Abstract
For the first time, specimens of Holothuria impatiens are reported from the Tunisian coast, supported by molecular (COI sequencing) and morphological (examination of ossicles) analyses. For comparative purposes, Holothuria impatiens samples were genetically analyzed with existing data from Spain, Italy, and the Red Sea. In addition to their external morphology, an external substance covering the individuals is described, besides the new ossicle structure described in the body wall of H. impatiens samples. The results of the morphometry of the specimens from the Tunisian coast revealed that, for two shapes of ossicles, the size of the individuals does not influence their structures. Low genetic diversity was observed in Tunisia, with two haplotypes; one of both haplotypes was reported for the first time exclusively in Tunisia. The phylogenetic tree showed that the haplotypes were shared across all locations, revealing that the Mediterranean and Red Sea individuals were clearly distinct. Further studies on the genetic diversity of H. impatiens are crucial for the evaluation of the singularity of Mediterranean populations.
1. Introduction
The class Holothuroidea has 6 orders with 1400 species (Pawson, 2007). Under this, the order the Holothuriida includes the Holothuriidae family, which comprises 5 genera and 187 species, namely 150 species of Holothuria (Linnaeus 1767), 17 species of Actinopyga (Bronn 1860), 11 species of Bohadschia (Jaeger 1833), 8 species of Labidodemas (Selenka 1867), and 1 species of Pearsonothuria [1]. At present, the genus Holothuria has 18 subgenera, with 7 subgenera comprising 13 species found in the north-eastern Atlantic and the Mediterranean Sea region [2].
As benthic invertebrates, Holothurians feed exclusively on marine deposits, such as micro-organisms, fecal matter, and detritus. This lends to them having a recycling role in coastal marine ecosystems in which they consume organic matter deposited on the sea floor, thereby driving the nutrient cycle by promoting bacterial activity [3]. Aside from this ecological contribution, sea cucumbers are commercially valuable and are targeted by several Asian fisheries. Consequently, there has been a severe decline in the populations traditionally targeted by these fisheries [4].
With the increasing fishing pressure in these regions, holothurian populations in tropical seas have begun to experience significant reductions, and even complete stock destruction [5]. The market demand has thus shifted to the Mediterranean Sea, with fisheries now targeting species such as Holothuria (Holothuria) tubulosa Gmelin, 1791, Holothuria (Holothuria) mammata Grube, 1840, and Holothuria (Roweothuria) polii (Delle Chiaje, 1824) [6,7,8]. These species, like H. polii and H. tubulosa, are now also becoming over-exploited, as indicated by the reduced sizes and weights found in several populations in Turkey [7].
Research on Mediterranean holothurian populations is scarce, and their population structures have not yet been well described. Furthermore, due to the inadequate taxonomic research delineating these species [9], most current work uses estimates derived from morphological characteristics to delimit the various species. In addition, existing species checklists for the southern areas of the Mediterranean Sea are vague and offer little detail [10,11]. To understand how commercial exploitation is affecting these species it is urgent to gain a better understanding of holothurians, particularly how to identify them taxonomically and what measures must be in place to prevent their depletion.
From a morphological perspective, the holothurian species are quite similar, with colors and forms varying with their environment [12]. Species can be identified by morphological techniques that examine their anatomy, such as the presentation of the body wall, podia, and calcareous ossicles (e.g., buttons, rods, and tables) [13,14,15,16]. Further useful morphological features include the calcareous ring, the tube feet arrangement, tentacles, anal teeth papillae, Cuvierian organs, and Polian vesicles [13].
As several species display similar external morphologies, taxonomic research has focused on their ecological characteristics [17]. For example, the three main ecological divisions of the Holothuria genus use the relationship between the ossicle shape and the ecological setting. However, ossicle shape and color differences have been found to be insufficient for the description of cryptic species [18]. Therefore, recent studies have provided more insight into the diversity and taxonomy of holothurians using molecular barcoding with mitochondrial DNA markers, e.g., cytochrome c oxidase subunit I (COI) and 16S rRNA [6,16].
Effectively managing commercial holothurian stocks and gaining a greater understanding of their diversity and taxonomy will become more feasible if genetic data become available, especially through the barcoding of mtDNA genes. This will allow proper species identification, as well as recognition of within-species complexes, enabling their evolutionary history to be described and preventing future mismanagement of the populations [19].
One of the most widespread holothurians, Holothuria (Thymiosycia) impatiens (Forsskål, 1775) can be found in the Mediterranean and Red Sea, with its range spanning from the Indo-West and East Pacific to the Caribbean Sea [20]. Being the representative type of the Thymiosycia subgenus (Pearson 1915, updated by Rowe 1969) [13], it has a relatively low commercial value, although it is the target of fisheries in Madagascar, Palau, and parts of the Eastern Pacific.
Michonneau, 2015 [19] conducted taxonomic research on this species, gathering specimens from across its known range. The author evaluated its temporal and spatial diversification dynamics via a molecular multi-locus dataset and morphological features, including coloration. Based on the results of this study, H. impatiens has a cryptic complex with a minimum of 13 separate entities.
Regarding the Mediterranean Sea, only three specimens from Italian waters and two samples from Spanish coastal waters are known [19]. No research has been conducted, so far, on H. impatiens populations off the coast of North Africa. With the aim to fill this gap, the current work performs a genetic validation of H. impatiens populations from the coastal waters of North Africa, providing a relevant dataset for the Mediterranean Sea. Specifically, H. impatiens specimens were collected from the waters of Tunisia based on the premise that further sampling areas from the Mediterranean Sea are required to gain more insight into the status of this species in this region.
During the present study, the external morphology, ossicle variability, and genetic structure of H. impatiens collected in Tunisia were also analyzed. In a subsequent step, an analysis of the combination of genetic and classic morphological identification was conducted both to detect potential correlations as well as to determine the efficacy of classic methods. The specific objectives of this study were: (1) to contribute to the knowledge of H. impatiens from the Mediterranean Sea and compare its population to individuals from the Red Sea; (2) to characterize the species using traditional criteria such as morphological identification and ossicle morphometry; and (3) to conduct a genetic study using mtDNA COI evaluation.
2. Materials and Methods
2.1. Sampling
Seventeen specimens were collected by hand in two sampling locations (the two sampling locations were visited more than once; the average number of individuals found every time was between 2 and 3 per visit) inside the small coastal lagoon of the Punic Port of Carthage (Gulf of Tunis; S1: 36°50′46.6″ N 10°19′32.2″ E, S2: 36°50′45.6″ N 10°19′35.6″ E). This lagoon is located near the ancient Carthaginian harbor near modern-day Tunis and is characterized by an abundance of algae (mainly Ulva sp.) on its edges and a high concentration of organic material on the bottom. The sampling period ran from March to June 2019. Four months were spent examining the lagoon’s edges to collect as many individuals of Holothuria (Thymiosycia) impatiens as possible. Sampling was carried out during the daytime, under and between rocks. A noteworthy find is that no specimens were found near the lagoon’s connection to the seaside. Due to the turbidity of the water in the lagoon, the maximum depth of collection was 1 m.
2.2. Morphological Identification
The taxonomical identification of the individuals started with the morphological characters and ossicles examination, based on the literature [9,13,15]. The morphology of the individuals was observed using two traditional methods. First, by analyzing the size, color, and shape, followed by dissection, according to the protocol described by Dabbagh and Kamrani, 2011 [21]. The maturity of the specimens was confirmed by checking the presence or absence of gametes. The ossicles are present in the body wall; therefore, the middle ventral and middle dorsal regions of each individual were carefully examined. A small piece of skin was placed on a slide, and ossicles were released of the body wall by adding a drop of house bleach. Once the soft tissue was completely dissolved, the samples were washed by adding some drops of distilled water, and the results were observed using an optical microscope performing an analytical study carried out for the two ossicles, tables and buttons, the most common ossicles using the protocol of identification [13,15,16]. The format, shape, and size of the ossicles were compared with those described by [13]. A piece of tissue from internal muscle bands was taken from each individual and preserved in 96% ethanol for molecular analysis.
Age-Related Variation of Ossicles
A total of eighty ossicles from the two selected shapes were photographed to be individually analyzed: 40 tables and 40 buttons, 20 from the dorsal side and 20 from the ventral side. The measurements of table and button ossicles were recorded using the software Image J 1.53e [22]. The measurements taken were the maximum and minimum diameter of the tables (lngT) and the max and min length of the buttons (lngB and lrgB). The selection of tables and buttons was random and based on automatic measurement.
To test and compare the changes throughout the stages of the life cycle of H. impatiens and to detect possible variation with growth, the individuals were divided into three size ranges (Table 1) estimating the aging of specimens from juveniles to mature adults expressed through their length [23]. The first range, Rg1, was from 0.5 to 4 cm (7 individuals), the second range, Rg2, was from 4 to 8 cm (4 individuals), and the third range, Rg3, was from 8 to 12 cm (6 individuals).

Table 1.
Size range of ossicles from the middle of Holothuria impatiens’ dorsal and ventral sides with different body sizes; Rg1 (0.5–4 cm); Rg2 (4–8 cm); Rg3 (8–12 cm).
2.3. Genetic Study
DNA Extraction and Amplification
Total genomic DNA was extracted from adductor muscle tissue using a RedExtract N-Amp Kit (Sigma-Aldrich) following the manufacturer’s instructions. A fragment of the cytochrome oxidase subunit I (COI) was amplified using COI universal primers, CO1eF (ACTGCCCACGCCCTAGTAATGATATTTTTTATGGTNATGCC), and CO1eR (TCGTGTGTCTACGTCCATTCCTACTGTRAACATRTG) [24,25].
The PCR reaction was: 3 min at 95 °C for denaturation followed by 35 cycles of denaturation at 94 °C during 4 s, annealing at 55 °C during 1:10 min, and an extension period of 1:10 min at 72 °C and a final extension at 72 °C for 5 min. The same primers were used for the sequencing reactions. PCR products were purified and sequenced in STABVIDA (http://www.stabvida.net/) (accessed on 6 March 2020). The sequences obtained were edited and aligned using Codoncode Aligner v8.0 ® software.
A Basic Local Alignment Search Tool (BLAST v 1.15.0) was performed on each DNA sequence on GenBank (www.ncbi.nlm.nih.gov/GenBank accessed on 25 December 2022). The software searches for regions of similarity between the input sequence and those in online databases and calculates statistical similarity scores. Emphasis was placed on the “Query cover” and “Percentage Identity” values. “Percentage Identity” represents the percentage of matching residues in the alignment, while “Query cover” indicates the percentage of the input sequence length that is aligned [26]. All sequences were deposited in GenBank (Accession numbers OQ561304-OQ561320, Table 2).

Table 2.
Holothuria (Thymiosycia) impatiens individuals included in this study and analyzed for the partial cytochrome oxidase subunit 1 (COI) with references, GenBank accession numbers, and sample location.
The median-joining network parsimony was created using PopART v. 1.7 [29] (Population Analysis with Reticulate Trees) estimating the connectivity within the haplotypes. The genetic diversity of the populations was estimated using analyses of molecular variance (AMOVA), and calculation of pairwise FSTs and nucleotide diversity (p) within species, as well as nucleotide divergence (Da) between populations, was performed using ARLEQUIN v. 3.5 software [30] based on the methods described by [31,32]. The remaining samples used for comparative purposes were retrieved from GenBank and the respective accession numbers can be found in (Table 2). The species of Cucumaria frondosa was used as an outreach to underline the genetic distance between the sequences studied compared to a species from a different order and the same class.
The phylogenetic relationships were created including sequences from Tunisia, Mediterranean samples, and the Red Sea using maximum parsimony (MP), maximum likehood (ML), and Bayesian inference (BI). Basic statistics were performed using MEGA7 (nucleotide and transversion frequencies) [32] and substitution models assessed using jModelTest [33]. Pairwise nucleotide distances (p-distance, 1000 bootstrap replicates) within and between groups were also computed in MEGA7.
2.4. PCA and Biplot Analysis
The data collected from both the genetic and morphometric analysis of the ossicles were used to check for a possible relationship between ossicle size and haplotypes, considering that buttons and tables are two shapes commonly used for the identification and characterization of this species [17]. The three variables used were (lngB, lrgB, and lngT) combined with 2 indices (two haplotypes) recorded and calculated from the 17 individuals. The first haplotype HYP is a group of 4 individuals and the second haplotype IndHP is represented by 13 individuals. Each point on the PCA represents an individual.
The values of the three variables studied were analyzed by principal component analysis (PCA) using the statistical software BioVinci v2.0 (Bio Turing Team). The purpose of this analysis is to verify the ordination at the PCA level between the haplotypes determined during the genetic analysis of our sample of H. impatiens and to evaluate if the traditional identification of this species can be used to detect the diversity within a species such as H. impatiens.
3. Results
3.1. Morphological Identification
Holothuria impatiens is a species with dark transverse stripes of variable shades of brown. The samples were small with a medium 5.7 cm length for 17 individuals deviating from 1 to 9 cm and presented a cylindrical vermiform body shape with a thin integument and soft body wall and white to yellowish papillae.
The individuals analyzed were yellow to dark brown, streaked with light brown, and often variegated pink and beige bands. The podia tubes were orange–white, and the anal papillae had an irregular yellow-transparent color (Figure 1). The average number of tentacles was 19 and the deviation was between 18 and 20, with yellow–orange colors. Eleven individuals out of the seventeen collected presented gametes (Table 3).

Figure 1.
A plate with morphological details of the species Holothuria impatiens in (A–G), external morphology of several individuals, (H) the anus, (I) the oral papillae, and (J) details of dorsal podia with (a) a layer adhering to the collected individuals.

Table 3.
Body weight without coelomic fluid (g) and the length (cm) of the samples of H. impatiens. (*) mature individuals with gametes.
Individuals varied in length from 3 to 11 cm, with the smallest mature individual (GHI13436) measuring 3 cm. Some individuals, as shown in (Table 3), were 5.5 (GHI14437) or 4.7 cm (GHI09431) have maturity lengths but lacked gametes while possessing gonads.
Analysis of both the length and weight of individuals suggests that the population includes individuals with varying lengths but without gametes in an advanced developmental stage. For example, GHI14437 (L = 5.5 cm; W = 3.87) is longer than GHI13436 (L = 3 cm; W = 3.55) which is mature, yet they have similar weights due to GHI13436’s maturity.
The weight of mature individuals was the most significant factor, making it a reliable indicator of the maturity of H. impatiens individuals (Table 3).
3.1.1. Special Observation
All individuals had their body walls covered with a second tissue-like substance that seems to play the role of protection or camouflage since it is particularly uniformly brownish, fused, and resistant, consisting of microparticles glued probably with mucus (Figure 1). This mucus covered all the individuals collected during our study and has the same structure for each individual. This substance was reported for the first time for the species H. impatiens.
3.1.2. Ossicles Morphology
It was possible to observe new, additional structures of ossicles in the body wall of the individuals. The newly observed ossicles, known as grid ossicles (Figure 2B), have never been previously mentioned as part of the ossicle list for this species. They are smaller in size compared to other ossicles (Figure 2B). Most of the tables have slightly square discs with eight holes. The stems of the tables end with short teeth of uniform size. (Figure 2C,E). The buttons (Figure 2A) are smooth with hole numbers fluctuating from 5 to 7 holes.
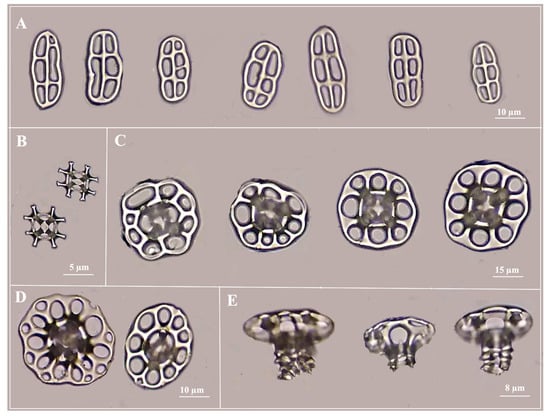
Figure 2.
Ossicles of the species Holothuria impatiens: (A) different button structures; (B) grid structure; (C) table with eight holes; (D) table with nine or more holes; and (E) table side view.
The button length deviations ranged from 69.5 to 97.2 µm for Rg1, from 63.8 to 77.4 µm for Rg2, and from 65.7 to 80.4 µm for Rg3. The widths of the buttons recorded deviations ranging from 78.1 to 83.6 µm for Rg1, from 84.7 to 91.9 µm for Rg2, and from 74.7 to 95.8 µm for Rg3 (Table 1). The disc diameter of the tables recorded a wider range of deviations, from 85.9 to 99 µm for Rg1, from 84.7 to 91.9 µm for Rg2, and from 74.7 to 95.8 µm for Rg3. No significant differences were detected in the morphometry of ossicles between the three age groups. There was no correlation between the size ranges and the sizes of ossicles in H. impatiens, as the values varied without showing a clear pattern of increase or decrease in relation to the size ranges of the individuals.
3.2. Genetic Study (Barcoding of the COI Marker)
BLAST comparisons of the 510 bp COI fragment from the 17 individuals collected at Tunisian localities confirmed the prelaminar taxonomical identification based on morphological characters (100% of the samples were assigned to the species Holothuria (Thymiosycia) impatiens).
The phylogenetic results based on the ML tree recognized two distinct clades. The first clade included 22 individuals from three localities of the Mediterranean Sea (Tunisia, Italy, and Spain) (Figure 3), composed of two groups: one group including 13 sequences from Tunisia, and a second group including 9 sequences from Tunisia, Italy and Spain.
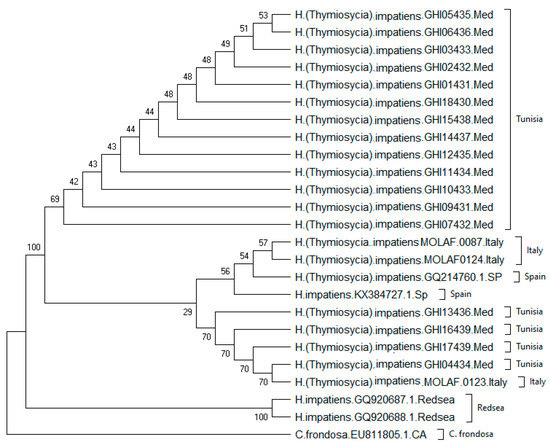
Figure 3.
Phylogenetic tree constructed with genetic distances between the individuals based on the method [30], showing the phylogenetic relationships between H. impatiens (a complex of 13 species) deduced from the sequences of the COI fragment of the mitochondrial DNA.
The second clade is composed exclusively of two sequences from the Red Sea. The greatest genetic distance between haplotypes was between GHI05435 from Tunisia and the two individuals from the Red Sea. There were similarities in the samples collected from three Mediterranean locations. Genetic differentiation was clear between Egypt and the Mediterranean locations, with a net average distance of 0.2019 between these groups (Table 4).

Table 4.
Pairwise nucleotide p-distances for the COI sequences of Holothuria (Thymiosycia) impatiens. The mean within = group (diagonal) and net average between-groups (below diagonal) divergence. Na: not applicable. Statistically significant pairs shown in bold (p < 0.05).
The estimation of pairwise distances for COI sequences showed that the Red Sea samples were distinct from those collected from other locations (Table 4). The Red Sea samples were significantly different from all the locations found in the Mediterranean Sea. The samples from the Red Sea were collected in Egypt, which is apparently not far from the Suez Canal.
The COI sequences obtained in this study reveal the presence of two haplotypes within the Tunisian individuals. One haplotype was shared with the samples previously described for the Mediterranean. The maximum number of mutations is four between the sequences. The second, representing most of our individuals was exclusive to the Tunisian samples (13 individuals). The most abundant haplotype of our samples was named IndHP. The second haplotype was composed of nine individuals mixing three locations: Tunisia, Spain, and Italy, and was named HYP.
A total of five haplotypes were identified, with four belonging to the species H. impatiens and one belonging to Cucumaria frondosa. Two haplotypes were found in seventeen individuals collected from Tunisia. The differences between the haplotypes included 93 transitions, 59 transversions, and 1 indel.
In the network, the Mediterranean population is represented by three haplotypes: one shared between Italy and Tunisia, another between Spain and Italy, and the third with the largest number of samples from Tunisia. The number of mutations within the Mediterranean clade shows a maximum of four mutations between the sequences (Figure 4).
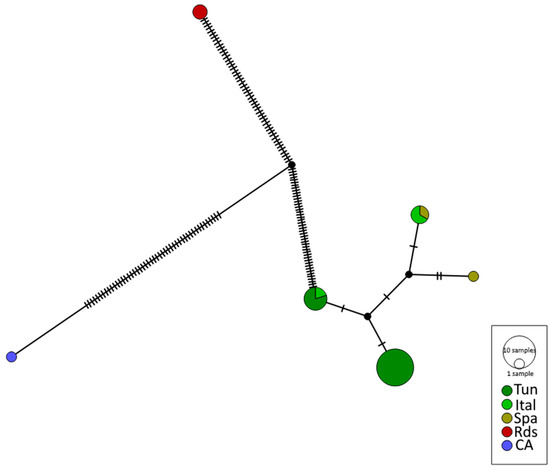
Figure 4.
Mitochondrial DNA haplotype network of Holothuria impatiens; the different colors correspond to the different geographical areas: (Tu) Tunisia, (Ital) Italy, (Spa) Spain, (Rds) Red Sea, and, as an external group, (CA) Canada for the species Cucumaria frondosa. The diameter of each circle is proportional to the frequencies of the haplotypes. In the case where the haplotypes are shared between the stations, the colors represented are proportional to the frequencies of the haplotypes of the stations.
Hence, the branch with the highest number of mutations for H. impatiens is between the Mediterranean and Red Sea populations. The results of the temporal differentiation tests showed a significant value of (FST = 0.273, p = 0.00050) between the Mediterranean and Red Sea populations according to AMOVA, indicating that the populations are genetically different confirming what was shown in the network. It is important to mention the significant difference between the Red Sea sequences and those from Tunisia with a molecular variability of 69.02 between the populations. Conversely, there was no significant difference between the Tunisia and Mediterranean Sea (Italy and Spain) samples with a molecular variability of 30.98.
3.3. Principal Component Analysis of the Ossicles
Our genetic data results were compared with the data collected from the ossicle measurements to test for a significant correlation. The distribution of values in the PCA space was mainly in the first two axes, with a balance between the top and bottom, with PC1 and PC2 values close to 58.71% and 36.43%, respectively. In general, only one individual was separated from the groups of the two haplotypes, which was likely due to an error. In conclusion, no relationship between ossicle morphometry and haplotypes was found.
PC1 was mostly influenced by the haplotype (IndHP), with values ranging from −10 to 9, and half of the haplotype (HPY) samples, with two individuals located on the positive part of the axis with PCA scores of 4 and 4.3, respectively. The other five individuals of the haplotype (IndHP) were located on the negative part, with the exception of one that was on the positive part. The two individuals of the haplotype (HPY) were also located on the negative part of PC1 (Figure 5). PC2 was influenced by 5 individuals of the haplotype (IndHP), with positive component scores ranging from 0 to 9. The other variables contributing to the positive axis of PC2 were the 2 individuals of the haplotype (HPY), with scores of 7.9 and 11.7, respectively. The negative part of PC2 was influenced by both haplotypes, with 8 individuals from (IndHP) having values between −3 and 0, and 2 individuals from (HPY) having scores of −6.9 and −9.
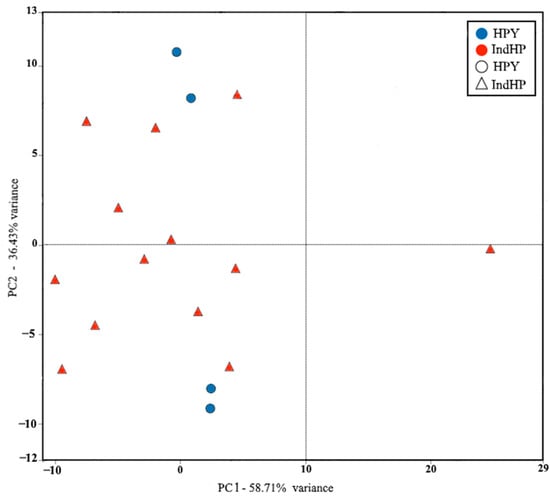
Figure 5.
Principal component analysis of the ossicles of the seventeen individuals from Tunisia; the values used are the means calculated from twenty ossicles per individual. Morphometric variables in the PCA: LngT: length of table-shaped ossicles, LngB: length of button-shaped ossicles, LagB: width of button-shaped ossicles. Distribution of the analyzed individuals on the space defined by the first two components of the PCA (blue circle: first haplotype HPY; red triangle: second haplotype IndHP).
Even though no significant structure was detected in the PCA, the haplotype HPY composed of four individuals showed two individuals close together in PC1 while the other two are on the opposite side in PC2. For the second haplotype IndHP, which includes most of the samples, only one individual was separated from the others. The results do not reveal any noticeable differentiation between individuals based on the ossicle sizes. Individuals belonging to the IndHP haplotype do not have differently shaped ossicles compared to those of the HPY haplotype.
Biplot Analysis of Ossicles
The results show similarity in “Button width” between the two haplotypes (IndHP and HPY), while the other two variables, “Button length” and “Table length,” show some differences in their range. Biplots were prepared to compare the length and width ranges of the buttons and the length of the tables for each haplotype separately. For “Button length”, the IndHP haplotype has a broader range, spanning from 50 to 110, compared to the narrower range of the HPY haplotype, which is between 70 and 80. In the case of “Table length”, the HPY haplotype has a wider range than the IndHP haplotype. (Figure 6).
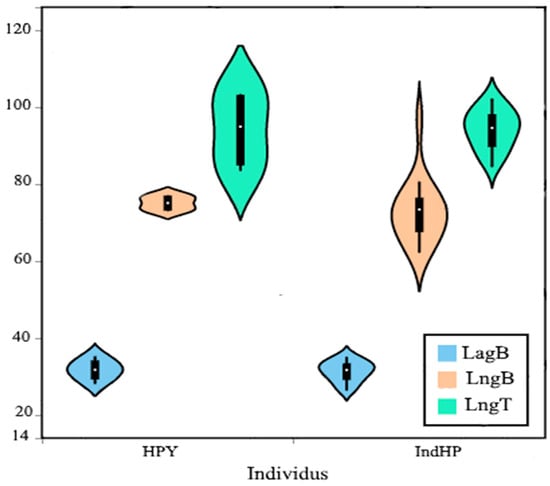
Figure 6.
Analysis of the variables in the form of a violin plot. (LngB) Length of the ossicles in the shape of buttons, (LagB) width of the ossicles in the shape of buttons, (LngT) length of the ossicles in the shape of tables, and the distribution of the analyzed individuals on the figure (left: haplotype (HPY) and right: haplotype (IndHP)).
4. Discussion
The population of H. impatiens in Tunisia was characterized through the examination of individual morphology and ossicle structure, comparing the genetic sequences of these individuals with samples from various locations in the Mediterranean and Red Sea. This work aims to enhance the understanding of populations of this species in the Mediterranean.
The Tunisian population of H. impatiens displays similar morphology to samples from Spain and Italy and a completely different morphology with the Red Sea samples. The external morphology of Holothuria impatiens from Tunisia resembles previous sample collections. It is noteworthy to mention the morphometry of the body wall’s ossicles, which seems comparable to earlier studies [17,34,35]. In addition, for the Tunisian population, the maximum length and weight were recorded for a mature sample with gametes. However, some samples showed a lighter weight with a considerable length but were not mature, suggesting that length does not significantly increase with individual maturity. The results indicate that weight is a better indicator of maturity for this species, suggesting two hypotheses: first, the ability of Holothurians to constantly stretch and shrink, as they have high body elasticity that can alter an individual’s length, and second, the significant increase in an individual’s weight with the growth of the gonads.
The second hypothesis explaining these results is based on the complex reproductive cycle of Holothurians in general, which is influenced by biotic and abiotic factors such as temperature [36], lunar rhythms [37], and food [38]. Some individuals with a significant length may have their reproductive cycle suppressed by one of these factors. Furthermore, samples from Tunisia exhibit a unique characteristic specific to this species: the presence of a substance resembling an external skin-like body wall, likely used for protection or camouflage. The individuals were covered with a layer made of very thin particles of sand from the lagoon that were joined together, forming a resilient second skin-like surface over the body.
Regarding ossicles, the shape of buttons found in the Tunisian population was similar to those from previous research [17,35,36]. Previous studies analyzing the button ossicles of H. impatiens [36] reported a similar number of holes as those found in this study, as well as for tables [35]. Deichmann [17] described the tables as oval-shaped, while James [34] described them as oval and ranging between 0.084 mm to 0.10 mm in length and 0.040 mm to 0.049 mm in width. These values are similar to the values obtained in the current study. Characterization ossicles of this species led to report the presence of a new ossicle shaped as grids found in the body wells of all the individuals collected from Tunisia. This also underlined the presence of tables with nine holes and buttons observed in the body wall, with a high abundance and an average of six parallel holes [34]. The combination of morphological results and ossicles morphometry showed that the maturity and size of individuals do not affect the length or width of the ossicles of H. impatiens. This is in contrast to findings in other species of Holothurians, where the shape or abundance of ossicles can change with maturity and can sometimes appear or disappear. [23].
The morphometry of the ossicles in this population was performed to assess the validity of the method for identifying Holothurian species. Using ossicles, in the form of buttons and tables [17], which are the most common tool to characterize Holothurian species, was a good starting point to identify individuals. However, some species may still be difficult to identify based solely on this method.
The ossicles are the most used tool to identify the morphology of H. impatiens individuals in the Mediterranean Sea [16] and different shapes and aspects of the calcareous ossicles in the body wall were used to determine the species [39]. Holothuria impatiens from the Mediterranean Sea appears to be a species with low differences in visible morphology and ossicle shape between individuals. However, its genetic diversity has remained unknown for decades. Moreover, this study has shown that ossicles can be useful as a preliminary tool for identification, but they should not be the sole tool used to classify a species.
To investigate further, the comparison of the results of the morphometry examination and the genetic structure of the Mediterranean populations, combining the present study with previous studies [34,39], showed that the taxonomic status of H. impatiens cannot solely rely on the examination of ossicle morphometry. This can be seen by the strong morphological similarities and the genetic diversity found between the Mediterranean and Red Sea populations. Additionally, despite its genetic complexity [19], H. impatiens has been inadequately studied in the Mediterranean. Based on the three samples studied from Italy [19], the species appears to be poorly diversified in the region. This raises questions about its status in the Mediterranean and whether it could potentially be a genetically diverse species with explicit differences between the Mediterranean and other locations.
Individuals that show similar morphologies may still be genetically diverse. The data from the difference between the two haplotypes, lngT and lngB, cannot confirm any real relationship between the haplotypes and the dimensions of their ossicles. This supports the hypothesis deduced earlier from the PCA graph that the dimensions of the calcareous structures are not sufficient to detect genetic diversity within a population.
Sequences of the COI fragment from our H. impatiens have provided two haplotypes found linked to the Mediterranean Sea and one is the first time reported in this area.
In this study, a single haplotype [HYP] was shared between Spain, Italy, and Tunisia. Likewise, over half of the Tunisian samples were represented by the second haplotype [IndHP], which has not been reported elsewhere. Low genetic diversity was observed and may be due to the limited number of samples (two from Spain and three from Italy) or the sequenced fragment. Michonneau [18] mentioned a single haplotype for the entire Mediterranean region; however, this study confirms the presence of a second haplotype.
From the genetic perspective, the two main findings of this study are: the low genetic diversity in the Mediterranean Sea, which may be underestimated, and the distinct separation between samples from the Red Sea and the Mediterranean Sea was observed and confirmed with a significant pairwise distance between both locations.
Based on the theory suggesting a correlation between mitochondrial and nuclear genomes [40], low genetic diversity using COI sequences can reflect the low allelic richness of microsatellites. Samples from the Mediterranean with low gene flow could indicate a genetic decline, as H. impatiens is a commercially exploited species. A similar situation was noted in the exploitation of the species Pagrus auratus, where COI was used to demonstrate a significant decline in nuclear genetic diversity in its population [41].
If the decline is confirmed, it supports the hypothesis that direct or indirect factors are affecting the diversity within isolated populations. The direct factors mentioned in this study include human impact, such as over-exploitation of species such as H. impatiens, which can significantly impact its genetic diversity. Indirect factors include global warming and changes in environmental parameters in the Mediterranean, which are changing faster than in other regions and may favor the selection or decline of certain genes. Another study showed negative impacts from anthropogenic pressures such as over-harvesting and coastal development which supposedly affected the diversity distribution of the species H. atra [42].
Further research is needed to either confirm or disprove the supposed decline in the diversity of H. impatiens species in the Mediterranean Sea. This is a potential early warning sign to update the conservation status of this species. Despite the low genetic diversity of the Mediterranean samples, they formed a separate clade from the samples collected from the Red Sea. The AMOVA analysis revealed significant differences between the groups, with the FST equal to 0.273 (p = 0.00050), indicating a high extent of genetic differentiation and potential genetic isolation. Comparing these two closely located areas with high genetic variability would be of great interest. There are several potential explanations for the observed pattern, one of the most intriguing being that the Mediterranean H. impatiens may include a new, cryptic species without any noticeable morphological difference.
For decades, species of the genus Holothuria have presented a complex taxonomic composition and high intraspecific variability, which has prompted researchers to investigate for better taxonomic characters to determine the boundaries between species. Currently, the best method for classifying species and subspecies is through molecular approaches. Although ossicle morphometry and morphological criteria can be used for identifying the genus or species, in some cases, such as H. impatiens, molecular approaches provide more reliable results. From a conservation perspective, it is necessary to conduct a more comprehensive study that takes into account factors such as environmental parameters and a wider range of genetic fragments, helping to increase the conservation efforts for this species in the Mediterranean Sea by potentially revealing its exclusive and specific characteristics. This could prevent potential over-exploitation of the Mediterranean and its species.
5. Conclusions
Despite its widespread distribution in shallow waters, H. impatiens has been found to exhibit high genetic diversity within its global populations due to its cryptic nature. However, accurately estimating the diversity of this species in the Mediterranean region remains challenging.
In fact, our data from Tunisia suggest that H. impatiens has a homogeneous population with minor differences. The ossicles do not show changes in shape, age, or morphology among individuals. To fully study this population, it is recommended to use various other methods to highlight the diversity within similar populations.
The slight increase in diversity observed in this study, which was based on analysis of only one location and one genetic fragment in the Mediterranean, suggests that a more comprehensive study using a larger number of samples from various locations in the region would be the perfect tool to answer some of the questions raised such as: is there any geographic isolation between the Mediterranean populations and other locations such as RedSea, Indo-West, East Pacific and the Caribbean Sea ? Is it legitimate to propose the hypothesis of an allopatric differentiation between the Mediterranean population of H. impatiens and other populations from different locations?
Author Contributions
Conceptualization, D.K., A.A., J.I.R. and S.M.F.; methodology, D.K., A.A., J.I.R. and S.M.F.; software, D.K. and S.M.F.; validation, D.K., A.A., J.I.R. and S.M.F.; investigation, D.K., A.A., J.I.R. and S.M.F.; resources, D.K., A.A., J.I.R. and S.M.F.; writing—original draft preparation, D.K., A.A., J.I.R. and S.M.F.; writing—review and editing, D.K., A.A., J.I.R., M.M., A.R.-C. and S.M.F.; supervision, A.A. and J.I.R.; project administration, M.M., A.A., A.R.-C. and J.I.R. All authors have read and agreed to the published version of the manuscript.
Funding
Expenses were covered by the University of Tunis ElManar, Faculty of Science of Tunis and the Portuguese national funds from FCT—Foundation for Science and Technology through projects MARE/UIDB/MAR/04292/2020, MARE/UIDP/04292/2020, and LA/P/0069/2020 granted to MARE/ARNET.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The first author is very grateful to the Ministry of Higher Education of Tunisia, the University of Tunis ElManar, Faculty Sciences Tunis (FST), the Marine and Environmental Sciences Centre (MARE), and Instituto Universitário Lisboa (ISPA) for their encouragement and financial support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
References
- O’Loughlin, P.M.; Paulay, G.; Vandenspiegel, D.; Samyn, Y. New Holothuria species from Australia (Echinodermata: Holothuroidea: Holothuriidae), with comments on the origin of deep and cool holothuriids. Mem. Mus. Vic. 2007, 64, 35–52. [Google Scholar] [CrossRef]
- Pérez, G.H.B. Sistemática y Filogeografía De Las Especies Del Subgénero Holothuria (Echinodermata: Holothuriidae: Holothuria) de la Región Atlanto-Mediterránea. Ph.D. Thesis, Universidad de Murcia, Murcia, Spain, 2010. [Google Scholar]
- MacTavish, T.; Stenton-Dozey, J.; Vopel, K.; Savage, C. Deposit-Feeding Sea Cucumbers Enhance Mineralization and Nutrient Cycling in Organically-Enriched Coastal Sediments. PLoS ONE 2012, 7, e50031. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.C.; Flemming, J.M.; Watson, R.; Lotze, H.K. Serial exploitation of global sea cucumber fisheries. Fish Fish. 2011, 12, 317–339. [Google Scholar] [CrossRef]
- Siegenthaler, A.; Cánovas, F.; González-Wangüemert, M. Spatial distribution patterns and movements of Holothuria arguinensis in the Ria Formosa (Portugal). J. Sea Res. 2015, 102, 33–40. [Google Scholar] [CrossRef]
- González-Wangüemert, M.; Borrero-Pérez, G. A new record of Holothuria arguinensis colonizing the Mediterranean Sea. Mar. Biodivers. Rec. 2012, 5, e105. [Google Scholar] [CrossRef]
- González-Wangüemert, M.; Aydin, M.; Conand, C. Assessment of sea cucumber populations from the Aegean Sea (Turkey): First insights to sustainable management of new fisheries. Ocean Coast. Manag. 2014, 92, 87–94. [Google Scholar] [CrossRef]
- González-Wangüemert, M.; Valente, S.; Henriques, F.; Domínguez-Godino, J.A.; Serrão, E.A. Setting preliminary biometric baselines for new target sea cucumbers species of the NE Atlantic and Mediterranean fisheries. Fish. Res. 2016, 179, 57–66. [Google Scholar] [CrossRef]
- Kœhler, R. Faune de France: Echinodermes. 1921. Available online: https://doris.ffessm.fr/Bibliographie/FAUNE-DE-FRANCE-ECHINODERMES-Koehler-R (accessed on 4 February 2023).
- Boudouresque, C.F. Critères de sélection et projet de liste des espèces en danger ou menacées. In Réunion D’experts sur Les espèces Menacées en Méditerranée; UNEP (OCA)/MED WG: Athens, Greece, 1995; Volume 100. [Google Scholar]
- Mustapha, K.B.; Hattour, A.; M’hetli, M. Etat de la bionomie benthique des étages infra et circalittoral du Golfe de Gabes. INSTM Bull. Mar. Freshw. Sci. 1999, 26, 5–48. [Google Scholar]
- Honey-Escandón, M.; Laguarda-Figueras, A.; Solís-Marín, F.A. Molecular phylogeny of the subgenus Holothuria (Selenkothuria) Deichmann, 1958 (Holothuroidea: Aspidochirotida). Zool. J. Linn. Soc. 2012, 165, 109–120. [Google Scholar] [CrossRef]
- Rowe, F.W.E. A review of the family Holothuriidae (Holothurioidea: Aspidochirotida). Bull. Br. Mus. (Nat. Hist.) Zool. 1969, 18, 117–170. [Google Scholar] [CrossRef]
- Samyn, Y.; Vandenspiegel, D. <strong>Sublittoral and bathyal sea cucumbers (Echinodermata: Holothuroidea) from the Northern Mozambique Channel with description of six new species</strong>. Zootaxa 2016, 4196, 451–497. [Google Scholar] [CrossRef]
- Hedingia Mediterranea (Bartolini Baldelli, 1914) Tortonese. 1965. Available online: https://www.gbif.org/species/4342673 (accessed on 4 February 2023).
- Borrero-Pérez, G.H.; Pérez-Ruzafa, A.; Marcos, C.; González-Wangüemert, M. The taxonomic status of some Atlanto-Mediterranean species in the subgenus Holothuria (Echinodermata: Holothuroidea: Holothuriidae) based on molecular evidence. Zool. J. Linn. Soc. 2009, 157, 51–69. [Google Scholar] [CrossRef]
- Deichmann, E. The Holothurioidea collected by the Velero III and IV during the years 1932 to 1954. Part II. Aspidochirota. Allan Hancock Pac. Exped. 1958, 11, 253–349. [Google Scholar]
- Michonneau, F.; McPherson, S.; O’Loughlin, P.M.; Paulay, G. More than meets the eye: Diversity and geographic patterns in sea cucumbers. arXiv 2015, arXiv:014282. [Google Scholar] [CrossRef]
- Michonneau, F. Cryptic and not-so-cryptic species in the complex “Holothuria (Thymiosycia) impatiens” (Forsskål, 1775) (Echinodermata: Holothuroidea: Holothuriidae). arXiv 2015, arXiv:014225. [Google Scholar] [CrossRef]
- WoRMS—World Register of Marine Species. Available online: https://www.marinespecies.org/aphia.php (accessed on 4 February 2023).
- Dabbagh, A.R.; Keshavarz, M. Three sea cucumbers from the Bandar-E Bostaneh coast (Persian Gulf, Iran). World Appl. Sci. J. 2011, 13, 1933–1937. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Rasolofonirina, R.; Jangoux, M. Appearance and development of skeletal structures in Holothuria scabra larvae and epibiont juveniles. SPC Beche-de-Mer Inf. Bull. 2005, 22, 5–10. [Google Scholar]
- Hoareau, T.B.; Boissin, E. Design of phylum-specific hybrid primers for DNA barcoding: Addressing the need for efficient COI amplification in the Echinodermata. Mol. Ecol. Resour. 2010, 10, 960–967. [Google Scholar] [CrossRef]
- Arndt, A.; Marquez, C.; Lambert, P.; Smith, M.J. Molecular Phylogeny of Eastern Pacific Sea Cucumbers (Echinodermata: Holothuroidea) Based on Mitochondrial DNA Sequence. Mol. Phylogenet. Evol. 1996, 6, 425–437. [Google Scholar] [CrossRef]
- Kerfeld, C.A.; Scott, K.M. Using BLAST to Teach “E-value-tionary” Concepts. PLoS Biol. 2011, 9, e1001014. [Google Scholar] [CrossRef] [PubMed]
- González-Wangüemert, M.; Domínguez-Godino, J.A.; Cánovas, F. New records of sea cucumbers inhabiting Mar Menor coastal lagoon (SE Spain). Mar. Biodiv. 2018, 48, 2177–2182. [Google Scholar] [CrossRef]
- So, J.J.; Mercier, A.; Hamel, J.-F. Habitat utilisation, growth and predation of Cucumaria frondosa: Implications for an emerging sea cucumber fishery. Fish. Manag. Ecol. 2010, 17, 473–484. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; p. 512. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- James, D.B. Taxonomic Studies of the Species of Holothuria (LINNAEUS, 1767) from the Seas Around India 505 1 Part I. 1992. Available online: http://eprints.cmfri.org.in/7434/1/449J._BOMBAY_NAT.HISTORY_SOC_1995.pdf (accessed on 1 January 2020).
- Raymund, D.J.; Victoria, T.N.; Vinya, M.A.; Frederick, G.T. The Body Wall Spicule Formations of Mature Holothuria impatiens Found in La Union, Philippines. J. Nat. Allied Sci. 2019, 3, 42–45. [Google Scholar]
- Ramofafia, C.; Battaglene, S.C.; Bell, J.D.; Byrne, M. Reproductive biology of the commercial sea cucumber Holothuria fuscogilva in the Solomon Islands. Mar. Biol. 2000, 136, 1045–1056. [Google Scholar] [CrossRef]
- Mercier, A.; Sun, Z.; Baillon, S.; Hamel, J.-F. Lunar rhythms in the deep sea: Evidence from the reproductive periodicity of several marine invertebrates. J. Biol. Rhythm. 2011, 26, 82–86. [Google Scholar] [CrossRef]
- Drumm, D.J.; Loneragan, N.R. Reproductive biology of Holothuria leucospilota in the Cook Islands and the implications of traditional fishing of gonads on the population. J. Mar. Freshw. Res. 2005, 39, 141–156. [Google Scholar] [CrossRef]
- Dabbagh, A.-R.; Keshavarz, M.; Mohammadikia, D.; Afkhami, M.; Nateghi, S.A. Holothuria scabra (Holothuroidea: Aspidochirotida): First record of a highly valued sea cucumber, in the Persian Gulf, Iran. Mar. Biodivers. Rec. 2012, 5, e69. [Google Scholar] [CrossRef]
- Wesselmann, M.; Gonzalez-Wanguemert, M.; Serrao, E.A.; Engelen, A.H.; Renault, L.; Garcia-March, J.R.; Hendriks, I.E. Genetic and oceanographic tools reveal high population connectivity and diversity in the endangered pen shell Pinna nobilis. Sci. Rep. 2018, 8, 4770. [Google Scholar] [CrossRef] [PubMed]
- Hauser, L.; Adcock, G.J.; Smith, P.J.; Ramírez, J.H.B.; Carvalho, G.R. Loss of Microsatellite Diversity and Low Effective Population Size in an Overexploited Population of New Zealand Snapper (Pagrus auratus) | PNAS. Available online: https://www.pnas.org/doi/full/10.1073/pnas.172242899 (accessed on 4 February 2023).
- Hamamoto, K.; Soliman, T.; Poliseno, A.; Fernandez-Silva, I.; Reimer, J.D. Higher Genetic Diversity of the Common Sea Cucumber Holothuria (Halodeima) atra in Marine Protected Areas of the Central and Southern Ryukyu Islands. Front. Conserv. Sci. 2021, 2, 736633. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).