Genome-Wide Insights into Intraspecific Taxonomy and Genetic Diversity of Argali (Ovis ammon)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Ethic Statement
2.2. Data Quality Control and Processing of Whole-Genome SNP Data
2.3. Processing of Complete Mitogenomes
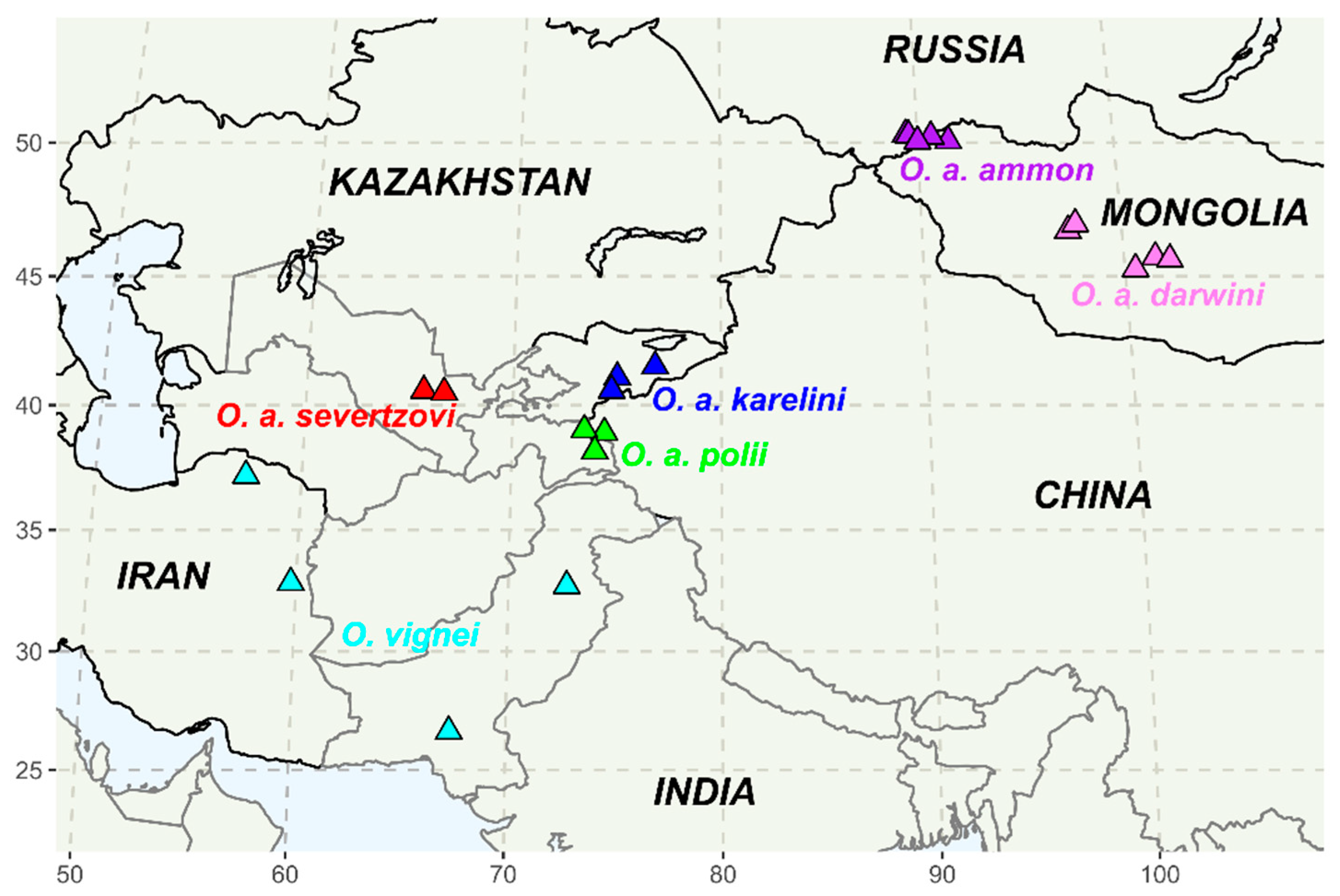
2.4. The map of Sampling Sites
3. Results
3.1. Whole-Genome SNP Analysis
3.2. Complete Mitochondrial Genomes Analysis
4. Discussion
4.1. The Phylogenetic Relationship between O. a. ammon and O. a. darwini
4.2. The phylogenetic Relationship between O. a. polii, O. a. karelini, and O. a. humei
4.3. The Evolution and Phylogeny of O. a. severtzovi
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Damm, G.R.; Franco, N. CIC Caprinae Atlas of the World; CIC International Council for Game and Wildlife Conservation: Budakeszi, Hungary; Rowland Ward Publications: Johannesburg, South Africa, 2014. [Google Scholar]
- Sanna, D.; Barbato, M.; Hadjisterkotis, E.; Cossu, P.; Decandia, L.; Trova, S.; Pirastru, M.; Leoni, G.; Naitana, S.; Francalacci, P.; et al. The First Mitogenome of the Cyprus Mouflon (Ovis gmelini ophion): New Insights into the Phylogeny of the Genus Ovis. PLoS ONE 2015, 10, e0144257. [Google Scholar] [CrossRef]
- Machová, K.; Málková, A.; Vostrý, L. Sheep Post-Domestication Expansion in the Context of Mitochondrial and Y Chromosome Haplogroups and Haplotypes. Genes 2022, 13, 613. [Google Scholar] [CrossRef]
- Lydekker, R. Wild Oxen, Sheep, and Goats of All Lands; Rowland Ward: London, UK, 1898; p. 239. [Google Scholar]
- Nasonov, N.V. Geographical Distribution of Old-World Wild Sheep; Printing house of Russian State Academy: Petrograd, Russia, 1923; p. 255. (In Russian) [Google Scholar]
- Tsalkin, V.I. Wild Sheep of Europe and Asia; Moskovskoe Obschestvo Ispytatelei Prirody: Moscow, Russia, 1951; p. 344. (In Russian) [Google Scholar]
- Nadler, C.F.; Hoffmann, R.S.; Woolf, A. G-band patterns as chromosomal markers, and the interpretation of chromosomal evolution in wild sheep (Ovis). Experientia 1973, 29, 117–119. [Google Scholar] [CrossRef]
- Schaller, B. Mountain Monarchs; University of Chicago Press: Chicago, IL, USA, 1977. [Google Scholar]
- Valdez, R. The Wild Sheep of the World; The Wild Goat and Sheep International: Mesilla, NM, USA, 1982; p. 186. [Google Scholar]
- Geist, V. On the taxonomy of giant sheep (Ovis ammon Linnaeus, 1766). Can. J. Zool. 1991, 69, 706–723. [Google Scholar] [CrossRef]
- Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E.; Reeder, D.M. (Eds.) The Johns Hopkins University Press: Baltimore, MD, USA, 2005; Volume 1–2, 2142p. [Google Scholar]
- Fedosenko, A.K.; Blank, D.A. Ovis ammon. Mamm. Species 2005, 773, 1–15. [Google Scholar] [CrossRef]
- Shackleton, D.M.; Lovari, S. Classification adopted for the Caprinae survey. In Wild Sheep and Goats and Their Relatives. Status Survey and Conservation Action Plan for Caprinae; Shackleton, D.M., Ed.; IUCN/SSC Caprinae Specialist Group: Gland, Switzerland; Cambridge, UK, 1997; pp. 9–14. [Google Scholar]
- Amgalanbaatar, S.; Reading, R.P. Altai argali. In Endangered Animals: Conflicting Issues; Reading, R.P., Miller, B., Eds.; Greenwood Press: Westport, CT, USA, 2000; pp. 5–9. [Google Scholar]
- Michel, S.; Muratov, R. Survey on Marco Polo Sheep and Other Mammal Species in the Eastern Pamir (Republic of Tajikistan, GBAO); Nature Protection Team, Khorog and Institute of Zoology and Parasitology of the Academy of Sciences of the Republic of Tajikistan: Dushanbe, Tajikistan, 2010. [Google Scholar]
- Reading, R.; Michel, S.; Amgalanbaatar, S. Ovis ammon. In The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2020; p. e.T15733A22146397. [Google Scholar] [CrossRef]
- Rezaei, H.R.; Naderi, S.; Chintauan-Marquier, I.C.; Taberlet, P.; Virk, A.T.; Naghash, H.R.; Rioux, D.; Kaboli, M.; Pompanon, F. Evolution and taxonomy of the wild species of the genus Ovis (Mammalia, Artiodactyla, Bovidae). Mol. Phylogenet. Evol. 2010, 54, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Ludt, C.J.; Schroeder, W.; Rottmann, O.; Kuehn, R. Mitochondrial DNA phylogeography of red deer (Cervus elaphus). Mol. Phylogenet. Evol. 2004, 31, 1064–1083. [Google Scholar] [CrossRef] [PubMed]
- Ropiquet, A.; Hassanin, A. Molecular evidence for the polyphyly of the genus Hemitragus (Mammalia, Bovidae). Mol. Phylogenet. Evol. 2005, 36, 154–168. [Google Scholar] [CrossRef]
- Dotsev, A.V.; Deniskova, T.E.; Okhlopkov, I.M.; Mészáros, G.; Sölkner, J.; Reyer, H.; Wimmers, K.; Brem, G.; Zinovieva, N.A. Genome-wide SNP analysis unveils genetic structure and phylogeographic history of snow sheep (Ovis nivicola) populations inhabiting the Verkhoyansk Mountains and Momsky Ridge (northeastern Siberia). Ecol. Evol. 2018, 8, 8000–8010. [Google Scholar] [CrossRef] [PubMed]
- Dotsev, A.V.; Kunz, E.; Kharzinova, V.R.; Okhlopkov, I.M.; Lv, F.-H.; Li, M.-H.; Rodionov, A.N.; Shakhin, A.V.; Sipko, T.P.; Medvedev, D.G.; et al. Mitochondrial DNA Analysis Clarifies Taxonomic Status of the Northernmost Snow Sheep (Ovis nivicola) Population. Life 2021, 11, 252. [Google Scholar] [CrossRef]
- Dotsev, A.V.; Rodionov, A.N.; Kharzinova, V.R.; Petrov, S.N.; Medvedev, D.G.; Bagirov, V.A.; Brem, G.; Zinovieva, N.A. An Assessment of Applicability of SNP Chip Developed for Domestic Goats in Genetic Studies of Caucasian Tur (Capra caucasica). Diversity 2021, 13, 312. [Google Scholar] [CrossRef]
- Wu, C.H.; Zhang, Y.P.; Bunch, T.D.; Wang, S.; Wang, W. Mitochondrial control region sequence variation within the argali wild sheep (Ovis ammon): Evolution and conservation relevance. Mammalia 2003, 67, 109–118. [Google Scholar] [CrossRef]
- Tserenbataa, T.; Ramey, R.R.; Ryder, O.A.; Quinn, T.W.; Reading, R.P. A population genetic comparison of argali sheep (Ovis ammon) in Mongolia using the ND5 gene of mitochondrial DNA; implications for conservation. Mol. Ecol. 2004, 13, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Frisina, M.R.; Webster, M.S.; Ulziimaa, G. Genetic Differentiation of Argali Sheep Ovis ammon in Mongolia Revealed by Mitochondrial Control Region and Nuclear Microsatellites Analyses. J. Bombay Nat. Hist. Soc. 2009, 106, 38–44. [Google Scholar]
- Luikart, G.; Amish, S.J.; Winnie, J.; Beja-Pereira, A.; Godinho, R.; Allendorf, F.W.; Harris, R.B. High connectivity among argali sheep from Afghanistan and adjacent countries: Inferences from neutral and candidate gene microsatellites. Conserv. Genet. 2011, 12, 921–931. [Google Scholar] [CrossRef]
- Delgerzul, B.; Unudbayasgalan, Z.; Bilguun, T.; Battsetseg, C.; Galbadrahk, B.; Tserendulam, B. Genetic comparison of Altai and Gobi argali sheep (Ovis ammon) populations using mitochondrial and microsatellite markers: Implication on conservation. Proc. Mong. Acad. Sci. 2019, 59, 54–61. [Google Scholar] [CrossRef]
- Miller, J.; Poissant, J.; Kijas, J.; Coltman, D. The International Sheep Genomics Consortium. A genome-wide set of SNPs detects population substructure and long range linkage disequilibrium in wild sheep. Mol. Ecol. Resour. 2010, 11, 314–322. [Google Scholar] [CrossRef]
- Sim, Z.; Hall, J.C.; Jex, B.; Hegel, T.M.; Coltman, D.W. Genome-wide set of SNPs reveals evidence for two glacial refugia and admixture from postglacial recolonization in an alpine ungulate. Mol. Ecol. 2016, 25, 3696–3705. [Google Scholar] [CrossRef]
- Sim, Z.; Coltman, D.W. Heritability of Horn Size in Thinhorn Sheep. Front. Genet. 2019, 10, 959. [Google Scholar] [CrossRef]
- Barbato, M.; Hailer, F.; Orozco-terWengel, P.; Kijas, J.; Mereu, P.; Cabras, P.; Mazza, R.; Pirastru, M.; Bruford, M.W. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci. Rep. 2017, 7, 7623. [Google Scholar] [CrossRef]
- Yang, C.; Xiang, C.; Qi, W.; Xia, S.; Tu, F.; Zhang, X.; Moermond, T.; Yue, B. Phylogenetic analyses and improved resolution of the family Bovidae based on complete mitochondrial genomes. Biochem. Syst. Ecol. 2013, 48, 136–143. [Google Scholar] [CrossRef]
- Kijas, J.W.; Porto-Neto, L.; Dominik, S.; Reverter, A.; Bunch, R.; Mcculloch, R.; Hayes, B.J.; Brauning, R.; McEwan, J. The International Sheep Genomics Consortium. Linkage disequilibrium over short physical distances measured in sheep using a high-density SNP chip. Anim. Genet. 2014, 45, 754–757. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.B.; Weir, B.S.; Cockerham, C.C. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics 1983, 105, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Pembleton, L.W.; Cogan, N.O.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef]
- Fitak, R.R. OptM: Estimating the optimal number of migration edges on population trees using Treemix. Biol. Methods Protoc. 2021, 6, bpab017. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. pophelper: An R package and web app to analyze and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Stoffel, M.A.; Esser, M.; Kardos, M.; Humble, E.; Nichols, H.; David, P.; Hoffman, J.I. inbreedR: An R package for the analysis of inbreeding based on genetic markers. Methods Ecol. Evol. 2016, 7, 1331–1339. [Google Scholar] [CrossRef]
- Edgar, R.C. Quality measures for protein alignment benchmarks. Nucleic Acids Res. 2010, 38, 2145–2153. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- FigTree. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 January 2023).
- Becker, R.A.; Wilks, A.R.; Brownrigg, R.; Minka, T.P.; Deckmyn, A. Maps: Draw Geographical Maps. R Package Version 3.3.0. 2018. Available online: https://CRAN.R-project.org/package=maps (accessed on 28 March 2023).
- Sopin, A.V. Intraspecific relationships in Ovis ammon (Artiodactyla, Bovidae). Zool. Zhurnal 1982, 61, 1882–1892. [Google Scholar]
- Kapitanova, D.V.; Lopatin, A.V.; Subbotin, A.E.; Wall, W.A. Cranial morphometry and taxonomy of argali, Ovis ammon (Artiodactyla, Bovidae), from former Soviet Union and Mongolia. Russ. J. Theriol. 2004, 3, 89–106. [Google Scholar] [CrossRef]
- Danilkin, A.A. Mammals of Russia and Adjacent Regions: Hollow-Horned Ruminants (Bovidae); KMK: Moscow, Russia, 2005. (In Russian) [Google Scholar]
- Nasonov, N. Űber Ovis severtzovi Nas. und über die Methode der Untersuchungen der Hörner der Wildschafe in systematischer Hinsicht. Bull. Académie Impériale Sci. St.-Pétersbourg 1914, 8, 761–778. [Google Scholar]
- Geptner, V.G.; Nasimovich, A.A.; Bannikov, A.G. Mammals of the Soviet Union. Ungulates and Odd-Toed Ungulates; Higher School: Moscow, Russia, 1961; Volume 1, p. 776. (In Russian) [Google Scholar]
- Groves, C.P.; Grubb, P. Ungulate Taxonomy; The John Hopkins University Press: Baltimore, MD, USA, 2011. [Google Scholar]
- Bunch, T.D.; Vorontsov, N.N.; Lyapunova, E.A.; Hoffmann, R.S. Chromosome number of Severtsov’s sheep (Ovis ammon severtzovi): G-banded karyotype comparisons within Ovis. J. Hered. 1998, 89, 266–269. [Google Scholar] [CrossRef] [PubMed]




| O. a. polii | O. a. karelini | O. a. severtzovi | O. a. ammon | O. a. darwini | O. vignei | |
|---|---|---|---|---|---|---|
| O. a. polii | 0.000 | 0.346 | 0.581 | 0.592 | 0.592 | 0.609 |
| O. a. karelini | 0.083 | 0.000 | 0.557 | 0.560 | 0.561 | 0.599 |
| O. a. severtzovi | 0.266 | 0.237 | 0.000 | 0.696 | 0.695 | 0.639 |
| O. a. ammon | 0.290 | 0.251 | 0.421 | 0.000 | 0.377 | 0.679 |
| O. a. darwini | 0.284 | 0.244 | 0.414 | 0.040 | 0.000 | 0.677 |
| O. vignei | 0.339 | 0.333 | 0.325 | 0.397 | 0.386 | 0.000 |
| Population | n | Ho ± se | uHe ± se | Fis [CI 95%] | AR ± se | MLH Mdn [Range] |
|---|---|---|---|---|---|---|
| O. a. polii | 12 | 0.277 ± 0.001 | 0.286 ± 0.001 | 0.024 [0.022; 0.026] | 1.685 ± 0.001 | 0.279 [0.212; 0.290] |
| O. a. karelini | 15 | 0.289 ± 0.001 | 0.295 ± 0.001 | 0.019 [0.017; 0.021] | 1.714 ± 0.001 | 0.288 [0.259; 0.299] |
| O. a. severtzovi | 4 | 0.242 ± 0.001 | 0.245 ± 0.001 | 0.011 [0.007; 0.015] | 1.570 ± 0.002 | 0.240 [0.237; 0.248] |
| O. a. ammon | 6 | 0.173 ± 0.001 | 0.203 ± 0.001 | 0.128 [0.124; 0.132] | 1.484 ± 0.002 | 0.179 [0.110; 0.191] |
| O. a. darwini | 5 | 0.185 ± 0.001 | 0.208 ± 0.001 | 0.092 [0.088; 0.096] | 1.499 ± 0.002 | 0.180 [0.171; 0.208] |
| O. vignei | 10 | 0.312 ± 0.001 | 0.356 ± 0.001 | 0.111 [0.108; 0.114] | 1.813 ± 0.001 | 0.313 [0.268; 0.329] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dotsev, A.; Koshkina, O.; Kharzinova, V.; Deniskova, T.; Reyer, H.; Kunz, E.; Mészáros, G.; Shakhin, A.; Petrov, S.; Medvedev, D.; et al. Genome-Wide Insights into Intraspecific Taxonomy and Genetic Diversity of Argali (Ovis ammon). Diversity 2023, 15, 627. https://doi.org/10.3390/d15050627
Dotsev A, Koshkina O, Kharzinova V, Deniskova T, Reyer H, Kunz E, Mészáros G, Shakhin A, Petrov S, Medvedev D, et al. Genome-Wide Insights into Intraspecific Taxonomy and Genetic Diversity of Argali (Ovis ammon). Diversity. 2023; 15(5):627. https://doi.org/10.3390/d15050627
Chicago/Turabian StyleDotsev, Arsen, Olga Koshkina, Veronika Kharzinova, Tatiana Deniskova, Henry Reyer, Elisabeth Kunz, Gábor Mészáros, Alexey Shakhin, Sergey Petrov, Dmitry Medvedev, and et al. 2023. "Genome-Wide Insights into Intraspecific Taxonomy and Genetic Diversity of Argali (Ovis ammon)" Diversity 15, no. 5: 627. https://doi.org/10.3390/d15050627
APA StyleDotsev, A., Koshkina, O., Kharzinova, V., Deniskova, T., Reyer, H., Kunz, E., Mészáros, G., Shakhin, A., Petrov, S., Medvedev, D., Kuksin, A., Bat-Erdene, G., Munkhtsog, B., Bagirov, V., Wimmers, K., Sölkner, J., Medugorac, I., & Zinovieva, N. (2023). Genome-Wide Insights into Intraspecific Taxonomy and Genetic Diversity of Argali (Ovis ammon). Diversity, 15(5), 627. https://doi.org/10.3390/d15050627











