Adaptive Genetic Management of a Reintroduction Program from Captive Breeding to Metapopulation Management of an Arboreal Marsupial
Abstract
:1. Introduction
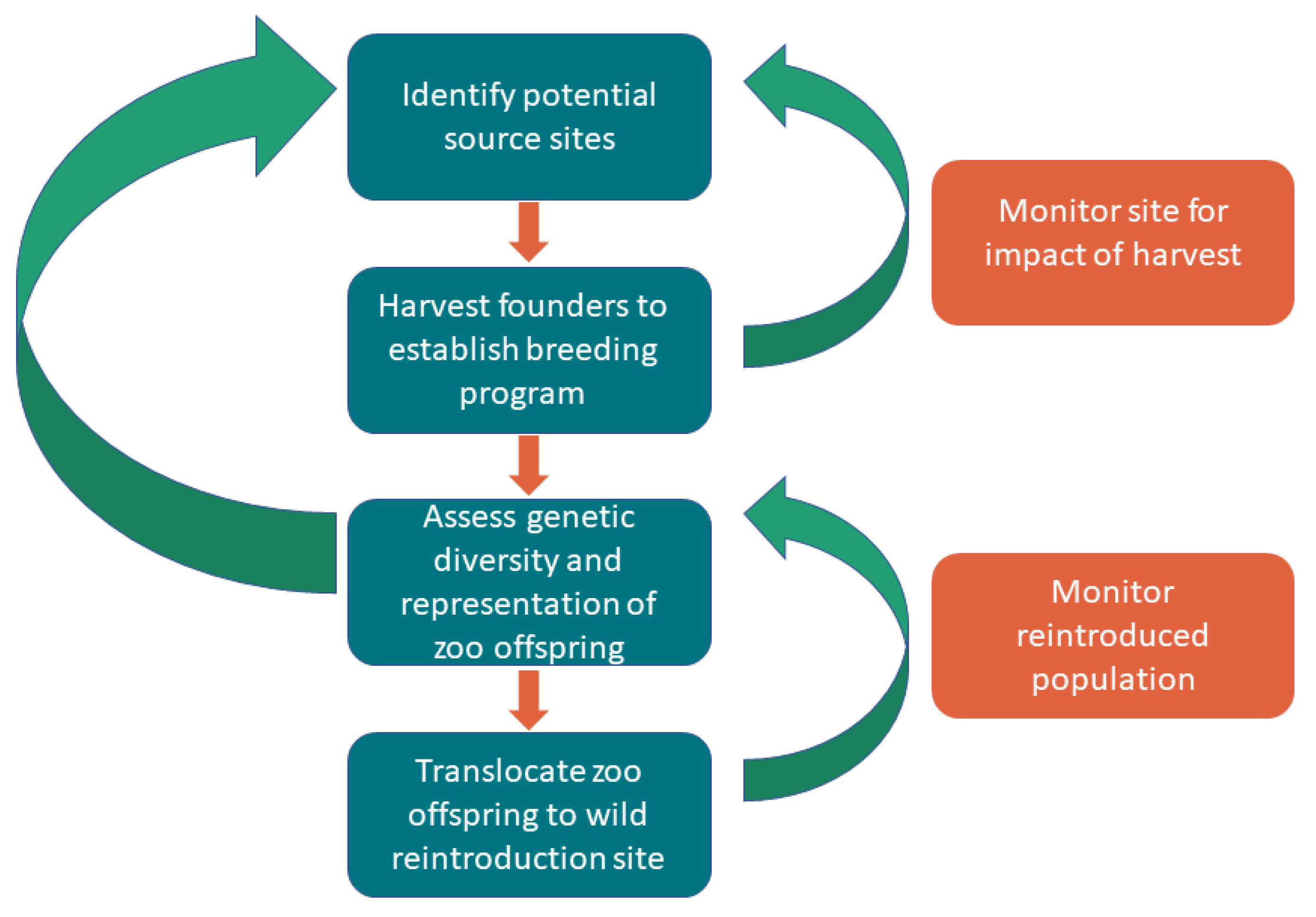
2. Materials and Methods
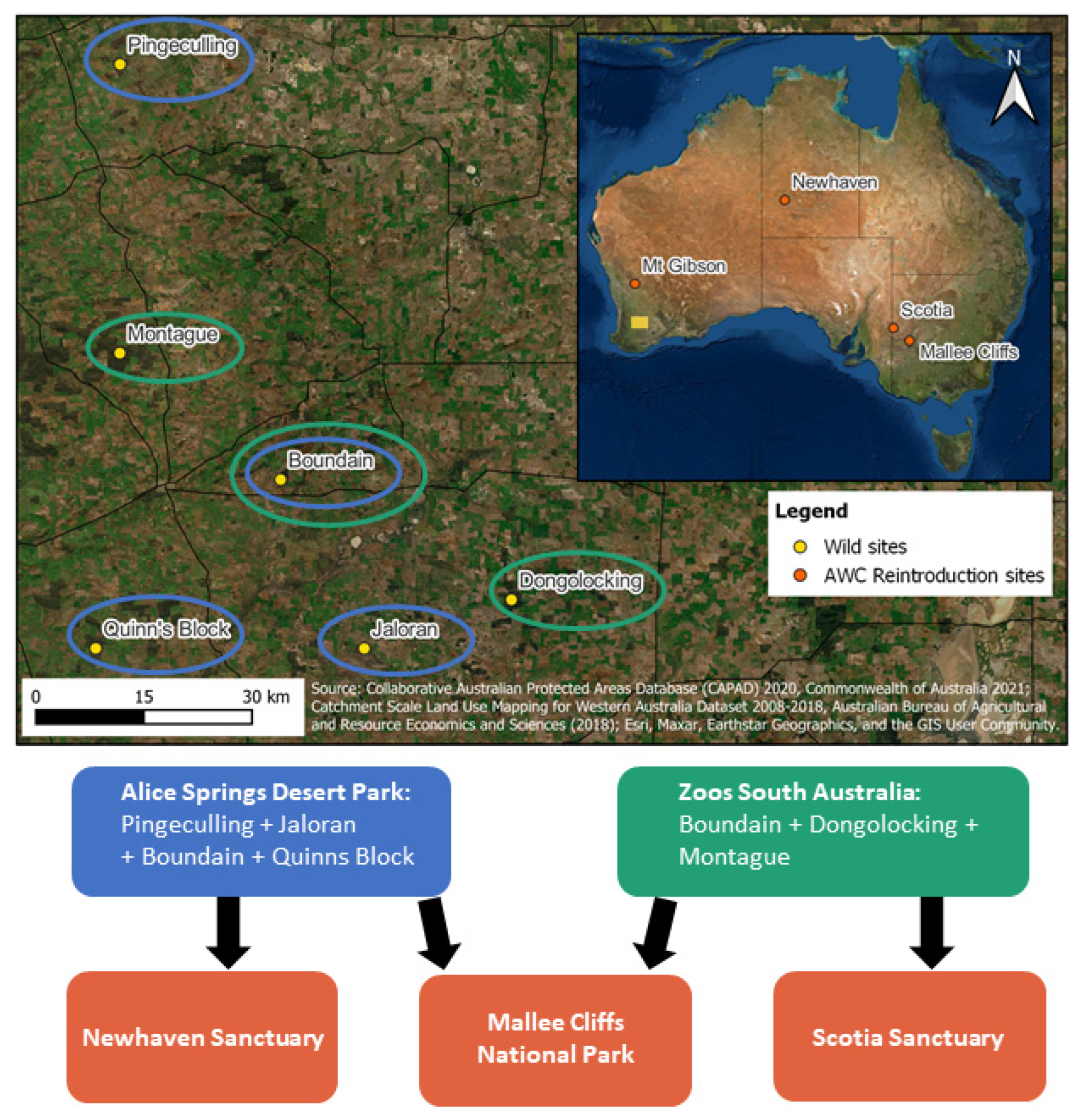
2.1. Wild Source Population Assessment
2.2. Translocation Methods
2.3. Captive Breeding Program
2.4. Genetic Methods
2.4.1. DNA Extraction and Sequencing
2.4.2. SNP Calling
2.4.3. Population Genetic Analyses
3. Results
3.1. Wild Assessment and Harvest
3.2. Captive Breeding and Translocations to Establish Metapopulation
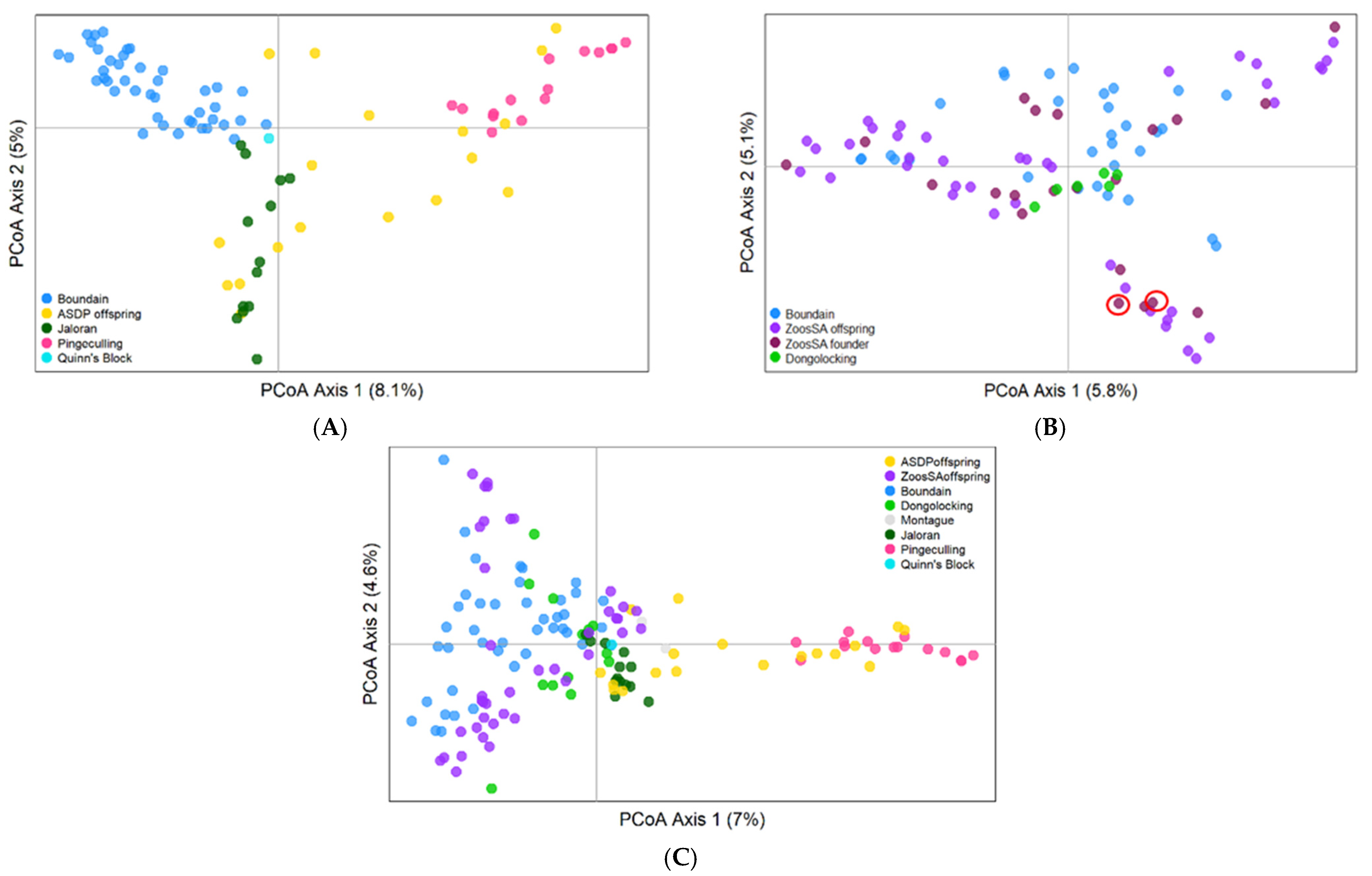
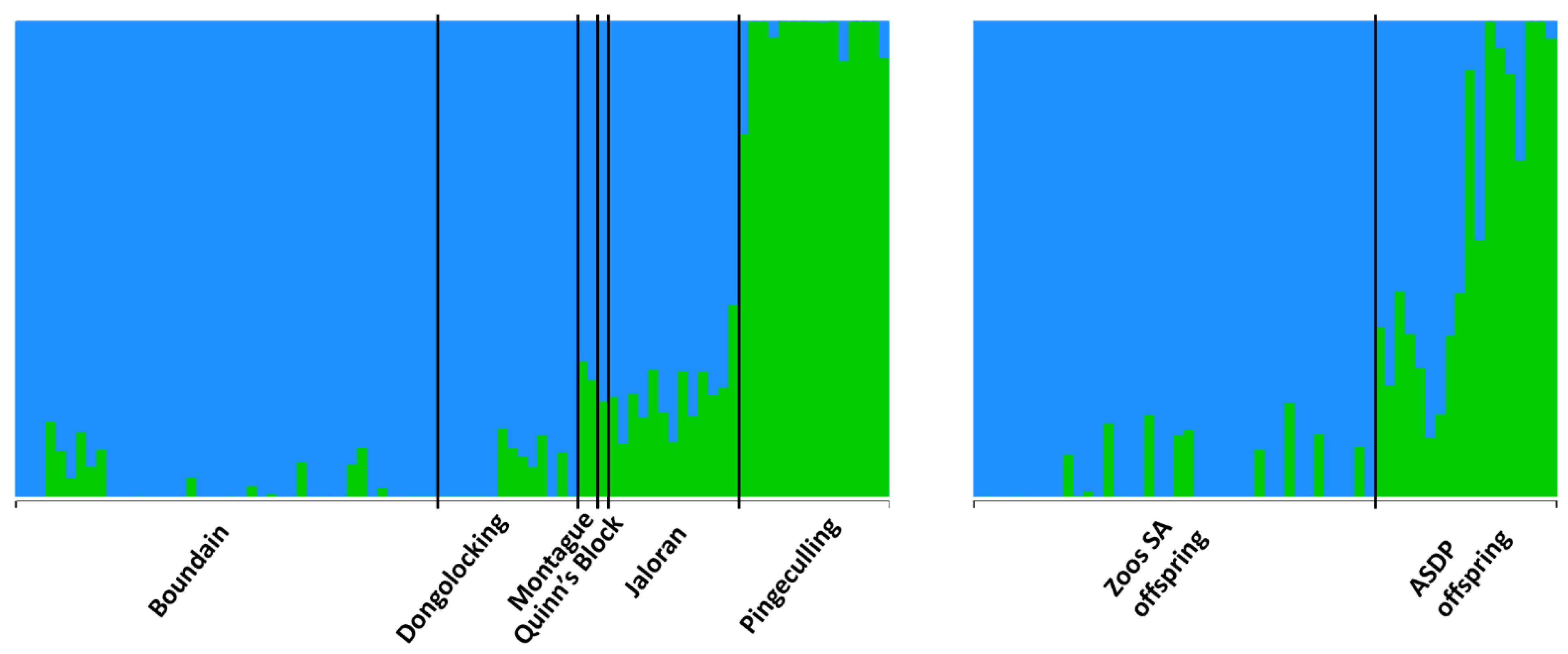
3.3. Genetic Data
3.3.1. Sequencing and SNP Calling
3.3.2. Population Genetic Analyses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berger-Tal, O.; Blumstein, D.T.; Swaisgood, R.R. Conservation Translocations: A Review of Common Difficulties and Promising Directions. Anim. Conserv. 2020, 23, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.J.; Pierson, J.C.; Neaves, L.E.; Gordon, I.J.; Ross, C.E.; Brockett, B.; Rapley, S.; Wilson, B.A.; Smith, K.J.; Andrewartha, T.; et al. Trends in Animal Translocation Research. Ecography 2023, 2023, e06528. [Google Scholar] [CrossRef]
- Morris, S.D.; Brook, B.W.; Moseby, K.E.; Johnson, C.N. Factors Affecting Success of Conservation Translocations of Terrestrial Vertebrates: A Global Systematic Review. Glob. Ecol. Conserv. 2021, 28, e01630. [Google Scholar] [CrossRef]
- Teitelbaum, C.S.; Sirén, A.P.K.; Coffel, E.; Foster, J.R.; Frair, J.L.; Hinton, J.W.; Horton, R.M.; Kramer, D.W.; Lesk, C.; Raymond, C.; et al. Habitat Use as Indicator of Adaptive Capacity to Climate Change. Divers. Distrib. 2021, 27, 655–667. [Google Scholar] [CrossRef]
- Forester, B.R.; Beever, E.A.; Darst, C.; Szymanski, J.; Funk, W.C. Linking Evolutionary Potential to Extinction Risk: Applications and Future Directions. Front. Ecol. Environ. 2022, 20, 507–515. [Google Scholar] [CrossRef]
- Silcock, J.L.; Simmons, C.L.; Monks, L.; Dillon, R.; Reiter, N.; Jusaitis, M.; Vesk, P.A.; Byrne, M.; Coates, D.J. Threatened Plant Translocation in Australia: A Review. Biol. Conserv. 2019, 236, 211–222. [Google Scholar] [CrossRef]
- Brockett, B.; Banks, S.; Neaves, L.E.; Gordon, I.J.; Pierson, J.C.; Manning, A.D. Establishment, Persistence and the Importance of Longitudinal Monitoring in Multi-source Reintroductions. Anim. Conserv. 2022, 25, 550–565. [Google Scholar] [CrossRef]
- Witzenberger, K.A.; Hochkirch, A. Ex Situ Conservation Genetics: A Review of Molecular Studies on the Genetic Consequences of Captive Breeding Programmes for Endangered Animal Species. Biodivers. Conserv. 2011, 20, 1843–1861. [Google Scholar] [CrossRef]
- Snyder, N.F.R.; Derrickson, S.R.; Beissinger, S.R.; Wiley, J.W.; Smith, T.B.; Toone, W.D.; Miller, B. Limitations of Captive Breeding in Endangered Species Recovery. Conserv. Biol. 1996, 10, 338–348. [Google Scholar] [CrossRef] [Green Version]
- Crates, R.; Stojanovic, D.; Heinsohn, R. The Phenotypic Costs of Captivity. Biol. Rev. 2022, 98, 434–449. [Google Scholar] [CrossRef]
- Conde, D.A.; Flesness, N.; Colchero, F.; Jones, O.R.; Scheuerlein, A. An Emerging Role of Zoos to Conserve Biodiversity. Science 2011, 331, 1390–1391. [Google Scholar] [CrossRef] [PubMed]
- Lacy, R.C. Analysis of Founder Representation in Pedigress: Founder Equivalents and Founder Genome Equivalents. Zoo Biol. 1989, 8, 111–123. [Google Scholar] [CrossRef]
- Robert, A. Captive Breeding Genetics and Reintroduction Success. Biol. Conserv. 2009, 142, 2915–2922. [Google Scholar] [CrossRef]
- Williams, S.E.; Hoffman, E.A. Minimizing Genetic Adaptation in Captive Breeding Programs: A Review. Biol. Conserv. 2009, 142, 2388–2400. [Google Scholar] [CrossRef]
- Ito, H.; Ogden, R.; Langenhorst, T.; Inoue-Murayama, M. Contrasting Results from Molecular and Pedigree-Based Population Diversity Measures in Captive Zebra Highlight Challenges Facing Genetic Management of Zoo Populations: Genetic Diversity of MtDNA in Grevy’s Zebra. Zoo Biol. 2017, 36, 87–94. [Google Scholar] [CrossRef]
- Schulte-Hostedde, A.I.; Mastromonaco, G.F. Integrating Evolution in the Management of Captive Zoo Populations. Evol. Appl. 2015, 8, 413–422. [Google Scholar] [CrossRef]
- Rabier, R.; Robert, A.; Lacroix, F.; Lesobre, L. Genetic Assessment of a Conservation Breeding Program of the Houbara Bustard (Chlamydotis Undulata Undulata) in Morocco, Based on Pedigree and Molecular Analyses. Zoo Biol. 2020, 39, 422–435. [Google Scholar] [CrossRef]
- Attard, C.R.M.; Möller, L.M.; Sasaki, M.; Hammer, M.P.; Bice, C.M.; Brauer, C.J.; Carvalho, D.C.; Harris, J.O.; Beheregaray, L.B. A Novel Holistic Framework for Genetic-Based Captive-Breeding and Reintroduction Programs. Conserv. Biol. 2016, 30, 1060–1069. [Google Scholar] [CrossRef]
- Williams, B.K. Adaptive Management of Natural Resources—Framework and Issues. J. Environ. Manag. 2011, 92, 1346–1353. [Google Scholar] [CrossRef]
- Westgate, M.J.; Likens, G.E.; Lindenmayer, D.B. Adaptive Management of Biological Systems: A Review. Biol. Conserv. 2013, 158, 128–139. [Google Scholar] [CrossRef]
- Pacioni, C.; Trocini, S.; Wayne, A.F.; Rafferty, C.; Page, M. Integrating Population Genetics in an Adaptive Management Framework to Inform Management Strategies. Biodivers. Conserv. 2020, 29, 947–966. [Google Scholar] [CrossRef]
- Short, J.; HIde, A. Distribution and Status of the Red-Tailed Phascogale (Phascogale Calura). Aust. Mammal. 2011, 34, 88–99. [Google Scholar] [CrossRef] [Green Version]
- Short, J.; HIde, A.; Stone, M. Habitat Requirements of the Endangered Red-Tailed Phascogale, Phascogale Calura. Wildl. Res. 2011, 38, 359–369. [Google Scholar] [CrossRef]
- Bradley, A.J. Reproduction and Life History in the Red-Tailed Phascogale Phascogale Calura (Marsupialia: Dasyuridae): The Adaptive-Stress Senescence Hypothesis. J. Zool. 1997, 241, 739–755. [Google Scholar] [CrossRef]
- Ringma, J.; Legge, S.; Woinarski, J.; Radford, J.; Wintle, B.; Bode, M. Australia’s Mammal Fauna Requires a Strategic and Enhanced Network of Predator-Free Havens. Nat. Ecol. Evol. 2018, 2, 410–411. [Google Scholar] [CrossRef]
- Legge, S.; Woinarski, J.C.Z.; Burbidge, A.A.; Palmer, R.; Ringma, J.; Radford, J.Q.; Mitchell, N.; Bode, M.; Wintle, B.; Baseler, M.; et al. Havens for Threatened Australian Mammals: The Contributions of Fenced Areas and Offshore Islands to the Protection of Mammal Species Susceptible to Introduced Predators. Wildl. Res. 2018, 45, 627. [Google Scholar] [CrossRef]
- Wright, B.; Farquharson, K.A.; McLennan, E.A.; Belov, K.; Hogg, C.J.; Grueber, C.E. From Reference Genomes to Population Genomics: Comparing Three Reference-Aligned Reduced-Representation Sequencing Pipelines in Two Wildlife Species. BMC Genom. 2019, 20, 453. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.M.; Amores, P.; Hohenlohe, P.A.; Cresko, W.A.; Postlethwait, J.H. Stacks: Building and Genotyping Loci de Novo from Short-Read Sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M. Usadel Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandies, P.A.; Tang, S.; Johnson, C.J.; Hogg, C.J.; Belov, K. The First Antechinus Reference Genome Provides a Resource for Investigating the Genetic Basis of Semelparity and Age-Related Neuro-Pathologies. Gigabyte 2020, 2020, gigabyte7. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennel, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Genome Project Data Processing Subgroup, 2009. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Knaus, B.J.; Grünwald, N.J. Vcfr: A Package to Manipulate and Visualize Variant Call Format Data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Coulon, A. Genhet: An Easy-to-Use R Function to Estimate Individual Heterozygosity. Mol. Ecol. Resour. 2010, 10, 167–169. [Google Scholar] [CrossRef]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. DiveRsity: An R Package for the Estimation and Exploration of Population Genetics Parameters and Their Associated Errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. HEIRFSTAT, a Package for R to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Pembleton, L.W.; Cogan, N.O.; Forster, J.W. St AMPP: An R Package for Calculation of Genetic Differentiation and Structure of Mixed-ploidy Level Populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Stephens, M.; Pritchard, J.K. FastSTRUCTURE: Variational Inference of Population Structure in Large SNP Data Sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Wang, J. COANCESTRY: A Program for Simulating, Estimating and Analysing Relatedness and Inbreeding Coefficients. Mol. Ecol. Resour. 2011, 11, 141–145. [Google Scholar] [CrossRef]
- Hogg, C.J.; Wright, B.; Morris, K.M.; Lee, A.V.; Ivy, J.A.; Grueber, C.E.; Belov, K. Founder Relationships and Conservation Management: Empirical Kinships Reveal the Effect on Breeding Programmes When Founders Are Assumed to Be Unrelated. Anim. Conserv. 2019, 22, 348–361. [Google Scholar] [CrossRef] [Green Version]
- Hoban, S. New Guidance for Ex Situ Gene Conservation: Sampling Realistic Population Systems and Accouting for Collection Attrition. Biol. Conserv. 2019, 235, 199–208. [Google Scholar] [CrossRef]
- Bussolini, L.T.; Crates, R.; Magrath, M.J.; Stojanovic, D. Identifying Factors Affecting Captive Breeding Success in a Critically Endangered Species. Emu—Austral Ornithol. 2023, 123, 161–169. [Google Scholar] [CrossRef]
- Mitchell, W.F.; Boulton, R.L.; Sunnucks, P.; Clarke, R.H. Are We Adequately Assessing the Demographic Impacts of Harvesting for Wild-Sourced Conservation Translocations. Conserv. Sci. Pract. 2022, 4, e569. [Google Scholar] [CrossRef]
- Wilson, B.A.; Evans, M.J.; Gordon, I.J.; Pierson, J.C.; Brockett, B.M.; Wimpenny, C.; Batson, W.G.; Newport, J.; Manning, A.D. Roadmap to Recovery Revealed through the Reintroduction of an IUCN Red List Species. Biodivers. Conserv. 2023, 32, 227–248. [Google Scholar] [CrossRef]
| Source | Males | Females | Total |
|---|---|---|---|
| (a) | |||
| Pingeculling | 3 | 7 | 10 |
| Boundain | 2 | 4 | 6 |
| Jaloran | 1 | 2 | 3 |
| Quinns Block | 0 | 2 | 2 |
| Total | 6 | 15 | 21 |
| (b) | |||
| Boundain | 2 | 10 | 12 |
| Dongolocking | 2 | 6 | 8 |
| Montague | 2 | 0 | 2 |
| Total | 6 | 16 | 22 |
| Remnant | 2016 | 2017 | 2019 | 2020 | 2021 |
|---|---|---|---|---|---|
| Boyagin | 1 | 1 | NA | NA | NA |
| Dryandra | 0 | 1 | NA | 1 | 1 |
| Pingeculling | 1 | 21 | 12 | 1 | NA |
| Boundain | 4 | 6 | 13 | 1 | 17 |
| East Yornaning | 3 | 2 | 0 | NA | 5 |
| Tutanning | 5 | 4 | 4 | 4 | NA |
| Dongolocking | 10 | 7 | 9 | 1 | 12 |
| Jaloran | 0 | 5 | 12 | 1 | NA |
| West Ashby | 9 | 13 | 6 | 7 | 3 |
| Quinn’s Block | NA | NA | 4 | 1 | 0 |
| Contine Block | NA | NA | NA | 1 | 2 |
| Montague | NA | NA | NA | NA | 7 |
| N | AR (±SE) | HO (±SE) | HE (±SE) | HS (±SE) | FIS (95% CI) | MK (95% CI) | |
|---|---|---|---|---|---|---|---|
| ASDP offspring | 18 | 1.575 (0.006) | 0.204 (0.003) | 0.202 (0.003) | 1.049 (0.014) | −0.009 (−0.051, 0.024) | 0.016 (0.012, 0.022) |
| Zoos SA offspring | 40 | 1.566 (0.005) | 0.200 (0.003) | 0.200 (0.003) | 0.996 (0.030) | −0.001 (−0.022, 0.020) | 0.024 (0.019, 0.033) |
| Boundain | 42 | 1.548 (0.006) | 0.188 (0.003) | 0.196 (0.003) | 0.963 (0.015) | 0.039 (0.012, 0.062) | 0.017 (0.013, 0.025) |
| Dongolocking | 14 | 1.545 (0.006) | 0.194 (0.003) | 0.194 (0.003) | 0.990 (0.025) | 0.001 (−0.054, 0.041) | 0.006 (0.005, 0.009) |
| Jaloran | 13 | 1.538 (0.006) | 0.198 (0.003) | 0.194 (0.003) | 1.014 (0.050) | −0.019 (−0.132, 0.050) | 0.017 (0.014, 0.023) |
| Pingeculling | 15 | 1.517 (0.006) | 0.189 (0.003) | 0.188 (0.003) | 0.960 (0.029) | −0.007 (−0.072, 0.040) | 0.020 (0.018, 0.025) |
| ASDP Offspring | Zoos SA Offspring | Boundain | Dongolocking | Jaloran | Pingeculling | |
|---|---|---|---|---|---|---|
| ASDP offspring | NA | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Zoos SA offspring | 0.055 (0.052, 0.059) | NA | <0.001 | <0.001 | <0.001 | <0.001 |
| Boundain | 0.053 (0.049, 0.057) | 0.020 (0.018, 0.022) | NA | <0.001 | <0.001 | <0.001 |
| Dongolocking | 0.061 (0.057, 0.066) | 0.021 (0.019, 0.024) | 0.057 (0.053, 0.062) | NA | <0.001 | <0.001 |
| Jaloran | 0.041 (0.037, 0.044) | 0.056 (0.052, 0.059) | 0.062 (0.058, 0.066) | 0.051 (0.047, 0.056) | NA | <0.001 |
| Pingeculling | 0.045 (0.042, 0.049) | 0.106 (0.100, 0.113) | 0.115 (0.109, 0.122) | 0.111 (0.105, 0.119) | 0.114 (0.108, 0.121) | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pierson, J.C.; Berry, L.; Alexander, L.; Anson, J.; Birkett, M.; Kemp, L.; Pascoe, B.A.; Farquharson, K.A.; Hogg, C.J. Adaptive Genetic Management of a Reintroduction Program from Captive Breeding to Metapopulation Management of an Arboreal Marsupial. Diversity 2023, 15, 848. https://doi.org/10.3390/d15070848
Pierson JC, Berry L, Alexander L, Anson J, Birkett M, Kemp L, Pascoe BA, Farquharson KA, Hogg CJ. Adaptive Genetic Management of a Reintroduction Program from Captive Breeding to Metapopulation Management of an Arboreal Marsupial. Diversity. 2023; 15(7):848. https://doi.org/10.3390/d15070848
Chicago/Turabian StylePierson, Jennifer C., Laurence Berry, Lauren Alexander, Jennifer Anson, Michelle Birkett, Leah Kemp, Bruce A. Pascoe, Katherine A. Farquharson, and Carolyn J. Hogg. 2023. "Adaptive Genetic Management of a Reintroduction Program from Captive Breeding to Metapopulation Management of an Arboreal Marsupial" Diversity 15, no. 7: 848. https://doi.org/10.3390/d15070848
APA StylePierson, J. C., Berry, L., Alexander, L., Anson, J., Birkett, M., Kemp, L., Pascoe, B. A., Farquharson, K. A., & Hogg, C. J. (2023). Adaptive Genetic Management of a Reintroduction Program from Captive Breeding to Metapopulation Management of an Arboreal Marsupial. Diversity, 15(7), 848. https://doi.org/10.3390/d15070848






