garson, M.J.; Marshall, N.J.; Cheney, K.L. Stabilizing Selection on Individual Pattern Elements of Aposematic Signals. Proc. R Soc. B 2017, 284, 20170926. [Google Scholar] [CrossRef] [Green Version]
- Wägele, H.; Ballesteros, M.; Avila, C. Defensive Glandular Structures In Opisthobranch Molluscs—From Histology To Ecology. Oceanogr. Mar. Biol. Annu. Rev. 2006, 44, 197–276. [Google Scholar] [CrossRef]
- Eisenbarth, J.-H.; Undap, N.; Papu, A.; Schillo, D.; Dialao, J.; Reumschüssel, S.; Kaligis, F.; Bara, R.; Schäberle, T.F.; König, G.M.; et al. Marine Heterobranchia (Gastropoda, Mollusca) in Bunaken National Park, North Sulawesi, Indonesia—A Follow-Up Diversity Study. Diversity 2018, 10, 127. [Google Scholar] [CrossRef] [Green Version]
- Kaligis, F.; Eisenbarth, J.-H.; Schillo, D.; Dialao, J.; Schäberle, T.F.; Böhringer, N.; Bara, R.; Reumschüssel, S.; König, G.M.; Wägele, H. Second Survey of Heterobranch Sea Slugs (Mollusca, Gastropoda, Heterobranchia) from Bunaken National Park, North Sulawesi, Indonesia—How Much Do We Know after 12 Years? Mar. Biodivers. Rec. 2018, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- Ackers, R.G.; Moss, D.; Picton, B.E.; Bt, B.; Stone, S.M.K.; Morrow, C.C. Sponges of the British Isles (“Sponge V”) —A Colour Guide and Working Document, 1992 Edition. Mar. Conserv. Soc. 2007, 165. [Google Scholar]
- Erpenbeck, D.; Galitz, A.; Ekins, M.; Cook, S.D.C.; van Soest, R.W.M.; Hooper, J.N.A.; Wörheide, G. Soft Sponges with Tricky Tree: On the Phylogeny of Dictyoceratid Sponges. J. Zool. Syst. Evol. Res. 2020, 58, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Bouschen, W.; Schulz, O.; Eikel, D.; Spengler, B. Matrix Vapor Deposition/Recrystallization and Dedicated Spray Preparation for High-Resolution Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry (SMALDI-MS) of Tissue and Single Cells. Rapid Commun. Mass Spectrom. 2010, 24, 355–364. [Google Scholar] [CrossRef]
- Koestler, M.; Kirsch, D.; Hester, A.; Leisner, A.; Guenther, S.; Spengler, B. A High-Resolution Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Ion Source for Imaging Analysis on an Ion Trap/Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Rapid Commun. Mass Spectrom. RCM 2008, 22, 3275–3285. [Google Scholar] [CrossRef]
- Guenther, S.; Koestler, M.; Schulz, O.; Spengler, B. Laser Spot Size and Laser Power Dependence of Ion Formation in High Resolution MALDI Imaging. Int. J. Mass Spectrom. 2010, 294, 7–15. [Google Scholar] [CrossRef]
- Römpp, A.; Guenther, S.; Schober, Y.; Schulz, O.; Takats, Z.; Kummer, W.; Spengler, B. Histology by Mass Spectrometry: Label-Free Tissue Characterization Obtained from High-Accuracy Bioanalytical Imaging. Angew. Chem. Int. Ed. 2010, 49, 3834–3838. [Google Scholar] [CrossRef]
- Römpp, A.; Spengler, B. Mass Spectrometry Imaging with High Resolution in Mass and Space. Histochem. Cell Biol. 2013, 139, 759–783. [Google Scholar] [CrossRef] [Green Version]
- Paschke, C.; Leisner, A.; Hester, A.; Maass, K.; Guenther, S.; Bouschen, W.; Spengler, B. Mirion—A Software Package for Automatic Processing of Mass Spectrometric Images. J. Am. Soc. Mass Spectrom. 2013, 24, 1296–1306. [Google Scholar] [CrossRef]
- Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [CrossRef] [Green Version]
- Karmeinski, D.; Meusemann, K.; Goodheart, J.A.; Schroedl, M.; Martynov, A.; Korshunova, T.; Wägele, H.; Donath, A. Transcriptomics Provides a Robust Framework for the Relationships of the Major Clades of Cladobranch Sea Slugs (Mollusca, Gastropoda, Heterobranchia), but Fails to Resolve the Position of the Enigmatic Genus Embletonia. BMC Ecology and Evolution 2021, 21, 226. [Google Scholar] [CrossRef]
- DesGroseillers, L.; Auclair, D.; Wickham, L.; Maalouf, M. A Novel Actin CDNA Is Expressed in the Neurons of Aplysia Californica. Biochim. Biophys. Acta 1994, 1217, 322–324. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- White, J.D.; Kawasaki, M. Total Synthesis of (+)-Latrunculin A, an Ichthyotoxic Metabolite of the Sponge Latrunculia Magnifica and Its C-15 Epimer. J. Org. Chem. 1992, 57, 5292–5300. [Google Scholar] [CrossRef]
- Fürstner, A.; De Souza, D.; Turet, L.; Fenster, M.D.B.; Parra-Rapado, L.; Wirtz, C.; Mynott, R.; Lehmann, C.W. Total Syntheses of the Actin-Binding Macrolides Latrunculin A, B, C, M, S and 16-Epi-Latrunculin B. Chem. Weinh. Bergstr. Ger. 2007, 13, 115–134. [Google Scholar] [CrossRef]
- Rudman, W.B.; Bergquist, P.R. A Review of Feeding Specificity in the Sponge-Feeding Chromodorididae (Nudibranchia: Mollusca). Molluscan Res. 2007, 27, 60–88. [Google Scholar]
- Putz, A.; König, G.M.; Wägele, H. Defensive Strategies of Cladobranchia (Gastropoda, Opisthobranchia). Nat. Prod. Rep. 2010, 27, 1386–1402. [Google Scholar] [CrossRef]
- Wertman, K.F.; Drubin, D.G.; Botstein, D. Systematic Mutational Analysis of the Yeast ACT1 Gene. Genetics 1992, 132, 337–350. [Google Scholar] [CrossRef]
- Rogers, S.D.; Paul, V.J. Chemical Defenses of Three Glossodoris Nudibranchs and Their Dietary Hyrtios Sponges. Mar. Ecol. Prog. Ser. 1991, 77, 221–232. [Google Scholar] [CrossRef]
- Cimino, G.; Fontana, A.; Giménez, F.; Marin, A.; Mollo, E.; Trivellone, E.; Zubía, E. Biotransformation of a Dietary Sesterterpenoid in the Mediterranean Nudibranch Hypselodoris Orsini. Experientia 1993, 49, 582–586. [Google Scholar] [CrossRef]
- Fontana, A.; Ciavatta, M.L.; DeSouza, L.; Mollo, E.; Naik, C.G.; Parameswaran, P.S.; Wahidullah, S.; Cimino, G. Selected Chemo-Ecological Studies of Marine Opisthobranchs from Indian Coasts. J. Indian Inst. Sci 2001, 81, 403–415. [Google Scholar]
- Gavagnin, M.; Mollo, E.; Docimo, T.; Guo, Y.-W.; Cimino, G. Scalarane Metabolites of the Nudibranch Glossodoris Rufomarginata and Its Dietary Sponge from the South China Sea. J. Nat. Prod. 2004, 67, 2104–2107. [Google Scholar] [CrossRef]
- Wahidullah, S.; Guo, Y.-W.; Fakhr, I.M.I.; Mollo, E. Chemical Diversity in Opisthobranch Molluscs from Scarcely Investigated Indo-Pacific Areas. In Molluscs: From Chemo-ecological Study to Biotechnological Application; Cimino, G., Gavagnin, M., Eds.; Progress in Molecular and Subcellular Biology; Springer: Berlin/Heidelberg, Germany, 2006; pp. 175–198. ISBN 978-3-540-30880-5. [Google Scholar]
- Manzo, E.; Gavagnin, M.; Somerville, M.J.; Mao, S.-C.; Ciavatta, M.L.; Mollo, E.; Schupp, P.J.; Garson, M.J.; Guo, Y.-W.; Cimino, G. Chemistry of Glossodoris Nudibranchs: Specific Occurrence of 12-Keto Scalaranes. J. Chem. Ecol. 2007, 33, 2325–2336. [Google Scholar] [CrossRef]
- Avila, C.; Durfort, M. Histology of Epithelia and Mantle Glands of Selected Species of Doridacean Molluscs with Chemical Defensive Strategies. Veliger-Berkeley- 1996, 39, 148–163. [Google Scholar]
- Fontana, A.; Mollo, E.; Ricciardi, D.; Fakhr, I.; Cimino, G. Chemical Studies of Egyptian Opisthobranchs: Spongian Diterpenoids from Glossodoris Atromarginata. J. Nat. Prod. 1997, 60, 444–448. [Google Scholar] [CrossRef]
- Haber, M.; Cerfeda, S.; Carbone, M.; Calado, G.; Gaspar, H.; Neves, R.; Maharajan, V.; Cimino, G.; Gavagnin, M.; Ghiselin, M.T.; et al. Coloration and Defense in the Nudibranch Gastropod Hypselodoris Fontandraui. Biol. Bull. 2010, 218, 181–188. [Google Scholar] [CrossRef]
- Sotka, E.E.; Forbey, J.; Horn, M.; Poore, A.G.B.; Raubenheimer, D.; Whalen, K.E. The Emerging Role of Pharmacology in Understanding Consumer–Prey Interactions in Marine and Freshwater Systems. Integr. Comp. Biol. 2009, 49, 291–313. [Google Scholar] [CrossRef] [Green Version]
- Davies, M.S.; Blackwell, J. Energy Saving through Trail Following in a Marine Snail. Proc. R. Soc. Lond. B Biol. Sci. 2007, 274, 1233–1236. [Google Scholar] [CrossRef] [Green Version]
- Winters, A.E.; White, A.M.; Cheney, K.L.; Garson, M.J. Geographic Variation in Diterpene-Based Secondary Metabolites and Level of Defence in an Aposematic Nudibranch, Goniobranchus Splendidus. J. Molluscan Stud. 2019, 85, 133–142. [Google Scholar] [CrossRef]
- Skruber, K.; Read, T.-A.; Vitriol, E.A. Reconsidering an Active Role for G-Actin in Cytoskeletal Regulation. J. Cell Sci. 2018, 131, 931–933. [Google Scholar] [CrossRef] [Green Version]
- Akıl, C.; Tran, L.T.; Orhant-Prioux, M.; Baskaran, Y.; Manser, E.; Blanchoin, L.; Robinson, R.C. Insights into the Evolution of Regulated Actin Dynamics via Characterization of Primitive Gelsolin/Cofilin Proteins from Asgard Archaea. Proc. Natl. Acad. Sci. USA 2020, 117, 19904–19913. [Google Scholar] [CrossRef]
- Boiero Sanders, M.; Antkowiak, A.; Michelot, A. Diversity from Similarity: Cellular Strategies for Assigning Particular Identities to Actin Filaments and Networks. Open Biol. 2020, 10, 200157. [Google Scholar] [CrossRef]
- Akıl, C.; Kitaoku, Y.; Tran, L.T.; Liebl, D.; Choe, H.; Muengsaen, D.; Suginta, W.; Schulte, A.; Robinson, R.C. Mythical Origins of the Actin Cytoskeleton. Curr. Opin. Cell Biol. 2021, 68, 55–63. [Google Scholar] [CrossRef]
- Vedula, P.; Kurosaka, S.; MacTaggart, B.; Ni, Q.; Papoian, G.; Jiang, Y.; Dong, D.W.; Kashina, A. Different Translation Dynamics of β- and γ-Actin Regulates Cell Migration. eLife 2021, 10, e68712. [Google Scholar] [CrossRef]
- Nèeman, I.; Fishelson, L.; Kashman, Y. Isolation of a New Toxin from the Sponge Latrunculia Magnifica in the Gulf of Aquaba (Red Sea). Mar. Biol. 1975, 30, 293–296. [Google Scholar] [CrossRef]
- Edmunds, M. Protective Mechanisms in the Eolidacea (Mollusca Nudibranchia). Zool. J. Linn. Soc. 1966, 46, 27–71. [Google Scholar] [CrossRef]
- Edmunds, M. On the Swimming and Defensive Response of Hexabranchus Marginatus (Mollusca, Nudibranchia). Zool. J. Linn. Soc. 1968, 47, 425–429. [Google Scholar] [CrossRef]
- Ghazali, S.R. Displays of Defense: Behavioral Differences in Antagonist Avoidance in Four Opisthobranch Mollusks. UC Berkeley: UCB Moorea Class: Biology and Geomorphology of Tropical Islands. 2006. Available online: https://escholarship.org/uc/item/9s6740fr (accessed on 30 November 2022).
- Kistler, H.C.; Broz, K. Cellular Compartmentalization of Secondary Metabolism. Front. Microbiol. 2015, 6, 68. [Google Scholar] [CrossRef] [Green Version]
- Blanco, J. Accumulation of Dinophysis Toxins in Bivalve Molluscs. Toxins 2018, 10, 453. [Google Scholar] [CrossRef] [Green Version]
- Johannes, F.J.; Gallwitz, D. Site-Directed Mutagenesis of the Yeast Actin Gene: A Test for Actin Function in Vivo. EMBO J. 1991, 10, 3951–3958. [Google Scholar] [CrossRef]
- Ujvari, B.; Casewell, N.R.; Sunagar, K.; Arbuckle, K.; Wüster, W.; Lo, N.; O’Meally, D.; Beckmann, C.; King, G.F.; Deplazes, E.; et al. Widespread Convergence in Toxin Resistance by Predictable Molecular Evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 11911–11916. [Google Scholar] [CrossRef] [Green Version]
- Dalla, S.; Dobler, S. Gene Duplications Circumvent Trade-Offs in Enzyme Function: Insect Adaptation to Toxic Host Plants. Evolution 2016, 70, 2767–2777. [Google Scholar] [CrossRef]
- Karageorgi, M.; Groen, S.C.; Sumbul, F.; Pelaez, J.N.; Verster, K.I.; Aguilar, J.M.; Hastings, A.P.; Bernstein, S.L.; Matsunaga, T.; Astourian, M.; et al. Genome Editing Retraces the Evolution of Toxin Resistance in the Monarch Butterfly. Nature 2019, 574, 409–412. [Google Scholar] [CrossRef]
- DesGroseillers, L.; Auclair, D.; Wickham, L. Nucleotide Sequence of an Actin CDNA Gene from Aplysia Californica. Nucleic Acids Res. 1990, 18, 3654. [Google Scholar] [CrossRef] [Green Version]
- Zappulla, J.P.; Angers, A.; Barbas, D.; Castellucci, V.F.; DesGroseillers, L. A Novel Actin Isoform Is Expressed in the Ovotestis of Aplysia Californica. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 140, 403–409. [Google Scholar] [CrossRef]
- Bryant, M.J.; Flint, H.J.; Sin, F.Y.T. Isolation, Characterization, and Expression Analysis of Three Actin Genes in the New Zealand Black-Footed Abalone, Haliotis Iris. Mar. Biotechnol. N. Y. N 2006, 8, 110–119. [Google Scholar] [CrossRef]
- Sin, F.Y.T.; Bryant, M.J.; Johnstone, A. Molecular Evolution and Phylogeny of Actin Genes in Haliotis Species (Mollusca: Gastropoda). Zool. Stud. 2007, 46, 734. [Google Scholar]
- Ivanov, M.; Todorovska, E.; Radkova, M.; Georgiev, O.; Dolashki, A.; Dolashka, P. Molecular Cloning, Characterization and Phylogenetic Analysis of an Actin Gene from the Marine Mollusk Rapana Venosa (Class Gastropoda). Int. J. Curr. Microbiol. App. Sci. 2015, 4, 687–700. [Google Scholar]
- Adema, C.M.; Hillier, L.W.; Jones, C.S.; Loker, E.S.; Knight, M.; Minx, P.; Oliveira, G.; Raghavan, N.; Shedlock, A.; do Amaral, L.R.; et al. Whole Genome Analysis of a Schistosomiasis-Transmitting Freshwater Snail. Nat. Commun. 2017, 8, 15451. [Google Scholar] [CrossRef] [Green Version]
- Matsunaga, S.; Fusetani, N.; Hashimoto, K.; Koseki, K.; Noma, M.; Noguchi, H.; Sankawa, U. Bioactive Marine Metabolites. 25. Further Kabiramides and Halichondramides, Cytotoxic Macrolides Embracing Trisoxazole, from the Hexabranchus Egg Masses. J. Org. Chem. 1989, 54, 1360–1363. [Google Scholar] [CrossRef]
- Klenchin, V.A.; Allingham, J.S.; King, R.; Tanaka, J.; Marriott, G.; Rayment, I. Trisoxazole Macrolide Toxins Mimic the Binding of Actin-Capping Proteins to Actin. Nat. Struct. Biol. 2003, 10, 1058–1063. [Google Scholar] [CrossRef]
- Tanaka, J.; Yan, Y.; Choi, J.; Bai, J.; Klenchin, V.A.; Rayment, I.; Marriott, G. Biomolecular Mimicry in the Actin Cytoskeleton: Mechanisms Underlying the Cytotoxicity of Kabiramide C and Related Macrolides. Proc. Natl. Acad. Sci. USA 2003, 100, 13851–13856. [Google Scholar] [CrossRef] [Green Version]
- Parrish, S.M.; Yoshida, W.; Yang, B.; Williams, P.G. Ulapualides C–E Isolated from a Hawaiian Hexabranchus Sanguineus Egg Mass. J. Nat. Prod. 2017, 80, 726–730. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Ojika, M.; Ishigaki, T.; Yoshida, Y.; Ekimoto, H.; Arakawa, M. Aplyronine A, a Potent Antitumor Substance and the Congeners Aplyronines B and C Isolated from the Sea Hare Aplysia Kurodai. J. Am. Chem. Soc. 1993, 115, 11020–11021. [Google Scholar] [CrossRef]
- Saito, S.; Watabe, S.; Ozaki, H.; Kigoshi, H.; Yamada, K.; Fusetani, N.; Karaki, H. Novel Actin Depolymerizing Macrolide Aplyronine A1. J. Biochem. 1996, 120, 552–555. [Google Scholar] [CrossRef]
- Ojika, M.; Kigoshi, H.; Yoshida, Y.; Ishigaki, T.; Nisiwaki, M.; Tsukada, I.; Arakawa, M.; Ekimoto, H.; Yamada, K. Aplyronine A, a Potent Antitumor Macrolide of Marine Origin, and the Congeners Aplyronines B and C: Isolation, Structures, and Bioactivities. Tetrahedron 2007, 63, 3138–3167. [Google Scholar] [CrossRef]
- Kigoshi, H.; Kita, M. Antitumor Effects of Sea Hare-Derived Compounds in Cancer. In Handbook of Anticancer Drugs from Marine Origin; Kim, S.-K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 701–739. ISBN 978-3-319-07145-9. [Google Scholar]
- Hughes, C.C.; Fenical, W. Antibacterials from the Sea. Chem. Weinh. Bergstr. Ger. 2010, 16, 12512–12525. [Google Scholar] [CrossRef] [Green Version]
- Ueoka, R.; Uria, A.R.; Reiter, S.; Mori, T.; Karbaum, P.; Peters, E.E.; Helfrich, E.J.N.; Morinaka, B.I.; Gugger, M.; Takeyama, H.; et al. Metabolic and Evolutionary Origin of Actin-Binding Polyketides from Diverse Organisms. Nat. Chem. Biol. 2015, 11, 705–712. [Google Scholar] [CrossRef]









| Experiment/Involved Species and Sizes | Number of Specimens Used in Respective Experiment | Locality | Fixation | Further Treatment |
|---|---|---|---|---|
| Chemical analysis | ||||
| Chromodoris annae (20–45 mm) | 58 | BNP 2015/16 | 96% EtOH | Whole animals |
| C. annae (20–27 mm) | 6 | Sangihe 2016 | 96% EtOH and stored at −20 °C | Before fixation mantle rim and rest of body separated |
| C. annae mucus | several | Bangka Island 2019 | 96% EtOH | |
| Chromodoris dianae (20–55 mm) | 56 | BNP 2015/16 | 96% EtOH | Whole animals |
| C. dianae (27–45 mm) | 6 | Sangihe 2016 | 96% EtOH and stored at −20 °C | Before fixation mantle rim and rest of body separated |
| Chromodoris lochi (15–50 mm) | 31 | BNP 2015/16 | 96% EtOH | Whole animals |
| Chromodoris willani (20–70 mm) | 32 | BNP 2015/16 | 96% EtOH | Whole animals |
| Chromodoris strigata (20–25 mm) | 2 | Bangka Island 2019 | 96% EtOH | Only preservation fluid |
| MALDI-MS Imaging | ||||
| C. annae | 2 | BNP 2017 | Frozen in seawater (−20 °C) | Stored at –20 °C |
| C. dianae | 1 | BNP 2017 | Brought alive to Germany, then snap-frozen in liquid nitrogen | |
| In Vivo Toxicity Assay | ||||
| C. annae (30 mm) | 1 | BNP 2018 | Brought alive to Germany | Kept in aquaria for 2 days before and after experiments |
| C. dianae (35, 43 mm) | 2 | BNP 2018 | Brought alive to Germany | Kept in aquaria for 2 days before and after experiments |
| Elysia viridis (10–12 mm) | 12 | Figueira da Foz, Portugal, 2018 | Brought alive to Germany | Kept in aquaria with C. fragile as food for 2 days before and after experiments, where appropriate |
| Species (Specimen Identifier) | Accession Number |
|---|---|
| Chromodoris annae (Chan16Sa-9, Chan16Sa-3, Chan16Bu-6, Chel16Sa-1) | OK074000 |
| Chromodoris dianae (Chdi16Sa-6, Chdi16Bu-6) | OK074001 |
| Chromodoris lochi (Chlo16Bu-1, Chlo16Bu-2) | OK074002 |
| Chromodoris strigata (Chmi16Bu-1, Chst16Sa-1, Chst17Ba-1) | OK074003 |
| Chromodoris willani (Chwi16Bu-1, Chwi16Bu-2) | OK074004 |
| Elysia viridis (gDNA by courtesy of G. Christa) | OK074005 |
| (1) | C. annae | C. dianae | C. strigata | C. lochi | C. willani | F. affinis | A. tigrina | E. pulchra | A. californica | E. viridis |
|---|---|---|---|---|---|---|---|---|---|---|
| C. annae | 99 | 99 | 99 | 99 | 70 | 69 | 70 | 69 | 69 | |
| C. dianae | 99 | 98 | 99 | 99 | 70 | 69 | 70 | 69 | 69 | |
| C. strigata | 99 | 98 | 98 | 99 | 70 | 69 | 70 | 70 | 69 | |
| C. lochi | 99 | 99 | 98 | 99 | 69 | 68 | 70 | 69 | 69 | |
| C. willani | 99 | 99 | 99 | 99 | 69 | 69 | 70 | 69 | 69 | |
| F. affinis | 70 | 70 | 70 | 69 | 69 | 82 | 87 | 85 | 87 | |
| A. tigrina | 69 | 69 | 69 | 68 | 69 | 82 | 87 | 87 | 85 | |
| E. pulchra | 70 | 70 | 70 | 70 | 70 | 87 | 87 | 88 | 89 | |
| A. californica | 69 | 69 | 70 | 69 | 69 | 85 | 87 | 88 | 91 | |
| E. viridis | 69 | 69 | 69 | 69 | 69 | 87 | 85 | 89 | 91 | |
| (2) | C. annae | C. dianae | C. strigata | C. lochi | C. willani | F. affinis | A. tigrina | E. pulchra | A. californica | E. viridis |
| C. annae | 99 | 99 | 99 | 100 | 76 | 77 | 77 | 76 | 77 | |
| C. dianae | 99 | 99 | 100 | 100 | 76 | 77 | 76 | 75 | 77 | |
| C. strigata | 99 | 99 | 99 | 100 | 76 | 77 | 77 | 76 | 77 | |
| C. lochi | 99 | 100 | 99 | 100 | 76 | 77 | 76 | 75 | 77 | |
| C. willani | 100 | 100 | 100 | 100 | 76 | 77 | 77 | 76 | 77 | |
| F. affinis | 76 | 76 | 76 | 76 | 76 | 92 | 93 | 93 | 93 | |
| A. tigrina | 77 | 77 | 77 | 77 | 77 | 92 | 95 | 95 | 96 | |
| E. pulchra | 77 | 76 | 77 | 76 | 77 | 93 | 95 | 97 | 97 | |
| A. californica | 76 | 75 | 76 | 75 | 76 | 93 | 95 | 97 | 96 | |
| E. viridis | 77 | 77 | 77 | 77 | 77 | 93 | 96 | 97 | 96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hertzer, C.; Undap, N.I.J.; Papu, A.; Bhandari, D.R.; Aatz, S.; Kehraus, S.; Kaligis, F.; Bara, R.; Schäberle, T.F.; Wägele, H.; et al. Is a Modified Actin the Key to Toxin Resistance in the Nudibranch Chromodoris? A Biochemical and Molecular Approach. Diversity 2023, 15, 304. https://doi.org/10.3390/d15020304
Hertzer C, Undap NIJ, Papu A, Bhandari DR, Aatz S, Kehraus S, Kaligis F, Bara R, Schäberle TF, Wägele H, et al. Is a Modified Actin the Key to Toxin Resistance in the Nudibranch Chromodoris? A Biochemical and Molecular Approach. Diversity. 2023; 15(2):304. https://doi.org/10.3390/d15020304
Chicago/Turabian StyleHertzer, Cora, Nani Ingrid Jacquline Undap, Adelfia Papu, Dhaka Ram Bhandari, Stefan Aatz, Stefan Kehraus, Fontje Kaligis, Robert Bara, Till F. Schäberle, Heike Wägele, and et al. 2023. "Is a Modified Actin the Key to Toxin Resistance in the Nudibranch Chromodoris? A Biochemical and Molecular Approach" Diversity 15, no. 2: 304. https://doi.org/10.3390/d15020304
APA StyleHertzer, C., Undap, N. I. J., Papu, A., Bhandari, D. R., Aatz, S., Kehraus, S., Kaligis, F., Bara, R., Schäberle, T. F., Wägele, H., & König, G. M. (2023). Is a Modified Actin the Key to Toxin Resistance in the Nudibranch Chromodoris? A Biochemical and Molecular Approach. Diversity, 15(2), 304. https://doi.org/10.3390/d15020304