Historical Landscape Evolution Shaped the Phylogeography and Population History of the Cyprinid Fishes of Acrossocheilus (Cypriniformes: Cyprinidae) According to Mitochondrial DNA in Zhejiang Province, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. Sequence Variations, Genetic Structure, and Phylogenetic Analyses
2.3. ABC Analyses Using DIYABC
3. Results
3.1. Genetic Diversity of A. fasciatus, A. wenchowensis and A. kreyenbergii
3.2. Genetic Structure and Phylogenetic Reconstruction
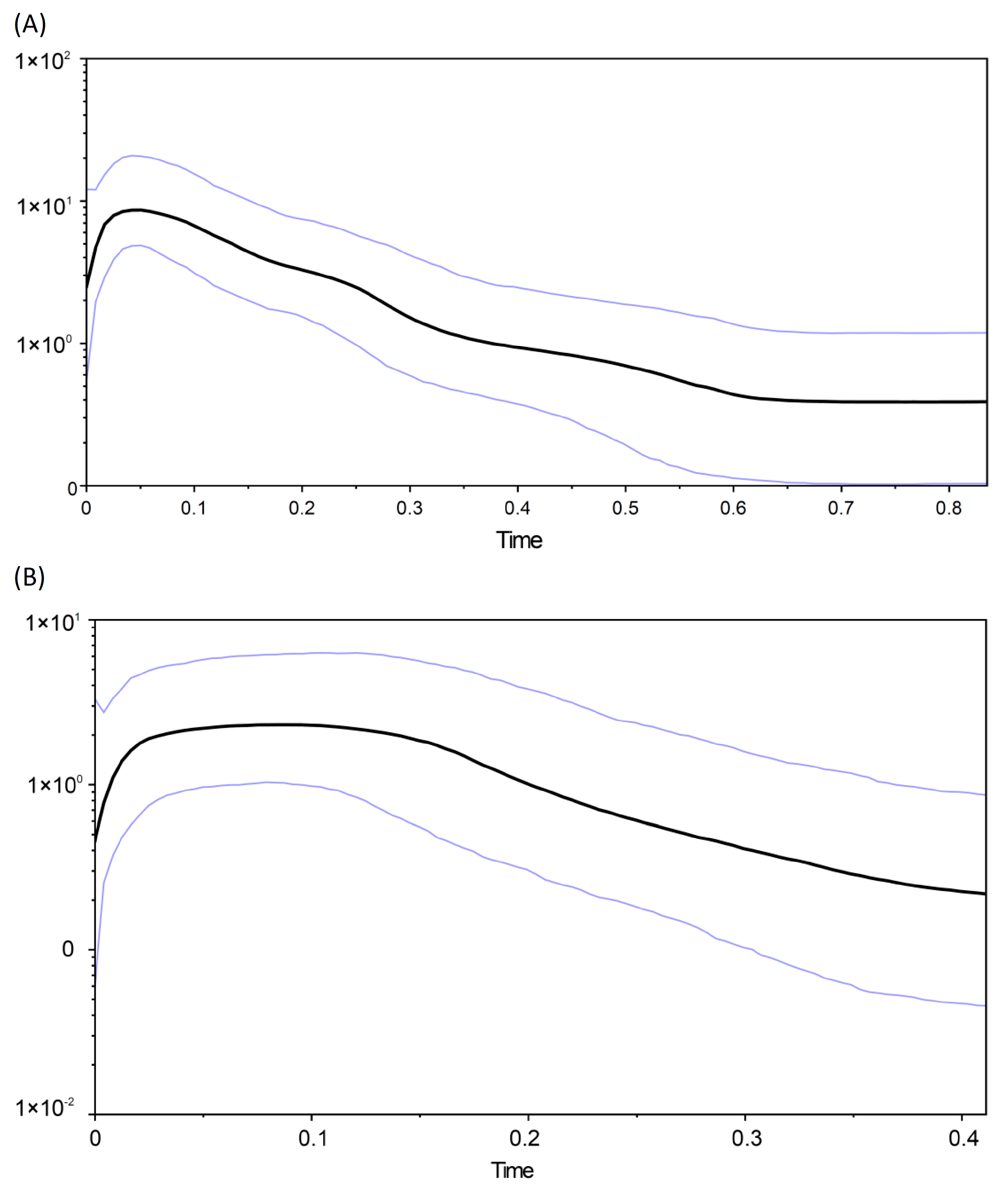
3.3. Historical Demography and Biogeographic Analysis
3.4. Approximate Bayesian Computation (ABC) Scenarios
4. Discussion
4.1. Genetic Diversity
4.2. Population Structure and Demographic History
4.3. Phylogeography of Acrossocheilus
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xing, Y.; Zhang, C.; Fan, E.; Zhao, Y. Freshwater fishes of China: Species richness, endemism, threatened species and conservation. Divers. Distrib. 2016, 22, 358–370. [Google Scholar] [CrossRef] [Green Version]
- WWF. Living Planet Report 2016. Risk and Resilience in a New Era; WWF: Gland, Switzerland, 2016. [Google Scholar]
- Li, M.; Yang, X.; Ni, X.; Fu, C. The Role of Landscape Evolution in the Genetic Diversification of a Stream Fish Sarcocheilichthys parvus from Southern China. Front. Genet. 2023, 13, 3764. [Google Scholar] [CrossRef]
- Yang, X.; Ni, X.; Fu, C. Phylogeographical Analysis of the Freshwater Gudgeon Huigobio chenhsienensis (Cypriniformes: Gobionidae) in Southern China. Life 2022, 12, 1024. [Google Scholar] [CrossRef]
- Yang, J.Q.; Tang, W.Q.; Liao, T.Y.; Sun, Y.; Zhou, Z.C.; Han, C.C.; Liu, D.; Lin, H.D. Phylogeographical analysis on Squalidus argentatus recapitulates historical landscapes and drainage evolution on the island of Taiwan and mainland China. Int. J. Mol. Sci. 2012, 13, 1405–1425. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Chenoweth, E.L.; Liu, Q. Population structure and genetic diversity of Sinibrama macrops from Ou River and Ling River based on mtDNA D-loop region analysis, China. Mitochondrial DNA Part A 2018, 29, 303–311. [Google Scholar] [CrossRef]
- Ding, X.H.; Hsu, K.C.; Tang, W.Q.; Liu, D.; Ju, Y.M.; Lin, H.D.; Yang, J.Q. Genetic diversity and structure of the Chinese lake gudgeon (Sarcocheilichthys sinensis). Mitochondrial DNA Part A 2020, 31, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Froese, R.; Pauly, D. (Eds.) FishBase; Version (08/2022). World Wide Web Electronic Publication. 2022. Available online: http://www.fishbase.org (accessed on 26 January 2023).
- Albert, J.S.; Craig, J.M.; Tagliacollo, V.A.; Petry, P. Upland and lowland fishes: A test of the river capture hypothesis. Mt. Clim. Biodivers. 2018, 273–294. [Google Scholar]
- Hou, X.J.; Lin, H.D.; Tang, W.Q.; Liu, D.; Han, C.C.; Yang, J.Q. Complete mitochondrial genome of the freshwater fish Acrossocheilus longipinnis (Teleostei: Cyprinidae): Genome characterization and phylogenetic analysis. Biologia 2020, 75, 1871–1880. [Google Scholar] [CrossRef]
- Kang, B.; Vitule, J.R.; Li, S.; Shuai, F.; Huang, L.; Huang, X.; Fang, J.; Shi, X.; Zhu, Y.; Xu, D.; et al. Introduction of non-native fish for aquaculture in China: A systematic review. Rev. Aquac. 2023, 15, 676–703. [Google Scholar] [CrossRef]
- Hall, T. BioEdit Version 7.0.0. Distributed by the Author. 2004. Available online: http://www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 26 January 2023).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. Mamm. Protein Metab. 1969, 3, 21–132. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pons, O.; Petit, R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, W.; Wu, J.; Li, C.; Ju, Y.M.; Lin, H.D.; Zhao, J. Multilocus Phylogeography and Population Genetic Analyses of Opsariichthys hainanensis Reveal Pleistocene Isolation Followed by High Gene Flow around the Gulf of Tonkin. Genes 2022, 13, 1908. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 26 March 2021).
- Rambaut, A.; Drummond, A.J. TreeAnnotator v1.8.2: MCMC Output Analysis. 2015. Available online: http://beast.bio.ed.ac.uk (accessed on 26 January 2023).
- Rambaut, A. Figtree, a Graphical Viewer of Phylogenetic Trees. 2016. Available online: http://tree.bio.ed.ac.uk/software/gtree (accessed on 20 December 2016).
- Tajima, F. The effect of change in population size on DNA polymorphism. Genetics 1989, 123, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Cornuet, J.M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.M.; Estoup, A. DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera, A.A.; Palsbøll, P.J. Inferring past demographic changes from contemporary genetic data: A simulation-based evaluation of the ABC methods implemented in diyabc. Mol. Ecol. Resour. 2017, 17, e94–e110. [Google Scholar] [CrossRef]
- Templeton, A.R. The “Eve” Hypotheses: A Genetic Critique and Reanalysis. Am. Anthropol. 1993, 95, 51–72. [Google Scholar] [CrossRef]
- Kang, B.; Deng, J.; Wu, Y.; Chen, L.; Zhang, J.; Qiu, H.; Liu, Y.; He, D. Mapping China’s freshwater fishes: Diversity and biogeography. Fish Fish. 2014, 15, 209–230. [Google Scholar] [CrossRef]
- Zheng, L.P.; Yang, J.X. Genetic diversity and population demography of the endemic species Acrossocheilus longipinnis (Teleostei, Cyprinidae) based on mtDNA COI and cyt b gene sequences. Mitochondrial DNA Part A 2018, 29, 403–408. [Google Scholar] [CrossRef]
- Zheng, L.P.; Yang, J.X. Genetic diversity and population structure of Acrossocheilus yunnanensis (Teleostei, Cyprinidae) inferred from four mitochondrial gene sequences. Mitochondrial DNA Part A 2018, 29, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.M.; Hsu, K.C.; Yang, J.Q.; Wu, J.H.; Li, S.; Wang, W.K.; Chen, C.W.; Lin, H.D. Mitochondrial diversity and phylogeography of Acrossocheilus paradoxus (Teleostei: Cyprinidae). Mitochondrial DNA Part A 2018, 29, 1194–1202. [Google Scholar] [CrossRef]
- Yu, D.; Chen, M.; Tang, Q.; Li, X.; Liu, H. Geological events and Pliocene climate fluctuations explain the phylogeographical pattern of the cold water fish Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 2014, 14, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prunier, J.G.; Dubut, V.; Loot, G.; Tudesque, L.; Blanchet, S. The relative contribution of river network structure and anthropogenic stressors to spatial patterns of genetic diversity in two freshwater fishes: A multiple-stressors approach. Freshw. Biol. 2018, 63, 6–21. [Google Scholar] [CrossRef]
- Ward, R.D.; Woodwark, M.; Skibinski, D.O.F. A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. J. Fish Biol. 1994, 44, 213–232. [Google Scholar] [CrossRef]
- Grant, W.S.; Bowen, B.W. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.Y. Monophyly, affinity and taxonomic revision of the Cyprinid Genus Acrossocheilus Oshima, 1919. Ph.D. Thesis, Graduate School of the Chinese Academy of Sciences, Beijing, China, 2009. [Google Scholar]
- Moritz, C. Defining ‘evolutionarily significant units’ for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef] [PubMed]
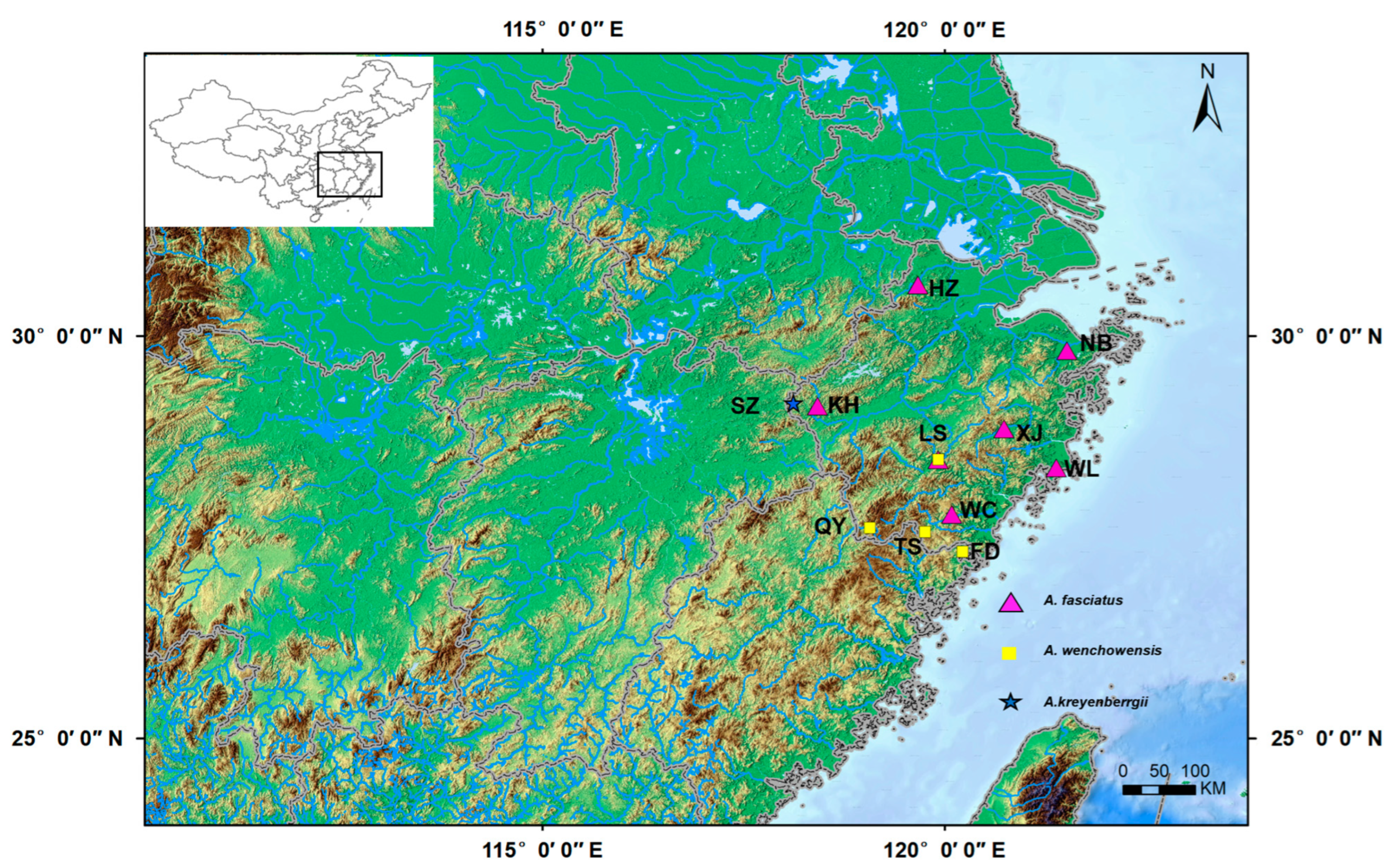



| Species | River System | Location (Abb.) | Haplotype | Latitude and Longitude | Sample Size (n) | Haplotype Number (Nh) | Haplotype Diversity (h) | Nucleotide Diversity (θπ) | Nucleotide Diversity (θw) |
|---|---|---|---|---|---|---|---|---|---|
| A. fasciatus | Tiaoxi River | Huzhou (AFHZ) | Hap1–4 | 30°38′ N, 119°40′ E | 8 | 4 | 0.643 | 0.00294 | 0.00454 |
| Yongjiang River | Ningbo (AFNB) | Hap56–68 | 29°49′ N, 121°31′ E | 15 | 13 | 0.971 | 0.01427 | 0.02380 | |
| Qiantang River | Kaihua (AFKH) | Hap5–44 | 29°08′ N, 118°25′ E | 49 | 40 | 0.990 | 0.01333 | 0.01831 | |
| Jiaojiang River | Xianju (AFXJ) | Hap77–88 | 28°51′ N, 120°44′ E | 16 | 12 | 0.942 | 0.01413 | 0.01940 | |
| Jinqing River | Wenling (AFWL) | Hap75–76 | 28°22′ N, 121°23′ E | 2 | 2 | 1.000 | 0.00490 | 0.00490 | |
| Oujiang Rive | Lishui (AFLS) | Hap45–55 | 28°28′ N, 119°55′ E | 14 | 11 | 0.934 | 0.00943 | 0.00895 | |
| Feiyun River | Wencheng (AFWC) | Hap69–74 | 27°47′ N, 120°05′ E | 6 | 6 | 1.000 | 0.00592 | 0.00580 | |
| 110 | 88 | 0.994 | 0.01835 | 0.03767 | |||||
| A. wenchowensis | Oujiang River | Lishui (AWLS) | Hap108–117 | 28°28′ N, 119°55′ E | 14 | 10 | 0.923 | 0.00712 | 0.00877 |
| Jiaoxi River | Qingyuan (AWQY) | Hap118–122 | 27°37′ N, 119°04′ E | 6 | 5 | 0.933 | 0.00124 | 0.00150 | |
| Feiyun River | Taishun (AWTS) | Hap123 | 27°34′ N, 119°45′ E | 4 | 1 | 0 | 0 | 0 | |
| Longshanxi River | Fuding (AWFD) | Hap98–107 | 27°19′ N, 120°13′ E | 11 | 10 | 0.982 | 0.01108 | 0.01423 | |
| 35 | 26 | 0.975 | 0.01346 | 0.02072 | |||||
| A. kreyenbergii | Yangtze River | Suzhuang (AKSZ) | Hap89–97 | 29°10′ N, 118°07′ E | 15 | 9 | 0.800 | 0.01622 | 0.01415 |
| Total | 160 | 123 | 0.994 | 0.03526 | 0.05524 |
| AFHZ | AFNB | AFKH | AFXJ | AFWL | AFLS | AFWC | AWLS | AWQY | AWTS | AWFD | AKSZ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFHZ | 0.000 | 0.000 | 0.000 | 0.063 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AFNB | 0.473 | 0.000 | 0.000 | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AFKH | 0.377 | 0.186 | 0.000 | 0.009 | 0.000 | 0.081 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AFXJ | 0.737 | 0.562 | 0.580 | 0.513 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AFWL | 0.900 | 0.735 | 0.751 | 0.098 | 0.000 | 0.045 | 0.009 | 0.054 | 0.072 | 0.000 | 0.009 | |
| AFLS | 0.386 | 0.240 | 0.131 | 0.630 | 0.802 | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AFWC | 0.554 | 0.258 | 0.119 | 0.657 | 0.839 | 0.198 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AWLS | 0.905 | 0.796 | 0.800 | 0.817 | 0.904 | 0.838 | 0.865 | 0.000 | 0.000 | 0.000 | 0.000 | |
| AWQY | 0.960 | 0.846 | 0.852 | 0.863 | 0.952 | 0.891 | 0.922 | 0.538 | 0.009 | 0.000 | 0.000 | |
| AWTS | 0.971 | 0.851 | 0.857 | 0.869 | 0.962 | 0.899 | 0.932 | 0.771 | 0.948 | 0.000 | 0.000 | |
| AWFD | 0.881 | 0.778 | 0.786 | 0.793 | 0.878 | 0.819 | 0.845 | 0.500 | 0.614 | 0.749 | 0.000 | |
| AKSZ | 0.822 | 0.734 | 0.733 | 0.736 | 0.821 | 0.765 | 0.790 | 0.820 | 0.858 | 0.859 | 0.804 | |
| Scheme Category Description | % Var. | Statistic | p |
|---|---|---|---|
| Scenario I: three species groups (A. fasciatus, A. wenchowensis and A. kreyenbergii) | |||
| Among groups | 63.31 | FCT = 0.633 | 0.000 |
| Among populations within groups | 16.02 | FSC = 0.436 | 0.000 |
| Within populations | 20.66 | FST = 0.793 | 0.000 |
| Scenario II: two geographical groups primarily divided by the Xianxia Mountains in A. fasciatus (HZ, KH, NB, XJ) (WL) | |||
| Among groups | 27.03 | FCT = −0.270 | 0.402 |
| Among populations within groups | 32.96 | FSC = 0.451 | 0.000 |
| Within populations | 40.02 | FST = 0.599 | 0.000 |
| Scenario III: two geographical groups primarily divided by the Qiantang and Tiaoxi Rivers and others in A. fasciatus (HZ, KH, LS, WC, WL) (NB, XJ) | |||
| Among groups | 10.15 | FCT = 0.101 | 0.189 |
| Among populations within groups | 34.32 | FSC = 0.381 | 0.000 |
| Within populations | 55.53 | FST = 0.444 | 0.000 |
| Scenario IV: two geographical groups primarily divided by the Feiyun River and others in A. wenchowensis (TS) (LS, FD, QY) | |||
| Among groups | 25.03 | FCT = 0.250 | 0.245 |
| Among populations within groups | 40.30 | FSC = 0.537 | 0.000 |
| Within populations | 34.67 | FST = 0.653 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.-Y.; Wang, J.-J.; Ren, J.-F.; Li, F.; Wu, J.-X.; Zhou, J.-J.; Li, J.-L.; Yang, J.-Q.; Lin, H.-D. Historical Landscape Evolution Shaped the Phylogeography and Population History of the Cyprinid Fishes of Acrossocheilus (Cypriniformes: Cyprinidae) According to Mitochondrial DNA in Zhejiang Province, China. Diversity 2023, 15, 425. https://doi.org/10.3390/d15030425
Zhou M-Y, Wang J-J, Ren J-F, Li F, Wu J-X, Zhou J-J, Li J-L, Yang J-Q, Lin H-D. Historical Landscape Evolution Shaped the Phylogeography and Population History of the Cyprinid Fishes of Acrossocheilus (Cypriniformes: Cyprinidae) According to Mitochondrial DNA in Zhejiang Province, China. Diversity. 2023; 15(3):425. https://doi.org/10.3390/d15030425
Chicago/Turabian StyleZhou, Mu-Yang, Jun-Jie Wang, Jian-Feng Ren, Fan Li, Jin-Xian Wu, Jia-Jun Zhou, Jia-Le Li, Jin-Quan Yang, and Hung-Du Lin. 2023. "Historical Landscape Evolution Shaped the Phylogeography and Population History of the Cyprinid Fishes of Acrossocheilus (Cypriniformes: Cyprinidae) According to Mitochondrial DNA in Zhejiang Province, China" Diversity 15, no. 3: 425. https://doi.org/10.3390/d15030425
APA StyleZhou, M.-Y., Wang, J.-J., Ren, J.-F., Li, F., Wu, J.-X., Zhou, J.-J., Li, J.-L., Yang, J.-Q., & Lin, H.-D. (2023). Historical Landscape Evolution Shaped the Phylogeography and Population History of the Cyprinid Fishes of Acrossocheilus (Cypriniformes: Cyprinidae) According to Mitochondrial DNA in Zhejiang Province, China. Diversity, 15(3), 425. https://doi.org/10.3390/d15030425








