Fungal Diversity Associated with Armadillidium Isopods: A Case Study in Central Park of Gwacheon, South Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Preparation for Illumina MiSeq
2.3. Bioinformatics and Statistics
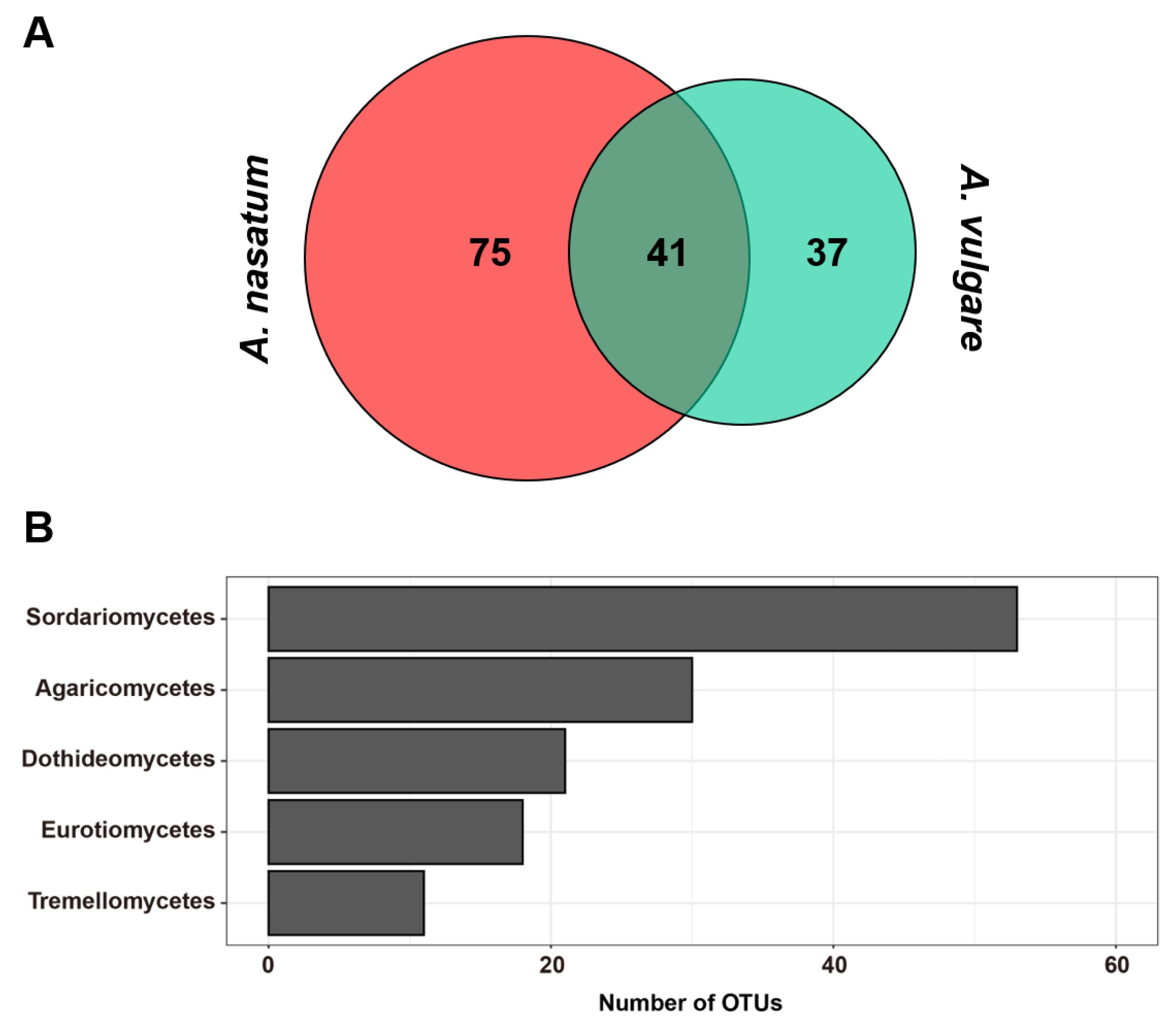
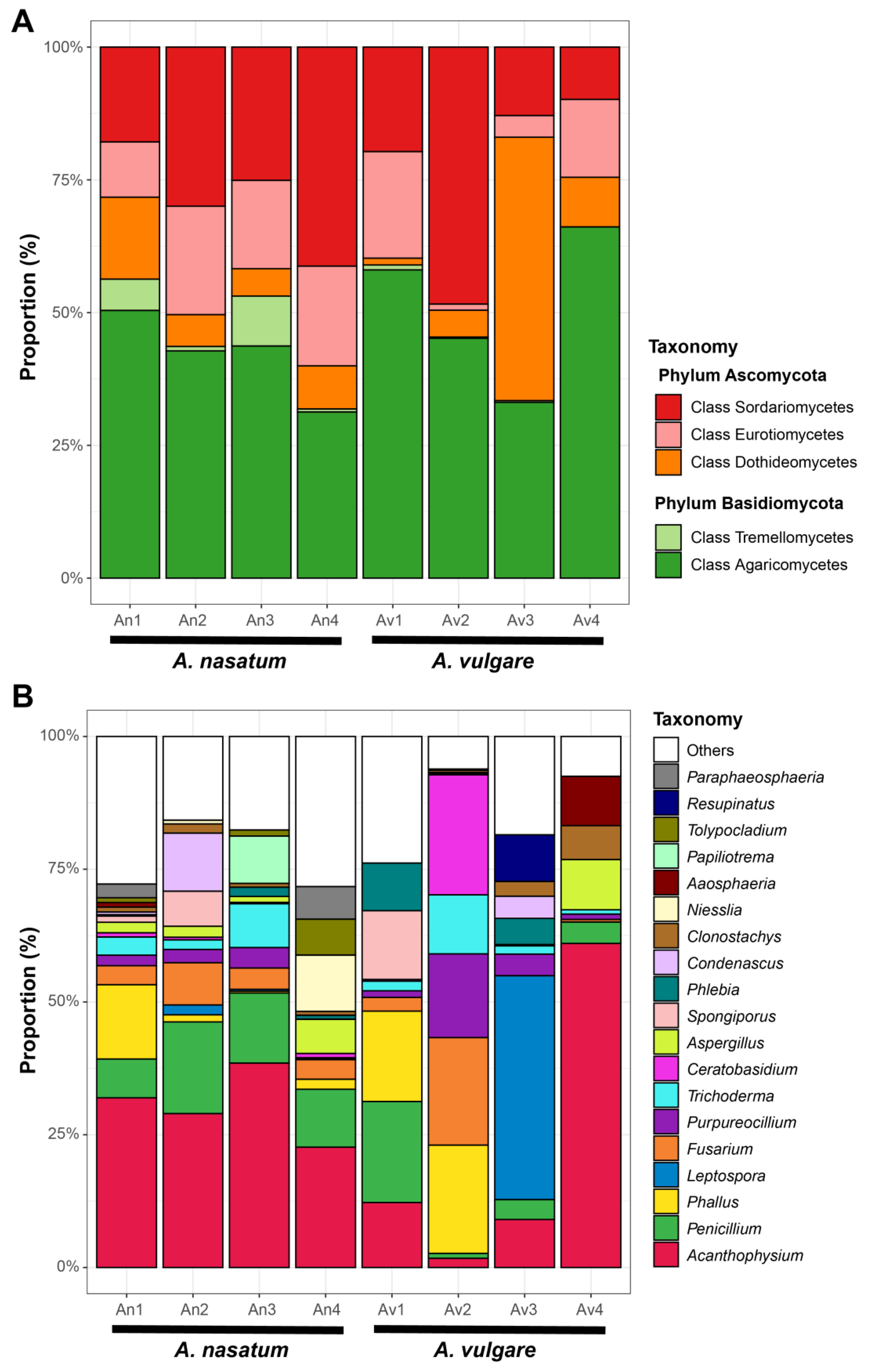
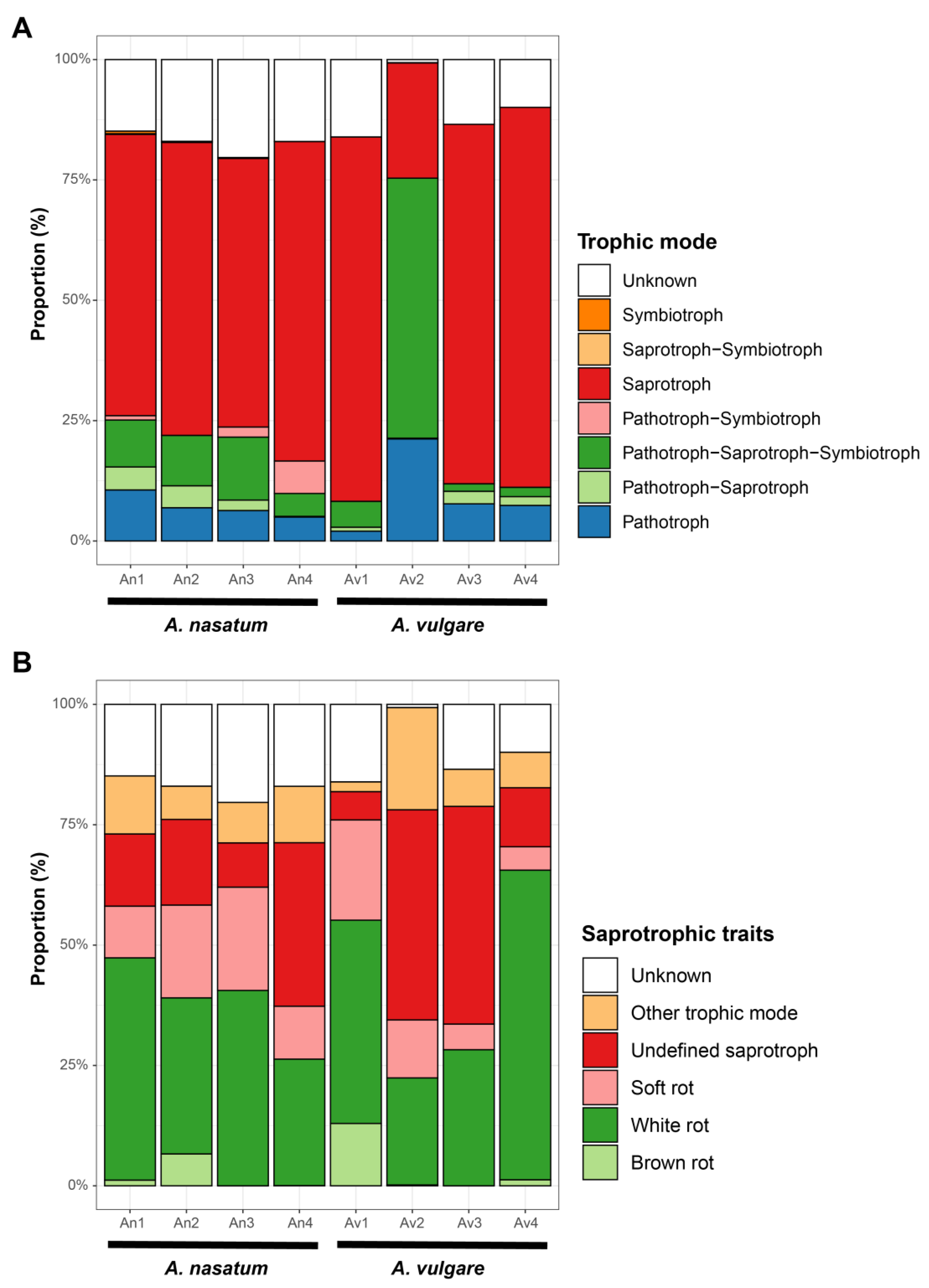
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zimmer, M. Nutrition in Terrestrial Isopods (Isopoda: Oniscidea): An Evolutionary-Ecological Approach. Biol. Rev. 2002, 77, 455–493. [Google Scholar] [CrossRef] [PubMed]
- Schmalfuss, H. World Catalog of Terrestrial Isopods (Isopoda: Oniscidea). Stuttgarter Beiträge zur Naturkunde, Serie A 2003, 654, 1–341. [Google Scholar]
- Sfenthourakis, S.; Taiti, S. Patterns of Taxonomic Diversity among Terrestrial Isopods. ZooKeys 2015, 515, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C. Phylogeny of the Terrestrial Isopoda (Oniscidea): A Review. Arthropod Syst. Phylogeny 2008, 66, 191–226. [Google Scholar]
- Faberi, A.J.; López, A.N.; Clemente, N.L.; Manetti, P.L. Importance of Diet in the Growth, Survivorship and Reproduction of the No-Tillage Pest Armadillidium vulgare (Crustacea: Isopoda). Rev. Chil. Hist. Nat. 2011, 84, 407–417. [Google Scholar] [CrossRef]
- Špaldoňová, A.; Frouz, J. The Role of Armadillidium vulgare (Isopoda: Oniscidea) in Litter Decomposition and Soil Organic Matter Stabilization. Appl. Soil Ecol. 2014, 83, 186–192. [Google Scholar] [CrossRef]
- Zimmer, M.; Pennings, S.C.; Buck, T.L.; Carefoot, T.H. Species-Specific Patterns of Litter Processing by Terrestrial Isopods (Isopoda: Oniscidea) in High Intertidal Salt Marshes and Coastal Forests. Funct. Ecol. 2002, 16, 596–607. [Google Scholar] [CrossRef] [Green Version]
- Zimmer, M.; Pennings, S.C.; Buck, T.L.; Carefoot, T.H. Salt Marsh Litter and Detritivores: A Closer Look at Redundancy. Estuaries 2004, 27, 753–769. [Google Scholar] [CrossRef]
- Elisabeth, H. Evolutionary Adaptation of Oniscidean Isopods to Terrestrial Life: Structure, Physiology and Behavior. Terr. Arthropod Rev. 2011, 4, 95–130. [Google Scholar] [CrossRef]
- Guo, S.; Ren, M.; Song, S.; Wei, P.; Luo, J. Evaluation of Antinociceptive and Anti-Inflammatory Effects of Aqueous Extract of Armadillidium Vulgare Latreille. Chin. J. Integr. Med. 2017, 23, 138–145. [Google Scholar] [CrossRef]
- Alves, R.R.; Alves, H.N. The Faunal Drugstore: Animal-Based Remedies Used in Traditional Medicines in Latin America. J. Ethnobiol. Ethnomedicine 2011, 7, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Liu, X.; Mikaye, M.S.; Zhao, H.; Zhang, Y. Traditional Chinese medicine in the treatment of high incidence diseases in cold areas: The thrombotic diseases. Frigid Zone Med. 2021, 1, 23–44. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Liu, Y.; Mi, H.; Jiang, X.; Sun, Y.; Zhao, H.; Chen, D.; Wang, L. A Comprehensive Evaluation of the Potential of Semiterrestrial Isopods, Ligia Exotica, as a New Animal Food. Sci. Rep. 2021, 11, 7213. [Google Scholar] [CrossRef] [PubMed]
- McMonigle, O. Pillbugs and Other Isopods: Cultivating Vivarium Clean-Up Crews and Feeders for Dart Frogs, Arachnids, and Insects; Coachwhip Publications: Greenville, OH, USA, 2013. [Google Scholar]
- Cordovez, V.; Dini-Andreote, F.; Carrion, V.; Raaijmakers, J. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevelline, B.K.; Kohl, K.D. The Gut Microbiome Influences Host Diet Selection Behavior. Proc. Natl. Acad. Sci. USA 2022, 119, e2117537119. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of Microorganisms in the Evolution of Animals and Plants: The Hologenome Theory of Evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Gilbert, S.F.; McDonald, E.; Boyle, N.; Buttino, N.; Gyi, L.; Mai, M.; Prakash, N.; Robinson, J. Symbiosis as a Source of Selectable Epigenetic Variation: Taking the Heat for the Big Guy. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Feldhaar, H. Bacterial Symbionts as Mediators of Ecologically Important Traits of Insect Hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. Speciation by Symbiosis. Trends Ecol. Evol. 2012, 27, 443–451. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. The Hologenomic Basis of Speciation: Gut Bacteria Cause Hybrid Lethality in the Genus Nasonia. Science 2013, 341, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Esposti, M.D.; Romero, E.M. The Functional Microbiome of Arthropods. PLoS ONE 2017, 12, e0176573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, T.-Z.; Qi, F.-H.; Wang, Z.-Y. Most Dominant Roles of Insect Gut Bacteria: Digestion, Detoxification, or Essential Nutrient Provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malassigné, S.; Minard, G.; Vallon, L.; Martin, E.; Valiente Moro, C.; Luis, P. Diversity and Functions of Yeast Communities Associated with Insects. Microorganisms 2021, 9, 1552. [Google Scholar] [CrossRef] [PubMed]
- Schapheer, C.; Pellens, R.; Scherson, R. Arthropod-Microbiota Integration: Its Importance for Ecosystem Conservation. Front. Microbiol. 2021, 12, 702763. [Google Scholar] [CrossRef]
- Dittmer, J.; Lesobre, J.; Moumen, B.; Bouchon, D. Host Origin and Tissue Microhabitat Shaping the Microbiota of the Terrestrial Isopod Armadillidium vulgare. FEMS Microbiol. Ecol. 2016, 92, fiw063. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, M.A.; Douglas, A.; Piertney, S.B. Microbiome Composition within a Sympatric Species Complex of Intertidal Isopods (Jaera albifrons). PLoS ONE 2018, 13, e0202212. [Google Scholar] [CrossRef] [Green Version]
- Delhoumi, M.; Catania, V.; Zaabar, W.; Tolone, M.; Quatrini, P.; Achouri, M.S. The Gut Microbiota Structure of the Terrestrial Isopod Porcellionides pruinosus (Isopoda: Oniscidea). Eur. Zool. J. 2020, 87, 357–368. [Google Scholar] [CrossRef]
- Newbound, M.; Mccarthy, M.A.; Lebel, T. Fungi and the Urban Environment: A Review. Landsc. Urban Plan. 2010, 96, 138–145. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in Aquatic Ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [Green Version]
- Bahram, M.; Netherway, T. Fungi as Mediators Linking Organisms and Ecosystems. FEMS Microbiol. Rev. 2022, 46, fuab058. [Google Scholar] [CrossRef] [PubMed]
- Cafaro, M.J. Baltomyces, a New Genus of Gut-Inhabiting Fungus in an Isopod. Mycologia 1999, 91, 517–519. [Google Scholar] [CrossRef]
- Cafaro, M.J. Gut Fungi of Isopods: The Genus Palavascia. Mycologia 2000, 92, 361–369. [Google Scholar] [CrossRef]
- White, M.M. Legerioides, a New Genus of Harpellales in Isopods and Other Trichomycetes from New England, USA. Mycologia 1999, 91, 1021–1030. [Google Scholar] [CrossRef]
- Jaber, S.; Mercier, A.; Knio, K.; Brun, S.; Kambris, Z. Isolation of Fungi from Dead Arthropods and Identification of a New Mosquito Natural Pathogen. Parasit. Vectors 2016, 9, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Wang, J.; Liu, J.; Zhu, H.; Sun, B.; Wang, J.; Zhang, J.; Luo, Z.; Yao, G.; Xue, Y.; et al. Armochaetoglobins A–J: Cytochalasan Alkaloids from Chaetomium globosum TW1-1, a Fungus Derived from the Terrestrial Arthropod Armadillidium vulgare. J. Nat. Prod. 2015, 78, 1193–1201. [Google Scholar] [CrossRef]
- Han, W.-B.; Wang, G.-Y.; Tang, J.-J.; Wang, W.-J.; Liu, H.; Gil, R.R.; Navarro-Vázquez, A.; Lei, X.; Gao, J.-M. Herpotrichones A and B, Two Intermolecular [4 + 2] Adducts with Anti-Neuroinflammatory Activity from a Herpotrichia Species. Org. Lett. 2020, 22, 405–409. [Google Scholar] [CrossRef]
- Zhai, Y.-J.; Huo, G.-M.; Wei, J.; Lin, L.-B.; Zhang, Q.; Li, J.-N.; Chen, X.; Han, W.-B.; Gao, J.-M. Structures and Absolute Configurations of Butenolide Derivatives from the Isopod-Associated Fungus Pidoplitchkoviella terricola. Phytochemistry 2022, 193, 112981. [Google Scholar] [CrossRef]
- Crowther, T.W.; Stanton, D.W.G.; Thomas, S.M.; A’Bear, A.D.; Hiscox, J.; Jones, T.H.; Voříšková, J.; Baldrian, P.; Boddy, L. Top-down Control of Soil Fungal Community Composition by a Globally Distributed Keystone Consumer. Ecology 2013, 94, 2518–2528. [Google Scholar] [CrossRef]
- Póss, A.M.; Bogdányi, F.T.; Tóth, F. Consumption of Fungi-Infected Fallen Pear Leaves by the Common Woodlouse. Acta Phytopathol. Entomol. Hung. 2022, 57, 79–91. [Google Scholar] [CrossRef]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.-M.; Huntemann, M.; et al. A Genomic Catalog of Earth’s Microbiomes. Nat. Biotechnol. 2021, 39, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshizawa, S. The OceanDNA MAG Catalog Contains over 50,000 Prokaryotic Genomes Originated from Various Marine Environments. Sci. Data 2022, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global Diversity and Geography of Soil Fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Anslan, S.; Nilsson, R.H.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L.; Bahram, M. Great Differences in Performance and Outcome of High-Throughput Sequencing Data Analysis Platforms for Fungal Metabarcoding. MycoKeys 2018, 39, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; de Farias, A.R.G.; Sun, Y.-R.; Wijesinghe, S.N.; Raza, M.; Bao, D.-F.; Lu, L.; Tibpromma, S.; et al. The Numbers of Fungi: Contributions from Traditional Taxonomic Studies and Challenges of Metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Kwon, D.H. Terrestrial Isopoda (Crustacea) from Korea. Korean J. Zool. 1993, 36, 133–158. [Google Scholar]
- Kwon, D.H. Terrestrial Isopoda (Crustacea) from Cheju Island, Korea. Anim. Syst. Evol. Divers. 1995, 11, 509–538. [Google Scholar]
- Song, J.-H. A New Record of Porcellio scaber (Isopoda: Oniscidea: Porcellionidae) from South Korea, with Notes on Its Variation. Anim. Syst. Evol. Divers. 2020, 36, 309–315. [Google Scholar] [CrossRef]
- Shultz, J.W. A Guide to the Identification of the Terrestrial Isopoda of Maryland, U.S.A. (Crustacea). ZooKeys 2018, 801, 207–228. [Google Scholar] [CrossRef]
- Kim, E.J.; Moon, D.H.; Jo, U.B. Taxonomic Study on the Four Species of Terrestrial Isopods, Oniscoidae, from the Pusan Area in Korea. J. Sci. 1990, 49, 225–251. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.L. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, H.; Hildebrand, F.; et al. Shotgun Metagenomes and Multiple Primer Pair-Barcode Combinations of Amplicons Reveal Biases in Metabarcoding Analyses of Fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Tedersoo, L.; Tooming-Klunderud, A.; Anslan, S. PacBio Metabarcoding of Fungi and Other Eukaryotes: Errors, Biases and Perspectives. New Phytol. 2018, 217, 1370–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Wickham, H. Package ‘Ggplot2′: Elegant Graphics for Data Analysis, Version 3.2.1.; Springer: New York, NY, USA, 2016. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, Version 2.6-4. 2020, 1–263. Available online: https://CRAN.R-project.org/package=vegan (accessed on 20 November 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Paris, O.H. The Ecology of Armadillidium vulgare (Isopoda: Oniscoidea) in California Grassland: Food, Enemies, and Weather. Ecol. Monogr. 1963, 33, 1–22. [Google Scholar] [CrossRef]
- Crowther, T.W.; Boddy, L.; Hefin Jones, T. Functional and Ecological Consequences of Saprotrophic Fungus–Grazer Interactions. ISME J. 2012, 6, 1992–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowther, T.W.; Thomas, S.M.; Maynard, D.S.; Baldrian, P.; Covey, K.; Frey, S.D.; van Diepen, L.T.A.; Bradford, M.A. Biotic Interactions Mediate Soil Microbial Feedbacks to Climate Change. Proc. Natl. Acad. Sci. USA 2015, 112, 7033–7038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Li, Y.; Wang, H.; Zu, Y. Differences in the Activities of Eight Enzymes from Ten Soil Fungi and Their Possible Influences on the Surface Structure, Functional Groups, and Element Composition of Soil Colloids. PLoS ONE 2014, 9, e111740. [Google Scholar] [CrossRef] [PubMed]
- de Passos, D.F.; Pereira, N.; Castro, A.M. de A Comparative Review of Recent Advances in Cellulases Production by Aspergillus, Penicillium and Trichoderma Strains and Their Use for Lignocellulose Deconstruction. Curr. Opin. Green Sustain. Chem. 2018, 14, 60–66. [Google Scholar] [CrossRef]
- Yoon, S.-M.; Kim, Y.-S.; Kim, Y.-K.; Kim, T.-J. A Novel Endo-β-1,4-Xylanase from Acanthophysium sp. KMF001, a Wood Rotting Fungus. J. Korean Wood Sci. Technol. 2018, 46, 670–680. [Google Scholar] [CrossRef]
- El-Shanshoury, A.R.; Mona, M.H.; Shoukr, F.A.; El-Bossery, A.M. The Enumeration and Characterization of Bacteria and Fungi Associated with Marine Wood-Boring Isopods, and the Ability of These Microorganisms to Digest Cellulose and Wood. Mar. Biol. 1994, 119, 321–326. [Google Scholar] [CrossRef]
- Banerjee, S.; Maiti, T.K.; Roy, R.N. Enzyme Producing Insect Gut Microbes: An Unexplored Biotechnological Aspect. Crit. Rev. Biotechnol. 2022, 42, 384–402. [Google Scholar] [CrossRef]
- Daniel, G.; Nilsson, T.; Cragg, S. Limnoria lignorum Ingest Bacterial and Fungal Degraded Wood. Holz Als Roh-Werkst. 1991, 49, 488–490. [Google Scholar] [CrossRef]
- Wong, D.W.S. Structure and Action Mechanism of Ligninolytic Enzymes. Appl. Biochem. Biotechnol. 2009, 157, 174–209. [Google Scholar] [CrossRef]
- Cragg, S.M.; Beckham, G.T.; Bruce, N.C.; Bugg, T.D.; Distel, D.L.; Dupree, P.; Etxabe, A.G.; Goodell, B.S.; Jellison, J.; McGeehan, J.E.; et al. Lignocellulose Degradation Mechanisms across the Tree of Life. Curr. Opin. Chem. Biol. 2015, 29, 108–119. [Google Scholar] [CrossRef] [Green Version]


| Taxonomy | Genus | An1 | An2 | An3 | An4 | Av1 | Av2 | Av3 | Av4 |
|---|---|---|---|---|---|---|---|---|---|
| Phylum Ascomycota | |||||||||
| unidentified; unidentified; unidentified | unidentified | <0.01 | <0.01 | <0.01 | <0.01 | 0.75 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Botryosphaeriales; Aplosporellaceae | Aplosporella | 5.49 | <0.01 | <0.01 | 0.20 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Cladosporiales; Cladosporiaceae | Cladosporium | 0.19 | <0.01 | 0.99 | 0.68 | 0.45 | 0.05 | 5.31 | <0.01 |
| Dothideomycetes; Dothideales; Saccotheciaceae | Aureobasidium | <0.01 | <0.01 | <0.01 | 0.09 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Dothideomycetes_inc * | Leptospora | 0.02 | 1.86 | 0.35 | <0.01 | <0.01 | <0.01 | 42.17 | <0.01 |
| Dothideomycetes; Mycosphaerellales; Mycosphaerellaceae | Cercospora | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 5.00 | <0.01 | <0.01 |
| Dothideomycetes; Mycosphaerellales; Teratosphaeriaceae | unidentified | 0.55 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Pleosporales; unidentified | unidentified | 0.52 | 0.38 | 0.86 | 0.14 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Pleosporales; Amorosiaceae | unidentified | <0.01 | 0.73 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Pleosporales; Didymellaceae | unidentified | <0.01 | 0.96 | <0.01 | <0.01 | <0.01 | <0.01 | 0.31 | <0.01 |
| Dothideomycetes; Pleosporales; Didymosphaeriaceae | Paraphaeosphaeria | 2.58 | <0.01 | <0.01 | 6.13 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Pleosporales; Nigrogranaceae | Nigrograna | 1.19 | 1.05 | 1.75 | 0.09 | 0.79 | <0.01 | 0.69 | <0.01 |
| Dothideomycetes; Pleosporales; Phaeosphaeriaceae | unidentified | 3.02 | <0.01 | <0.01 | 0.11 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Pleosporales; Pleosporaceae | Curvularia | 0.77 | 0.80 | <0.01 | 0.05 | <0.01 | <0.01 | <0.01 | <0.01 |
| Dothideomycetes; Pleosporales; Pleosporales_inc | Aaosphaeria | 0.84 | <0.01 | <0.01 | 0.03 | <0.01 | <0.01 | <0.01 | 9.31 |
| Dothideomycetes; Pleosporales; Thyridariaceae | Xenoroussoella | <0.01 | <0.01 | 0.94 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Eurotiomycetes; Chaetothyriales; Cyphellophoraceae | Cyphellophora | <0.01 | 0.36 | 1.48 | <0.01 | <0.01 | 0.09 | <0.01 | 1.14 |
| Eurotiomycetes; Chaetothyriales; Herpotrichiellaceae | unidentified | 0.94 | <0.01 | <0.01 | 0.02 | <0.01 | <0.01 | <0.01 | <0.01 |
| Eurotiomycetes; Chaetothyriales; Herpotrichiellaceae | Exophiala | <0.01 | <0.01 | <0.01 | <0.01 | 0.82 | <0.01 | <0.01 | <0.01 |
| Eurotiomycetes; Eurotiales; unidentified | unidentified | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Eurotiomycetes; Eurotiales; Aspergillaceae | Aspergillus | 1.99 | 2.06 | 1.10 | 6.41 | <0.01 | 0.13 | 0.24 | 9.47 |
| Eurotiomycetes; Eurotiales; Aspergillaceae | Penicillium | 7.31 | 17.26 | 13.21 | 10.90 | 19.02 | 0.93 | 3.75 | 4.04 |
| Lecanoromycetes; unidentified; unidentified | unidentified | <0.01 | <0.01 | <0.01 | 0.01 | <0.01 | 0.01 | <0.01 | <0.01 |
| Lecanoromycetes; Lecanorales; Lecanorineae | Xanthoparmelia | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.04 | <0.01 | <0.01 |
| Leotiomycetes; Helotiales; unidentified | unidentified | 0.73 | <0.01 | <0.01 | 0.02 | 0.33 | <0.01 | <0.01 | <0.01 |
| Leotiomycetes; Leotiomycetes_inc; Myxotrichaceae | Oidiodendron | <0.01 | <0.01 | 0.96 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Lichinomycetes; Lichinales; Lichinaceae | Phylliscum | 0.54 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; unidentified; unidentified | unidentified | <0.01 | <0.01 | <0.01 | <0.01 | 0.47 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Coniochaetales; Coniochaetaceae | Coniochaeta | 1.67 | <0.01 | 0.56 | 0.07 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Diaporthales; Gnomoniaceae | Ophiognomonia | <0.01 | <0.01 | <0.01 | <0.01 | 0.09 | 0.01 | <0.01 | 0.65 |
| Sordariomycetes; Hypocreales; unidentified | unidentified | 0.18 | <0.01 | <0.01 | <0.01 | 3.39 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Bionectriaceae | Clonostachys | 0.93 | 1.72 | 0.71 | 0.73 | <0.01 | 0.43 | 2.80 | 6.37 |
| Sordariomycetes; Hypocreales; Hypocreaceae | Trichoderma | 3.43 | 1.81 | 8.24 | 0.09 | 1.81 | 11.13 | 1.57 | 0.81 |
| Sordariomycetes; Hypocreales; Hypocreales_inc | Acremonium | <0.01 | 0.21 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Hypocreales_inc | Xenoacrodontium | <0.01 | <0.01 | <0.01 | 1.38 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Nectriaceae | unidentified | 0.17 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Nectriaceae | Cosmospora | <0.01 | 0.37 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Nectriaceae | Fusarium | 3.58 | 7.95 | 4.01 | 3.67 | 2.59 | 20.23 | <0.01 | 0.48 |
| Sordariomycetes; Hypocreales; Nectriaceae | Mariannaea | 0.08 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Niessliaceae | Niesslia | <0.01 | 0.73 | <0.01 | 10.60 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Ophiocordycipitaceae | Purpureocillium | 1.99 | 2.49 | 3.86 | 0.26 | 1.24 | 15.77 | 4.05 | 1.01 |
| Sordariomycetes; Hypocreales; Ophiocordycipitaceae | Tolypocladium | 0.91 | <0.01 | 1.14 | 6.75 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Hypocreales; Sarocladiaceae | Sarocladium | <0.01 | <0.01 | <0.01 | <0.01 | 1.48 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Microascales; Microascaceae | Microascus | 0.05 | <0.01 | <0.01 | <0.01 | 0.58 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Sordariales; Cephalothecaceae | Phialemonium | <0.01 | 0.23 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Sordariales; Chaetomiaceae | unidentified | <0.01 | 0.07 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Sordariales; Chaetomiaceae | Chaetomium | 0.73 | <0.01 | 1.13 | 0.03 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Sordariales; Chaetomiaceae | Collariella | <0.01 | <0.01 | 1.14 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Sordariales; Chaetomiaceae | Condenascus | 0.51 | 10.93 | <0.01 | 0.02 | <0.01 | 0.18 | 4.18 | <0.01 |
| Sordariomycetes; Sordariales; Schizotheciaceae | Schizothecium | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.56 | <0.01 | <0.01 |
| Sordariomycetes; Sordariomycetes_inc; Phomatosporaceae | Phomatospora | 1.91 | 0.13 | <0.01 | 0.08 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; unidentified | unidentified | 0.63 | <0.01 | 1.65 | 0.03 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Apiosporaceae | Apiospora | 0.27 | <0.01 | 0.69 | 0.02 | <0.01 | <0.01 | <0.01 | 0.53 |
| Sordariomycetes; Xylariales; Diatrypaceae | Peroneutypa | <0.01 | <0.01 | <0.01 | 3.65 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Hypoxylaceae | Daldinia | 0.07 | 0.01 | 0.04 | 4.97 | <0.01 | 0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Sporocadaceae | Discosia | <0.01 | <0.01 | <0.01 | <0.01 | 7.79 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Sporocadaceae | Pestalotiopsis | 0.22 | 0.46 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Xylariaceae | Biscogniauxia | 0.26 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Xylariaceae | Nemania | <0.01 | 1.73 | <0.01 | 5.73 | <0.01 | 0.04 | <0.01 | <0.01 |
| Sordariomycetes; Xylariales; Xylariaceae | Nodulisporium | <0.01 | <0.01 | 0.67 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Phylum Basidiomycota | |||||||||
| unidentified | unidentified | 5.74 | <0.01 | <0.01 | 4.78 | 0.12 | <0.01 | <0.01 | <0.01 |
| Agaricomycetes | unidentified | <0.01 | 0.25 | 0.45 | <0.01 | <0.01 | <0.01 | 3.43 | <0.01 |
| Agaricomycetes; Agaricales; unidentified | unidentified | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.05 | <0.01 | <0.01 |
| Agaricomycetes; Agaricales; Omphalotaceae | Gymnopus | <0.01 | <0.01 | <0.01 | <0.01 | 3.42 | <0.01 | <0.01 | <0.01 |
| Agaricomycetes; Agaricales; Tricholomataceae | Resupinatus | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 8.78 | <0.01 |
| Agaricomycetes; Cantharellales; Ceratobasidiaceae | Ceratobasidium | 0.78 | 0.50 | 0.27 | 0.82 | 0.32 | 22.59 | <0.01 | <0.01 |
| Agaricomycetes; Corticiales; Corticiaceae | Laetisaria | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.17 | <0.01 |
| Agaricomycetes; Hymenochaetales; Hymenochaetaceae | Hydnoporia | 0.14 | <0.01 | 0.18 | <0.01 | <0.01 | 0.01 | <0.01 | <0.01 |
| Agaricomycetes; Hymenochaetales; Tubulicrinaceae | Tubulicrinis | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.03 | <0.01 | 1.07 |
| Agaricomycetes; Phallales; Phallaceae | Phallus | 13.98 | 1.34 | 0.34 | 1.90 | 17.02 | 20.41 | <0.01 | <0.01 |
| Agaricomycetes; Polyporales; unidentified | unidentified | 0.14 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.47 |
| Agaricomycetes; Polyporales; Dacryobolaceae | Dacryobolus | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.05 | <0.01 | 1.26 |
| Agaricomycetes; Polyporales; Dacryobolaceae | Spongiporus | 1.18 | 6.63 | <0.01 | 0.05 | 12.95 | 0.13 | <0.01 | <0.01 |
| Agaricomycetes; Polyporales; Irpicaceae | unidentified | 1.12 | 1.40 | <0.01 | 0.04 | 0.22 | 0.06 | <0.01 | <0.01 |
| Agaricomycetes; Polyporales; Irpicaceae | Efibula | <0.01 | <0.01 | <0.01 | 1.71 | <0.01 | <0.01 | <0.01 | <0.01 |
| Agaricomycetes; Polyporales; Irpicaceae | Flavodon | <0.01 | 1.79 | <0.01 | 0.99 | <0.01 | 0.03 | 0.15 | 0.66 |
| Agaricomycetes; Polyporales; Irpicaceae | Meruliopsis | <0.01 | <0.01 | <0.01 | <0.01 | 0.57 | <0.01 | <0.01 | <0.01 |
| Agaricomycetes; Polyporales; Meruliaceae | Phlebia | 0.25 | <0.01 | 1.76 | 0.71 | 8.98 | <0.01 | 4.91 | <0.01 |
| Agaricomycetes; Polyporales; Phanerochaetaceae | Phaeophlebiopsis | <0.01 | <0.01 | <0.01 | <0.01 | 1.63 | <0.01 | <0.01 | <0.01 |
| Agaricomycetes; Polyporales; Phanerochaetaceae | Phanerochaete | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.05 | <0.01 | 1.55 |
| Agaricomycetes; Polyporales; Steccherinaceae | Cabalodontia | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 5.40 | <0.01 |
| Agaricomycetes; Russulales; Stereaceae | unidentified | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.46 | <0.01 |
| Agaricomycetes; Russulales; Stereaceae | Acanthophysium | 31.95 | 28.99 | 38.49 | 22.66 | 12.24 | 1.72 | 9.04 | 61.03 |
| Agaricomycetes; Trechisporales; Trechisporales_inc | Sistotremastrum | <0.01 | 0.29 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Cystobasidiomycetes; Cystobasidiales; unidentified | unidentified | <0.01 | 0.25 | <0.01 | 0.20 | <0.01 | <0.01 | <0.01 | <0.01 |
| Cystobasidiomycetes; Cystobasidiomycetes_inc; Symmetrosporaceae | Symmetrospora | 0.21 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Exobasidiomycetes; Golubeviales; Golubeviaceae | Golubevia | <0.01 | <0.01 | <0.01 | 2.69 | <0.01 | <0.01 | <0.01 | <0.01 |
| Exobasidiomycetes; Microstromatales; Microstromatales_inc | Jaminaea | <0.01 | <0.01 | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Malasseziomycetes; Malasseziales; Malasseziaceae | Malassezia | <0.01 | 3.25 | <0.01 | <0.01 | <0.01 | 0.05 | 2.29 | <0.01 |
| Microbotryomycetes; Sporidiobolales; Sporidiobolaceae | Rhodotorula | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.16 |
| Tremellomycetes; Filobasidiales; Filobasidiaceae | Naganishia | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.02 | <0.01 |
| Tremellomycetes; Tremellales; Bulleribasidiaceae | Dioszegia | <0.01 | 0.54 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Tremellomycetes; Tremellales; Bulleribasidiaceae | Hannaella | 0.29 | 0.26 | <0.01 | 0.51 | <0.01 | <0.01 | 0.28 | <0.01 |
| Tremellomycetes; Tremellales; Rhynchogastremataceae | Papiliotrema | <0.01 | <0.01 | 8.92 | <0.01 | <0.01 | 0.22 | <0.01 | <0.01 |
| Tremellomycetes; Tremellales; Trimorphomycetaceae | Saitozyma | <0.01 | <0.01 | <0.01 | <0.01 | 0.92 | <0.01 | <0.01 | <0.01 |
| Phylum Chytridiomycota | |||||||||
| Chytridiomycetes; Rhizophydiales; Rhizophydiales_inc | Operculomyces | <0.01 | <0.01 | 4.09 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Phylum Mucoromycota | |||||||||
| Mortierellomycetes; Mortierellales; Mortierellaceae | Mortierella | <0.01 | 0.23 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Mucoromycetes; Mucorales; Mucoraceae | Mucor | <0.01 | <0.01 | 0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cha, Y.; Oh, S.-Y. Fungal Diversity Associated with Armadillidium Isopods: A Case Study in Central Park of Gwacheon, South Korea. Diversity 2023, 15, 533. https://doi.org/10.3390/d15040533
Cha Y, Oh S-Y. Fungal Diversity Associated with Armadillidium Isopods: A Case Study in Central Park of Gwacheon, South Korea. Diversity. 2023; 15(4):533. https://doi.org/10.3390/d15040533
Chicago/Turabian StyleCha, Yehyeon, and Seung-Yoon Oh. 2023. "Fungal Diversity Associated with Armadillidium Isopods: A Case Study in Central Park of Gwacheon, South Korea" Diversity 15, no. 4: 533. https://doi.org/10.3390/d15040533
APA StyleCha, Y., & Oh, S.-Y. (2023). Fungal Diversity Associated with Armadillidium Isopods: A Case Study in Central Park of Gwacheon, South Korea. Diversity, 15(4), 533. https://doi.org/10.3390/d15040533









