Abstract
Cartusia hunanesis sp. nov. was isolated from a stream in China, and two strains (ZJJ02 and ZJJ03) of which were inquired using morphological features, ecological evidence, and molecular data consisting of the 16S rRNA gene and 16S–23S rRNA intergenic transcribed spacer (ITS) region. Cartusia hunanesis varies from the type species Cartusia fontana by having only a single trichome in the sheath and large granules near the cross wall. The investigated strains of C. hunanesis were revealed to be a sister clade of C. fontana, according to the phylogenetic analysis based on the 16S rRNA gene. In addition, the Cartusia was clustered with the family Oculatellaceae members, genera Pegethrix and Elainella. These two strains displayed 97.6% similarity to C. fontana. The ITS region of C. hunanesis was found to be considerably distinct from that of C. fontana in terms of the secondary structure, which demonstrated that C. hunanesis is a novel species owing to the divergences in its morphological and genetic data compared with the related C. fontana.
1. Introduction
An integral part of the Earth’s biosphere, cyanobacteria are found in mountains, deserts, hot springs, and polar regions [1,2]. As primary producers worldwide, cyanobacteria have lived throughout most of the Earth’s history and play a significant role in the nitrogen cycle, as well as the oxygen and carbon cycles [3,4]. Moreover, it is widely believed that the chloroplasts of plants and eukaryotic algae originated from the ancestors of cyanobacteria [5]. There is no doubt that cyanobacteria are key to understanding the evolutionary history of the Earth [6]. Additionally, cyanobacteria can form dense blooms that affect the water quality of both freshwater and marine environments [7]. Previous studies have shown that the formation of cyanobacterial blooms is related to co-occurring microorganisms [8]; however, few pure isolates are available for studying the interactions between cyanobacteria and co-occurring microorganisms. The isolation of pure and new cyanobacterial species can help with understanding the mechanisms of cyanobacterial bloom development. With the discovery and exploration of various cyanobacteria worldwide, knowledge of their taxa is constantly expanding. The classification of cyanobacteria has always been difficult; traditionally, it was based on morphological characteristics [9,10,11]. However, traditional classification leads to false identification owing to the lack of definite morphological features [12,13,14,15,16,17]. On the other hand, some primitive or advolution morphological characteristics, for instance, multicellular morphology, rod-shaped monocellular morphology, filamentous branching, false branching, mucus sheath production, and gas vesicle, appear repeatedly in the evolution of cyanobacteria [18,19]. With the development of modern molecular analyses, more and more studies have used 16S rRNA gene sequencing to classify cyanobacteria [20,21,22,23]. The taxonomic description of microorganisms based on 16S rRNA gene sequencing has improved their classification and is considered to be the most commonly used method for prokaryote identification based on molecular markers [24,25]. The employ of the polyphasic method in the classification of cyanobacteria has been repeatedly emphasized and is based on phylogenetic classification, which combines and adds all cytomorphological data and ecological markers [9,11,24,26]. Thus, the polyphasic approach has become the basis and best methodology for cyanobacterial taxonomic revisions. And under these conditions, the widespread use of polyphasic approach method stimulated creation of a large number of new taxa of over 80 new genera from 2014 to 2022 [27].
Synechococcales is a large polyphyletic group consisting of no less than 90 genera of both thin filamentous and small unicellular cyanobacteria with a parietal thylakoid arrangement. This order is challenging because it comprises phylogenetically intermixed groups. Komárek et al. (2014) [24] divided Synechococcales into 11 families: Synechococcaceae, Merismopediaceae, Prochloraceae, Coelosphaeriaceae, Acaryochloridaceae, Chamaesiphonaceae, Heteroleibleiniaceae, Leptolyngbyaceae, Pseudanabaenaceae, Romeriaceae, and Schizotrichaceae. As the largest family in the order Synechococcales, Leptolyngbyaceae have many genera, including Nodosilinea Perkerson et al. 2011, Oculatella Zammit et al. 2012, Johansen et al. 2013, Pantanalinema Vaz et al. 2015, Alkalinema Vaz et al. 2015, Timaviella Sciuto et al. 2017, Albertania Zammitt 2018, etc. [14,21,28,29,30]. This family contains considerable number of strains that have been inadequately placed taxonomically because of the lack of diagnostic morphological characteristics [16,17]. In the last decade or so, members of Leptolyngbyaceae have been classified from various habitats, but the introduction of phylogenetic analysis of DNA sequences into the classification of cyanobacteria, together with ecological characteristics, was able to revise the diversity of this polyphyletic family in more detail [12,13,14,15,16,25,26,27]. Using the polyphasic approach, the Leptolyngbyaceae family was broken down into four monophyletic families: two redefined families (Prochlorotrichaceae and Leptolyngbyaceae) and two newly proposed families (Oculatellaceae and Trichocoleaceae), in 2018 [30]. As a sister taxon of the Leptolyngbayae, the Oculatellaceae contain a variety of subaerophytic taxa from soils and moist rocks. Some groups in this newly described family have been isolated from Leptolyngbaya sensu lato [30]. And this newly family contained the genera Cartusia, Elainella, Drouetiella, Komarkovaea, Tildeniella, Kaiparowitsia, Oculatella, Pegethrix, Thermoleptolyngbya, Timaviella, Trichotorquatus, etc. [30].
However, because of the recent surge in the number of new species and genomic sequences, the current classification system now appears to have limited coverage. In the latest cyanobacteria taxonomy, to achieve monophyly at the level of order, Strunecký (2023) [31] divided the Cyanophyceae into 20 orders: Gloeobacterales, Thermostichales, Aegeococcales, Acaryochloridales, Pseudanabaenales, Gloeomargaritales, Prochlorotrichales, Synechococcales, Nodosilineales, Oculatellales, Leptolyngbyales, Geitlerinematales, Desertifilales, Oscillatoriales, Coleofasciculales, Spirulinales, Chroococcales, Gomontiellales, Chroococcidiopsidales, and Nostocales. The family Oculatellaceae, which contains 17 genera, has been transferred to the Oculatellales as the only family in this order [31].
The newly described genus Cartusia Mai, Johansen et Pietrasiak in Mai et al. [30], contains only one species, Cartusia fontana. The type species, C. fontana, was separated from a pale green biofilm on the wall of the interior of a church in Slovakia. Morphologically, C. fontana is characterized by flexuous or straight filaments, sometimes with over one trichome in a sheath, sometimes forming a bundle of trichomes, without false branching, and with a single large central granule that can be seen in the cells.
Species diversity within the genus Cartusia is still poorly understood, as only one species is described, suggesting that more work is required to enrich the clade. In this study, we separated two cyanobacteria strains from Zhangjiajie, Hunan Province, China. Morphologically, we identified them as Leptolyngbya-related morphological types and a molecular analysis verified their placement in the Cartusia clade. Phylogenetic analysis and internal transcribed spacer (ITS) secondary structures were used to determine the exact taxonomic status. Overall, morphological and molecular analyses proclaimed that the two strains did not match the type species of Cartusia. Using a polyphasic approach, a new Cartusia species is herein described.
2. Material and Methods
2.1. Isolation and Cultivation
The samples used in the present study were harvested on 3 October 2019, from Zhangjiajie National Forest Park, Hunan Province, China. Samples were collected using a 20 µm mesh plankton net and stored in a 50 mL blue-cap bottle. During transportation, the bottles were placed in foam boxes containing ice packs. On arrival at the laboratory, samples were stored in a 4 °C refrigerator. Unialgal filaments were isolated using a laboratory-made Pasteur pipette under a microscope (NEXCOPE, Ningbo, China) on 5 October 2019. The samples were rinsed several times with sterile water until clean and no other cyanobacteria were present. The cells were then transferred to 24-well plates (Corning Costar, Glendale, AZ, USA) containing 1 mL of BG11 medium [32]. The strains were maintained at a temperature of 25 ± 1 °C during a 12:12 h light-dark cycle with low light (35 μmol·m−2·s−1) from the white fluorescent lamps. After three weeks of growth in the 24-well plates, the cyanobacterial isolates were cultured in spiral tubes containing 7 mL of BG11 medium. The tubes were manually shaken two to three times a day. Living cyanobacterial strains were maintained at Wuhan Polytechnic University, Wuhan, China. The studied strains were ZJJ02 and ZJJ03. The dry specimen of strain ZJJ02 was freeze-dried at −50 °C and stored in the Freshwater Algal Culture Collection of the Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan.
2.2. Morphological and Ultrastructural Characterization
The microscopic observation of all live strains was performed using a microscope (NEXCOPE, Ningbo, China). Microphotographs of the filaments were obtained using a Nikon NIS-Elements 3.2D (Nikon, Tokyo, Japan) attached to the microscope. The lengths and widths of more than 60 vegetative cells were measured and recorded by the aid of microscope-carried auxiliary software (Imageview). The ultrastructure of strain ZJJ02 was observed by using transmission electron microscopy (TEM). The fresh sample of strain ZJJ02 was fixed in 2.5% glutaraldehyde and 0.1 M phosphate buffer with a pH 7.2 for three days, at 4 °C. The sample was then cleaned with 0.1 M phosphate buffer and fixed with 1% osmium tetroxide for 2 hours. Reuse the same buffer solution for cleaning. The sample was dehydrated with successive ethanol gradients (30, 50, 70, 90, and 100%) and embedded in Spurr’s resin, then stained with 2% uranyl acetate and lead citrate [33]. Finally, a TEM (Hitachi HT-7700, Tokyo, Japan) was used to capture images of the treated sample at an acceleration voltage of 80 kV.
2.3. Molecular Analysis
The total genomic DNA was extracted from the cultured strains by using a FastDNATM Spin Kit (MP Biomedicals, Irvine, CA, USA). The extracted DNA samples were obtained at −20°C for subsequent experiments. Amplification of the 16S rRNA gene and 16S–23S ITS region was executed by using the primers PA and B23S (Table 1) [34,35]. The polymerase chain reaction (PCR), with a final volume of 50 µL, comprised 1 µL of each primer (10 μmol·L−1), 2 μL of genomic DNA (100 ng μL−1), 21 μL of sterile water, and 25 μL of PCR mix with Taq polymerase (No. RR901Q, TaKaRa Bio Inc., Otsu, Japan). The program for the 16S rRNA gene ran for 34 cycles of 30 s at 94 °C, 30 s at 58 °C, and 2 min at 72 °C. The PCR amplification was carried out using an MJ Mini Personal Thermal Cycler (Bio–Rad, Hercules, CA, USA). The PCR products were visualized on 1% agarose gel. Successful amplified products were purified using the E.Z.N.A Gel Extraction Kit (Omega, Georgia, USA) and cloned into the pMD-18T vector (Takara, Kusatsu, Japan) following the procedure described by Sambrook and Russell (2001) [36]. The target genes were sequenced using an ABI 3730 Automated Sequencer (PerkinElmer, Waltham, MA, USA).

Table 1.
Information on the primers PA and B23S used in this study.
2.4. Examination of the 16S rRNA Gene Sequence and Phylogenetic Analysis
All cloned gene sequences of isolated strains were examined using Basic Local Alignment Search Tool (BLAST) and compared with other sequences in the database of the National Center for Biotechnology Information (NCBI, http:/www.ncbi.nlm.nih.gov/genbank/, accessed on 12 September 2022). In order to determine the phylogenetic placement of the studied strains, the 16S rRNA gene sequences obtained in the present study and several cyanobacterial sequences available from NCBI were used for the phylogenetic analyses. The taxa used for phylogenetic analysis of the 16S rRNA genes consisted of 189 sequences, Gloeobacter violaceus VP3-01 and Gloeobacter kilaueensis JS1 were selected as the outgroup to establish a justified evolutionary relationship. Sequences were aligned with the auto-selected strategy FFT-NS-I (with default parameters) using MAFFT v7.312 [37] and visually inspected using MEGA v.7.0 [38]. After multiple-sequence alignment, 189 sequences with nucleotide sites were obtained. The consensus phylogenetic trees used in the present study were inferred using the Bayesian inference (BI), neighbor-joining (NJ), and maximum likelihood (ML) methods. MEGA software v7.0 [38] was used for the NJ analysis. The robustness of the branch was tested by analyzing 1,000 replicated bootstraps. The ML algorithms were inferred using PAUP v4.0b10 [39]. The best fit model GTR+I+G was selected for the BI and ML analyses using the Akaike Information Criterion in ModelFinder [40]. The BI analysis was calculated using MrBayes v3.2.6 [41,42] in CIPRES Science Gateway V.3.3 [43] (http://www.phylo.org/, accessed on 12 September 2022). The Markov Chain Monte Carlo process was set to four chains for 10 million generations, with a sampling frequency of once every 100 generations. The obtained phylogenetic trees were displayed using TreeView 1.6.6 and edited using Adobe Photoshop CS6 Version 13.0. The p-distance of the 16S rRNA gene was caculated using MEGA software v.7.0 [38] to determine the sequence identity (100 × (1 − p)) of the 16S rRNA data. The 16S rRNA and 16S–23S ITS gene sequences of strains ZJJ02 and ZJJ03 have been deposited to the NCBI GenBank database, with the accession numbers ON306331, ON306332, ON306364, and ON306365.
2.5. 16S–23S ITS Secondary Structure Analysis
Sequences in the 16S-23S ITS region were selected for the taxonomic analysis of the studied strains. The complete ITS sequence of Cartusia fontana Kovacik 1999/1-LC was obtained from NCBI. The ITS secondary structures of D1–D1′, Box–B and V3 helices of these two strains were determined using “RNAstructure” ver. 6.4 [44]. The secondary structures were created under ideal conditions, with the temperature fixed at 37 °C by default and the structure assigned as untangle loop fix. The secondary structure was then redrawn using Adobe Illustrator 2020.
3. Results
3.1. Morphology
Cartusia hunanesis F. Cai et R. Li sp. nov. (Figure 1).
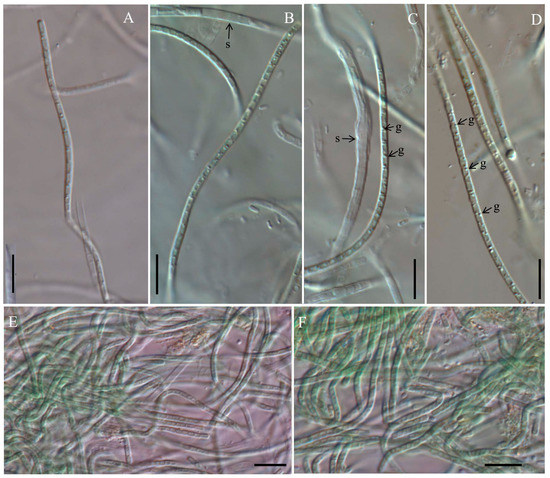
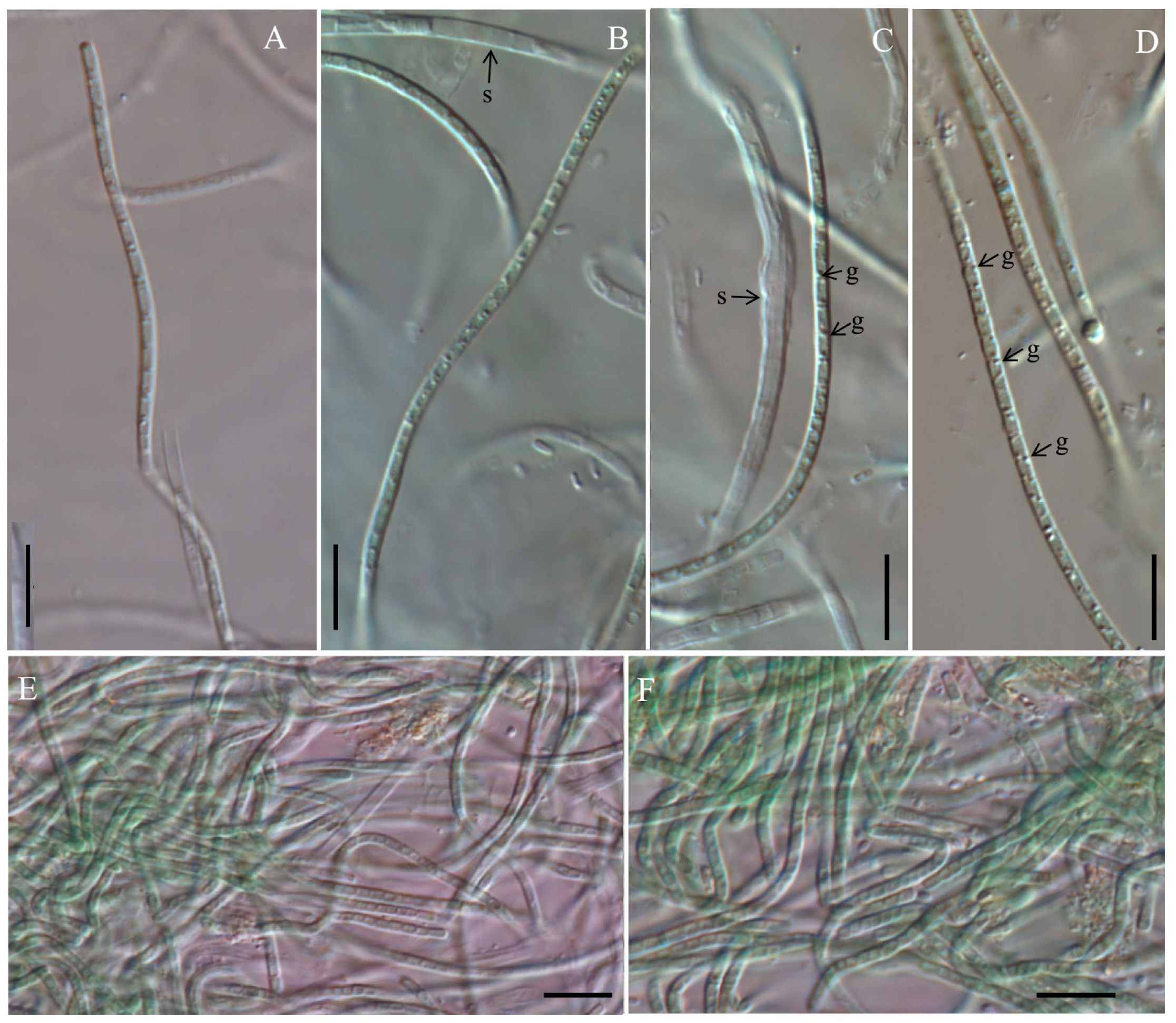
Figure 1.
Micrographs of Cartusia hunanesis by light microscopy (LM): (A,B,E) Cartusia hunanesis ZJJ02; (C,D,F) Cartusia hunanesis ZJJ03. s: sheath, g: granules. Scale bar = 10 µm.
Description: Colony spherical and blue-green, becoming yellow-green with age. Filaments straight or flexuous, freely entangled, without false branching, with only a single trichome in a sheath. Sheath thin, firm, colorless, occasionally widened. Trichomes not narrowing, not or slightly constricted at the cross-walls. Cells in young trichomes longer than wide, in mature trichomes shorter than wide, 0.82–2.81 μm long (X¯: 1.47 ± 0.51 μm) and 1.36–2.43 μm wide (X¯: 1.83 ± 0.26 μm), with large granules visible in the cell cross walls; with parietal thylakoids. End cells rounded, untapered, without calyptra. Reproduction by hormogonia or trichome breakage.
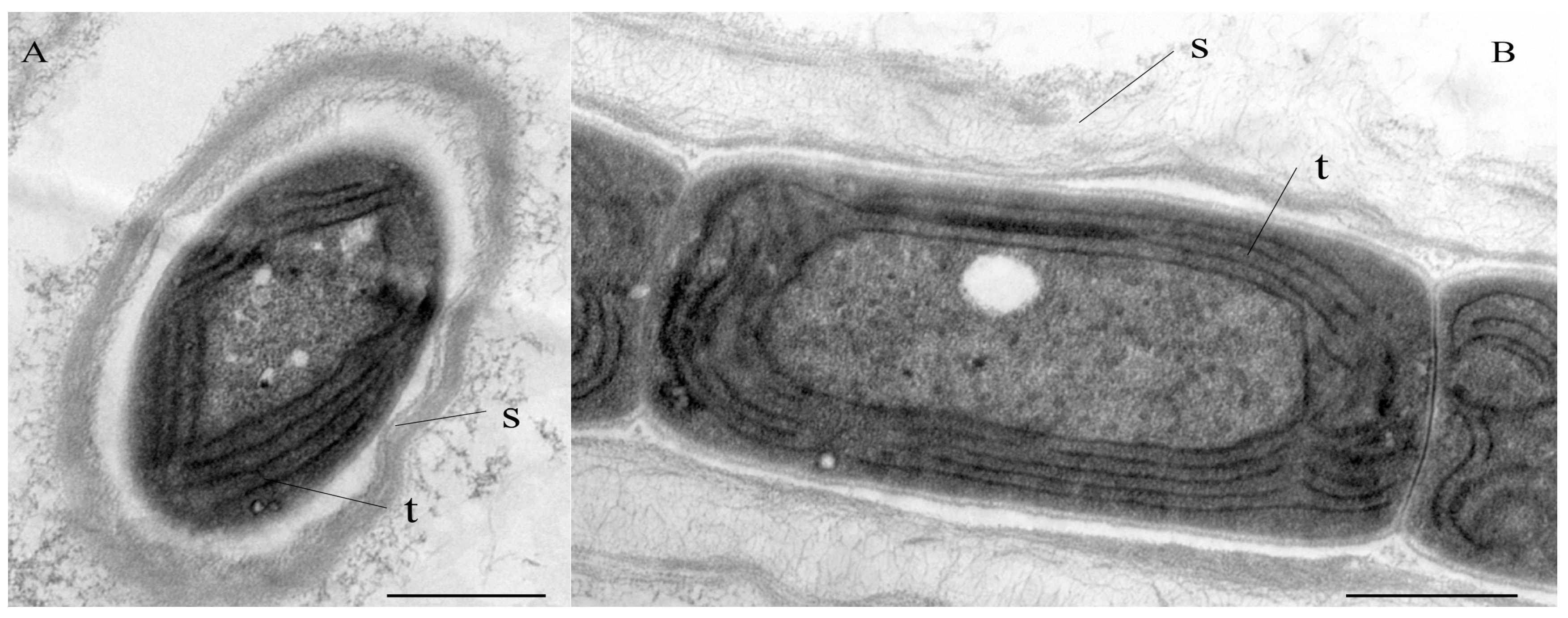
Diagnosis: The ultrastructure showed parietal arrangement of the thylakoids (4–5 per cell), like the order Synechococcales (Figure 2). Compared with C. fontana, the type species C. hunanesis forms a single trichome in a common sheath, and the granules are visible near the cell wall, as shown in Table 2. The phylogeny based on the 16S rRNA gene exhibited that C. hunanesis forms a separate node in the Cartusia clade. In addition, the low similarity of the 16S rRNA gene sequence of this species to that of C. fontana and significant differences in the structure and length of the secondary structures of the 16S–23S ITS region support it as a new Cartusia species.
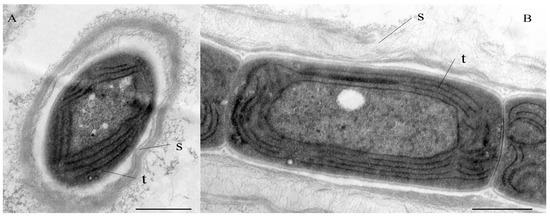
Figure 2.
TEM micrographs of Cartusia hunanesis ZJJ02. (A) Cross section of a cell in its surrounding sheath. (B) Longitudinal section of a filament. Scale bar = 0.5 μm. S = sheath; t = thylakoids. The thylakoids are arranged roughly parallel in a parietal position.

Table 2.
Comparison of the characteristics of Cartusia hunanesis and Cartusia fontana.
Reference strain: ZJJ02, culture maintained at Wuhan Polytechnic University, Wuhan, China.
Additional strain: ZJJ03, culture maintained at Wuhan Polytechnic University, Wuhan, China.
Type locality: The edge of a stream in Zhangjiajie National Forest Park, Hunan Province, China (41°28′00.00′′ N, 124°08′00.00′′ E).
Holotype here designated: Dry specimen of the strain ZJJ02 stored in the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Science, Wuhan, China, with specimen number of No. HN201911.
Etymology: hunanesis refers to Hunan province, where the strains were collected, transliterated into Latin.
Habitat: Planktic.
3.2. Molecular and Phylogenetic Analysis
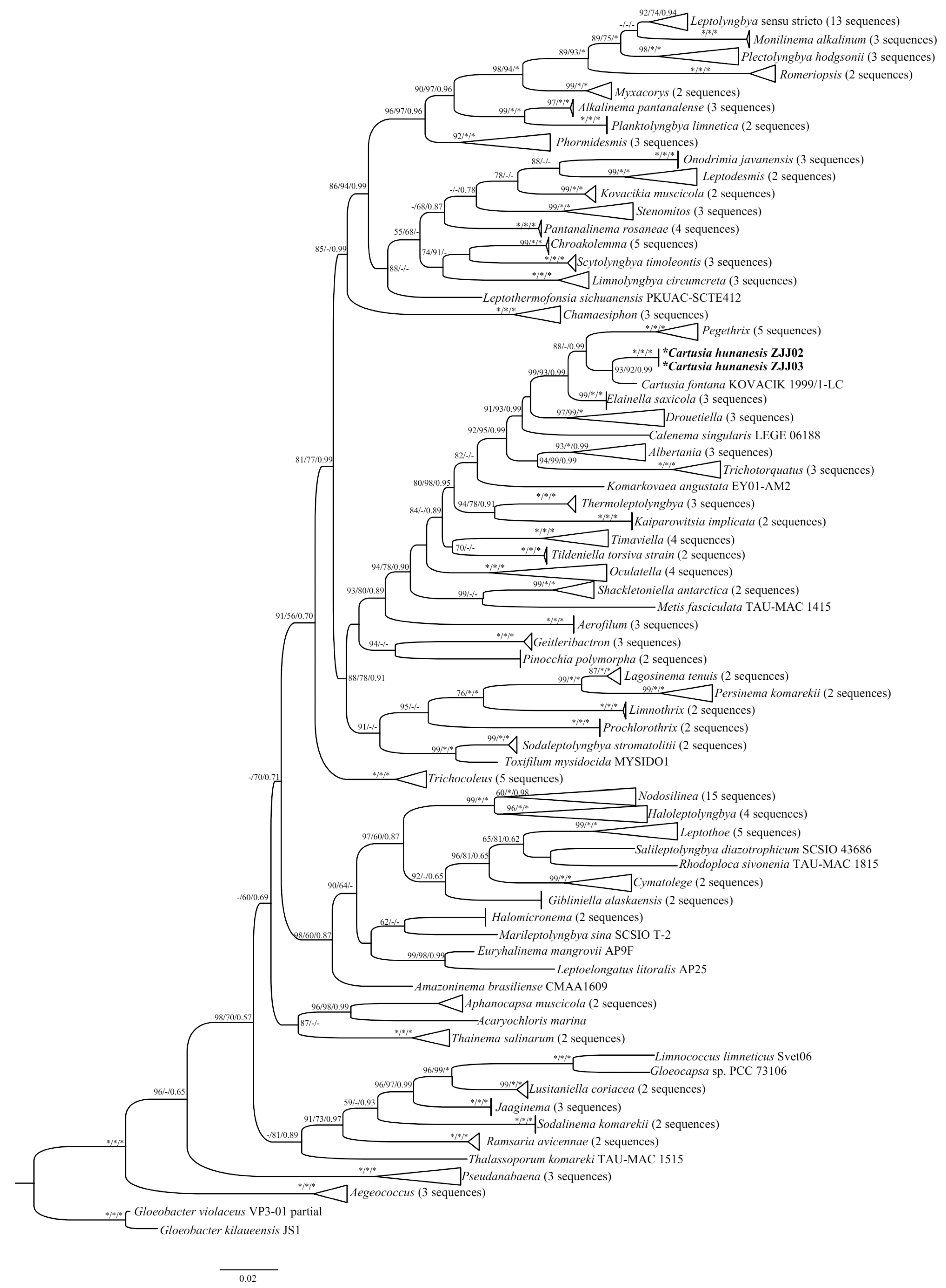
The 16S rRNA gene sequences of these two strains were acquired and assessed with BLAST analyses in NCBI. The p-distance matrix based on the 16S rRNA gene revealed that the two Cartusia strains shared 99.7% similarity with each other and 97.6% similarity with C. fontana. Phylogenetic trees based on the 16S rRNA gene sequences from 189 cyanobacterial taxa, including two Cartusia strains, were constructed using the ML, NJ, and Bayesian methods. The phylogenetic analysis (ML tree) (Figure 3) indicated that our strains were nested within the genus Cartusia (93% and 92% ML and NJ bootstrap percentage (BP) and 0.99 posterior probability (PP)). And the 16S rRNA gene sequence phylogeny showed that strains of Cartusia form a monophyletic lineage among other thin filamentous cyanobacteria genera, most related were Pegethrix, Elainella, Drouetiella, Calenema, Albertania, and Trichotorquatus. Consistent with the results of previous studies, Oculatellaceae forms a monophyletic system at the family level in our study.
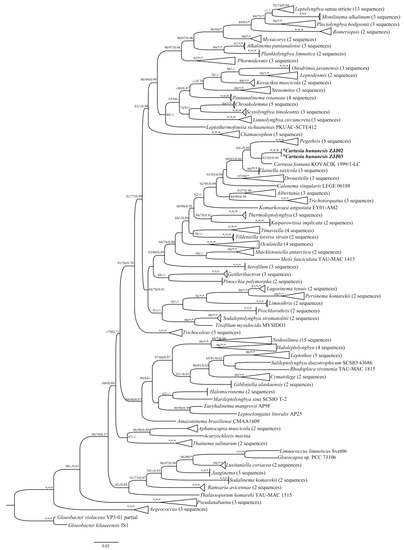
Figure 3.
Maximum likelihood (ML) phylogenetic tree based on 16S rDNA sequences (1144 bp) of the studied strains and other cyanobacterial strains. Bootstrap values greater than 50% with the ML/NJ/Mrbayes methods are indicated on the tree, the asterisks and - at the nodes mean 100 and bootstrap value less than 50%. The novel species are in bold font.
3.3. 16S–23S ITS Region
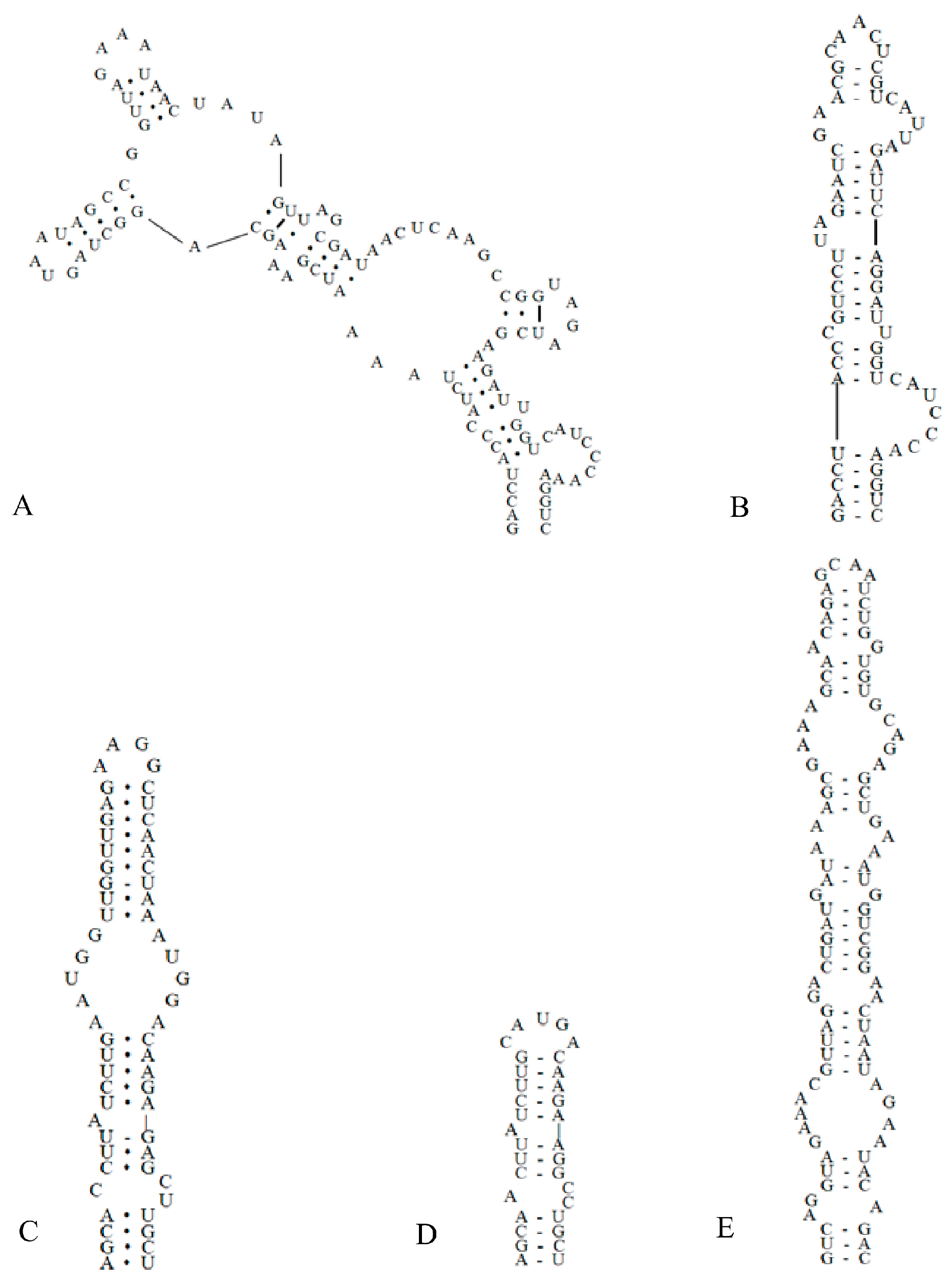
The ITS secondary structure was constructed from the ITS regions of the two studied strains combined with the sequence of C. fontana. Analyses of the secondary structures of the D1–D1′ helix (Figure 4) showed that C. hunanesis ZJJ02 and ZJJ03 possessed a unique structure. C. fontana had 107 nucleotides, the base stem contained a 5 bp helix, followed by an 8 bp side loop (5′-CAUCCCAA-3′) on the 3′ side, and further followed by two side branches from two large internal loops (Figure 4A). In contrast, The D1–D1′ helix of C. hunanesis had 65 nucleotides, with a basal 3′ unilateral bulge of 7 nucleotides (8 bases in C. fontana), and was then followed by a 1:1 base (C:U) bilateral bulge. This bilateral bulge was connected by a side loop with two bases (UA) on the 5ʹ side, and following this 5ʹ side loop was a bilateral bulge of 2:5 bases. The terminal loop consisted of five bases (Figure 4B).
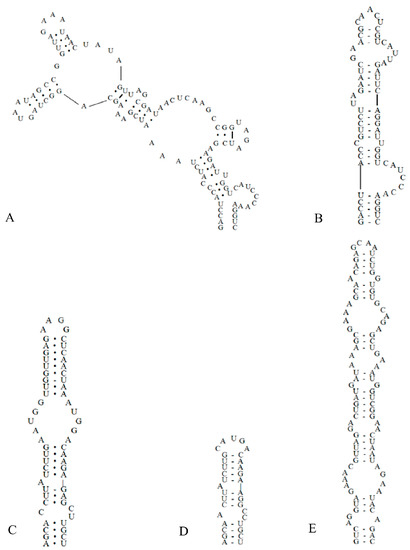
Figure 4.
Secondary structures of the 16S–23S ITS region in Cartusia hunanesis and Cartusia fontana. (A). D1–D1′ helix of Cartusia fontana Kovacik 1999/1. (B). D1–D1′ helix of Cartusia hunanesis ZJJ02 and ZJJ03. (C). Box–B helix of Cartusia fontana Kovacik 1999/1. (D). Box–B helix of Cartusia hunanesis ZJJ02 and ZJJ03. (E). V3 helix of Cartusia hunanesis ZJJ02 and ZJJ03.
As shown in Figure 4, there is an obvious contrast between the Box–B helix of C. hunanesis and C. fontana. The base of the stem of C. hunanesis contained a 4 bp (AGCA) helix, following this base stem was a 1:2 base (A:CC) bilateral bulge, and then further followed a side loop with a single base (A) on the 5′ side. The terminal loop consisted of 5 bp bases (CAUGA) (Figure 4D). In contrast, C. fontana contained a 4 bp helix at the base of the stem, followed by a bilateral bulge of 1:2 bases (C:CU), and following this bilateral bulge was a side loop with a single base on the 5ʹ side; this side loop was further connected by a 5:5 bases bilateral bulge, and the terminal loop consisted of four nucleoside bases (AAGG) (Figure 4C).
As shown in Figure 4E, the V3 helix of C. hunanesis had 94 nucleotides, the basal stem contained a 3 bp helix, followed by seven bilateral bulges, and the terminal loop contained four nucleoside bases (GCAA). The V3 helix of C. fontana was not identified because of its short end sequence.
4. Discussion
The largest family Leptolyngbyaceae has attracted significantly more research attention in recent years [24]. Mai et al. 2018 [30] proposed that this large family should be broken down into four monophyletic clades (Leptolyngbyaceae, Oculatellaceae, Prochlorotrichaceae, and Trichocoleaceae). According to 16S rRNA gene sequence phylogeny, the newly defined family Oculatellaceae Mai et Johansen fam. nov. is a group consisting of several monophyletic genera, which has no sheath in active growing populations, but sometimes develops facultatively sheaths in the established populations, false branching are absent in some genera, but are usually present in most genera. However, the latest study of cyanobacterial taxonomy by Strunecký et al. (2023) [31] kept Oculatellales as a separate order with only one family, the Oculatellaceae. The filaments of the Oculatellales are bent, coiled, or straight, flexuous, or firm, with rounded terminal cells surrounded by a thin and colorless sheath, sometimes with nodules and false branching [31]. Taxonomically, C. fontana belongs to the genus Leptolyngbya [30]. Leptolyngbya belongs to the family Leptolyngbyaceae. To date, only one species, the type species C. fontana, has been described.
In the present study, two cyanobacterial strains were classified as novel species within the genus Cartusia based on the polyphasic approach to taxonomy. The two strains are morphologically similar, and the molecular data also revealed high similarities in 16S rRNA gene sequence (99.7%) and the ITS region (100%) between both strains, therefore, it is impossible to distinguish these two strains into different species. The morphological characteristics of C. hunanesis were compared with those of C. fontana (Table 2), indicating the morphological differences between these two species. Ecologically, the habitats of the two species are quite different. Cartusia fontana occurred as a pale green biofilm on the wall in the interior of a church in Slovakia as a subaerophytic species, whereas C. hunanesis was collected from a stream in China as plankton. Hence, C. hunanesis and C. fontana are geographically distinct.
In the 16S rRNA gene-based phylogeny, strains ZJJ02 and ZJJ03, along with C. fontana Kovacik 1999/1-LC, formed a unique clade, that is, the Cartusia clade described in 2018, which clearly indicates that these two strains belong to the genus Cartusia. Additionally, the strains ZJJ02 and ZJJ03 formed a separate lineage within the Cartusia branch, indicating that the two strains are new members of the genus Cartusia. The description of a new species of Cartusia in the present study is well supported by molecular evidence. The similarity of 16S rRNA gene sequences between C. hunanesis and C. fontana is approximately 97.6%, which is below the recommended value for ascertaining interspecific differentiation [45]. Other studies [14,30,42,46,47] also followed the same threshold set by Yarza et al. (2014) [45] for species delineation below 98.7% genetic similarity.
The molecular analysis used to describe species also involves the study of 16S–23S ITS secondary structures, which was considered to be an important evaluation criterion for alpha-level taxonomy [48,49,50]. The ITS secondary structure analysis was also well-supported to validate that strains ZJJ02 and ZJJ03 are the second new species in the genus Cartusia. The output of the ITS secondary structure of the D1–D1′, Box–B, and V3 helices revealed that the structures of the D1–D1′ and Box–B helices were significantly different in the investigated strains ZJJ02 and ZJJ03 when compared with C. fontana. The D1–D1′ region of ZJJ02 and ZJJ03 was significantly shorter than the D1–D1′ region of C. fontana (Figure 4); the latter has two side branches from two large internal loops. Additionally, the Box–B region of C. fontana was significantly longer than the Box–B region in ZJJ02 and ZJJ03 (Figure 4), with varying bilateral bulges and internal stems.
To sum up, ZJJ02 and ZJJ03 were described as a new species Cartusia hunanesis sp. nov. based on the combination of morphology, ultrastructure, 16S rRNA phylogeny, 16S rRNA gene threshold, and ITS secondary structures. And this new species is furthermore supported by the ecological data, which is different from the type species C. fontana. This is the first new species of Cartusia found in China. The recognition of Leptolyngbya-like cyanobacteria in China is limited, and further studies are needed to fully document the Leptolyngbya-like cyanobacteria flora of Zhangjiajie National Forest Park and China more broadly. We hope that the publication of one new species will attract more scholars to study species richness in Zhangjiajie Forest Park, which is locally and internationally famous, and may lead to more interesting discoveries. With increasingly advanced biotechnological tools, the more we know about cyanobacteria, the better we can treat cyanobacterial blooms. Finally, we anticipate that the descriptions of Leptolyngbya-like taxa will increase in the coming years.
Author Contributions
Conceptualization, S.L. and F.C.; methodology, F.C.; software, S.L. and F.C.; formal analysis, S.L.; investigation, F.C.; data curation, S.L.; writing—original draft preparation, S.L. and F.C.; writing—review and editing, F.C. and R.L.; visualization, D.Y.; funding acquisition, H.Z. and F.C. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by the National Natural Science Foundation of China (32000166) and the Monitoring Project of Groundwater and Important Lakes Along Main Stream of Yangtze River in Hubei Province (420000-2022-218-006-003).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Zhenfei Xing for the TEM images technical support.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Whitton, B.A. The Ecology of Cyanobacteria II: Their Diversity in Time and Space; Springer: Dordrecht, The Netherlands, 2012; 669p. [Google Scholar]
- Tomitani, A.; Knoll, A.H.; Cavanaugh, C.M.; Ohno, T. The evolutionary diversification of cyanobacteria: Molecular–phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. USA 2006, 103, 5442–5447. [Google Scholar] [CrossRef] [PubMed]
- Brocke, H.J.; Wenzhoefer, F.; de Beer, D.; Mueller, B.; van Duyl, F.C.; Nugues, M.M. High dissolved organic carbon release by benthic cyanobacterial mats in a Caribbean reef ecosystem. Sci. Rep. 2015, 5, 8852. [Google Scholar] [CrossRef] [PubMed]
- Brocke, H.J.; Piltz, B.; Herz, N.; Abed, R.M.M.; Palinska, K.A.; John, U.; den Haan, J.; de Beer, D.; Nugues, M.M. Nitrogen fixation and diversity of benthic cyanobacterial mats on coral reefs in Curaçao. Coral Reefs 2018, 37, 861–874. [Google Scholar] [CrossRef]
- Delwiche, C.F.; Palmer, J.D. The origin of plastids and their spread via secondary symbiosis. Plant Syst. Evol. 1997, 11, 53–86. [Google Scholar]
- Houk, K. Phylogenetic placement of seven strains of Synechococcales (Cyanobacteria) isolated from desert soils in Zion National Park. Sr. Honor. Proj. 2021, 129. Available online: https://collected.jcu.edu/honorspapers/129 (accessed on 19 September 2022).
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef]
- Wang, K.; Mou, X. Coordinated Diel Gene Expression of Cyanobacteria and Their Microbiome. Microorganisms 2021, 9, 1670. [Google Scholar] [CrossRef] [PubMed]
- Komárek, J. A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. Eur. J. Phycol. 2016, 51, 346–353. [Google Scholar] [CrossRef]
- Dvořák, P.; Casamatta, D.A.; Hašler, P.; Jahodářová, E.; Norwich, A.R.; Poulíčková, A. Diversity of the Cyanobacteria. In Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects; Hallenbeck, P.C., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–46. [Google Scholar]
- Komárek, J. Quo vadis, taxonomy of cyanobacteria (2019). Fottea 2020, 20, 104–110. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota 2. Tiel: Oscillatoriales. In Süsswasserflora von Mitteleuropa 19/1; Ettl, H., Gärtner, G., Heynig, H., Mollenhauer, D., Eds.; Gustav Fischer: Ulm, Germany, 2005; pp. 1–548. [Google Scholar]
- Osorio-Santos, K.; Pietrasiak, N.; Bohunická, M.; Miscoe, L.H.; Kováčik, L.; Martin, M.P.; Johansen, J.R. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria). Eur. J. Phycol. 2014, 49, 450–470. [Google Scholar] [CrossRef]
- Vaz, M.G.M.V.; Bonaldo-Genuário, D.; Dini-Andreote, A.P.; Silva-Malone, C.F.; Sant’Anna, C.L.; Barbiero, L.; Fátima-Fiore, M. Pantanalinema gen. nov. and Alkalinema gen. nov.: Two novel pseudanabaenacean genera (Cyanobacteria) isolated from saline-alkaline lakes. Int. J. Syst. Evol. Micr. 2015, 65, 298–308. [Google Scholar] [CrossRef]
- Bagchi, S.N.; Dubey, N.; Singh, P. Phylogenetically distant clade of Nostoc-like taxa with the description of Aliinostoc gen. nov. and Aliinostoc morphoplasticum sp. nov. Int. J. Syst. Evol. Micr. 2017, 67, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Absalón, I.; Johansen, J.R.; Muñoz-Martín, M.A.; Montejano, G. Chroakolemma gen. nov. (Leptolyngbyaceae, Cyanobacteria) from soil biocrusts in the semi-desert Central Region of Mexico. Phytotaxa 2018, 367, 201–218. [Google Scholar] [CrossRef]
- Chakraborty, S.; Maruthanayagam, V.; Achari, A.; Pramanik, A.; Jaisankar, P.; Mukherjee, J. Aerofilum fasciculatum gen. nov., sp. nov. (Oculatellaceae) and Euryhalinema pallustris sp. nov. (Prochlorotrichaceae) isolated from an Indian mangrove forest. Phytotaxa 2021, 522, 165–186. [Google Scholar] [CrossRef]
- Dvořák, P.; Casamatta, D.A.; Poulíčková, A.; Hašler, P.; Ondřej, V.; Sanges, R. Synechococcus: 3 billion years of global dominance. Mol. Ecol. 2014, 23, 5538–5551. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeister, B.E.; de Vos, J.M.; Antonelli, A.; Bagheri, H.C. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc. Natl. Acad. Sci. USA 2013, 110, 1791–1796. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.R.; Casamatta, D.A. Recognizing cyanobacterial diversity through adoption of a new species paradigm. Algol. Stud. 2005, 117, 71–93. [Google Scholar] [CrossRef]
- Perkerson, R.B.; Johansen, J.R.; Kováčik, L.; Brand, J.; Kaštovský, J.; Casamatta, D.A. A unique pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar]
- Silva, C.S.P.; Genuário, D.B.; Vaz, M.G.M.V.; Fiore, M.F. Phylogeny of culturable cyanobacteria from Brazilian mangroves. Syst. Appl. Microbiol. 2014, 37, 100–112. [Google Scholar] [CrossRef]
- Bravakos, P.; Kotoulas, G.; Skaraki, K.; Pantazidou, A.; Economou-Amilli, A. A polyphasic taxonomic approach in isolated strains of Cyanobacteria from thermal springs of Greece. Mol. Phylogenet. Evol. 2016, 98, 147–160. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Palinska, K.; Surosz, W. Taxonomy of cyanobacteria: A contribution to consensus approach. Hydrobiologia 2014, 740, 1–11. [Google Scholar] [CrossRef]
- Genuário, D.B.; Vaz, M.; Hentschke, G.S.; Sant’Anna, C.L.; Fiore, M.F. Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. Int. J. Syst. Evol. Micr. 2015, 65, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2023, 59, 12–51. [Google Scholar] [CrossRef] [PubMed]
- Zammit, G.; Billi, D.; Albertano, P.B. The subaerophytic cyanobacterium Oculatella subterranea (Oscillatoriales, Cyanophyceae) gen. et sp. nov.: A cytomorphological and molecular description. Eur. J. Phycol. 2012, 47, 341–354. [Google Scholar] [CrossRef]
- Sciuto, K.; Moschin, E.; Moro, I. Cryptic cyanobacterial diversity in the Giant Cave (Trieste, Italy): The new genus Timaviella (Leptolyngbyaceae). Cryptogam. Algol. 2017, 38, 285–323. [Google Scholar] [CrossRef]
- Zammit, G. Systematics and biogeography of sciophilous cyanobacteria; an ecological and molecular description of Albertania skiophila (Leptolyngbyaceae) gen. & sp. nov. Phycologia 2018, 57, 481–491. [Google Scholar]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 001–059. [Google Scholar]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Geng, R.; Wang, Y.; Cai, F.; Zhang, Y.; Yang, P.; Dai, G.; Yu, G. Neochroococcus gongqingensis gen. et sp. nov., a new member of coccoid cyanobacteria from a watercourse, Eastern China. Fottea 2021, 21, 44–52. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar]
- Gkelis, S.; Rajaniemi, P.; Vardaka, E.; Moustaka-Gouni, M.; Lanaras, T.; Sivonen, K. Limnothrix redekei (Van Goor) Meffert (Cyanobacteria) strains from Lake Kastoria, Greece form a separate phylogenetic group. Microb. Ecol. 2005, 49, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; CSHL Press: New York, NY, USA, 2001; 2344p. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. Phylogenetic Analysis Using Parsimony, Version 4.0; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [PubMed]
- Cai, F.; Yu, G.; Liu, Y.; Sun, Y.; Li, R. Description of two new species of Nostoc from China based on the polyphasic approach. Fottea 2021, 21, 259–271. [Google Scholar] [CrossRef]
- Miller, M.; Schwartz, T.; Pickett, B.; He, S.; Klem, E.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A. A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evol. Bioinform. 2015, 11, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Mathews Lab. RNA Structure Version 6.4. 2021. Available online: http://rna.urmc.rochester.edu/RNAstructure.html (accessed on 20 September 2022).
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Witman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Vinogradova, O.; Mikhailyuk, T.; Glaser, K.; Holzinger, A.; Karsten, U. New species of Oculatella (Synechococcales, Cyanobacteria) from terrestrial habitats of Ukraine. Ukranian Bot. J. 2017, 74, 509–520. [Google Scholar] [CrossRef]
- Cai, F.; Yu, G.; Li, R. Description of two new species of Pseudoaliinostoc (Nostocales, Cyanobacteria) from China based on the polyphasic approach. J. Oceanol. 2022, 40, 1233–1244. [Google Scholar] [CrossRef]
- Boyer, S.L.; Flechtner, V.R.; Johansen, J.R. Is the 16S–23S rRNA Internal Transcribed Spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 2001, 18, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.R.; Kováčik, L.; Casamatta, D.A.; Fučiková, K.; Kaštovský, J. Utility of 16S-23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwig. 2011, 92, 283–302. [Google Scholar] [CrossRef]
- Chakraborty, S.; Maruthanayagam, V.; Achari, A.; Mahansaria, R.; Pramanik, A.; Jaisankar, P.; Mukherjee, J. Oxynema aestuarii sp. nov. (Microcoleaceae) isolated from an Indian mangrove forest. Phytotaxa 2018, 374, 24–40. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).