Morphological and Molecular Evaluation of Pseudanabaena epilithica sp. nov. and P. suomiensis sp. nov. (Pseudanabaenaceae, Cyanobacteria) from Finland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains, Microbiological Media and Growth Conditions
2.1.1. Origin of Cyanobacterial Strains, Isolation and Maintenance
2.1.2. Bacteria and Fungi
2.2. Morphological Evaluation and Type Material Preparation
2.2.1. Microscopy
2.2.2. Type Material Preparation
2.3. Molecular Characterization
2.3.1. DNA Extraction, PCR Amplification and Sequencing
2.3.2. Phylogenetic Analysis
2.3.3. Study of 16S–23S Internal Transcribed Spacer (ITS) Region
2.4. Biochemical Evaluation
2.4.1. Antimicrobial Susceptibility Tests
2.4.2. Extraction of Phycobiliproteins
2.4.3. Liquid Chromatography
3. Results
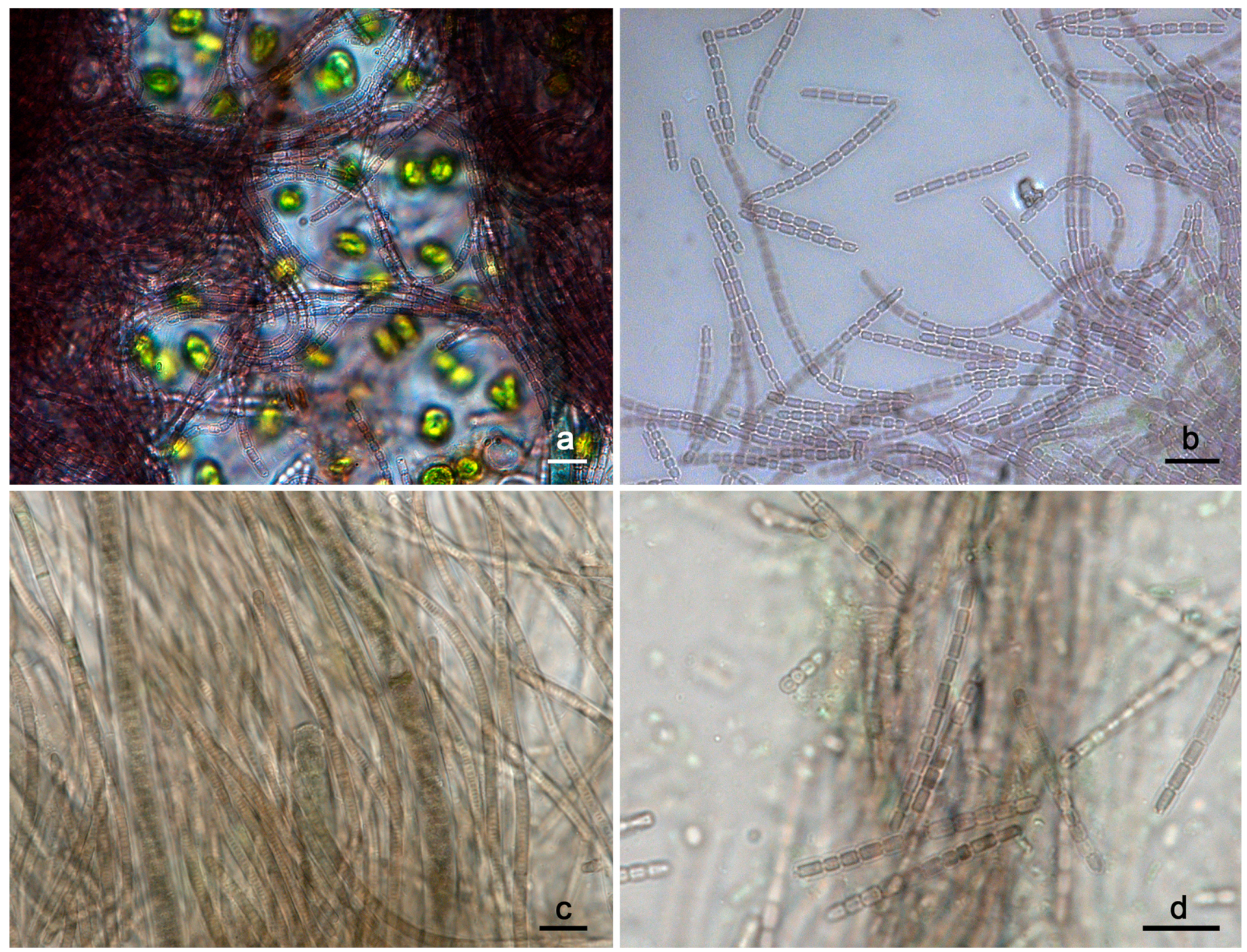
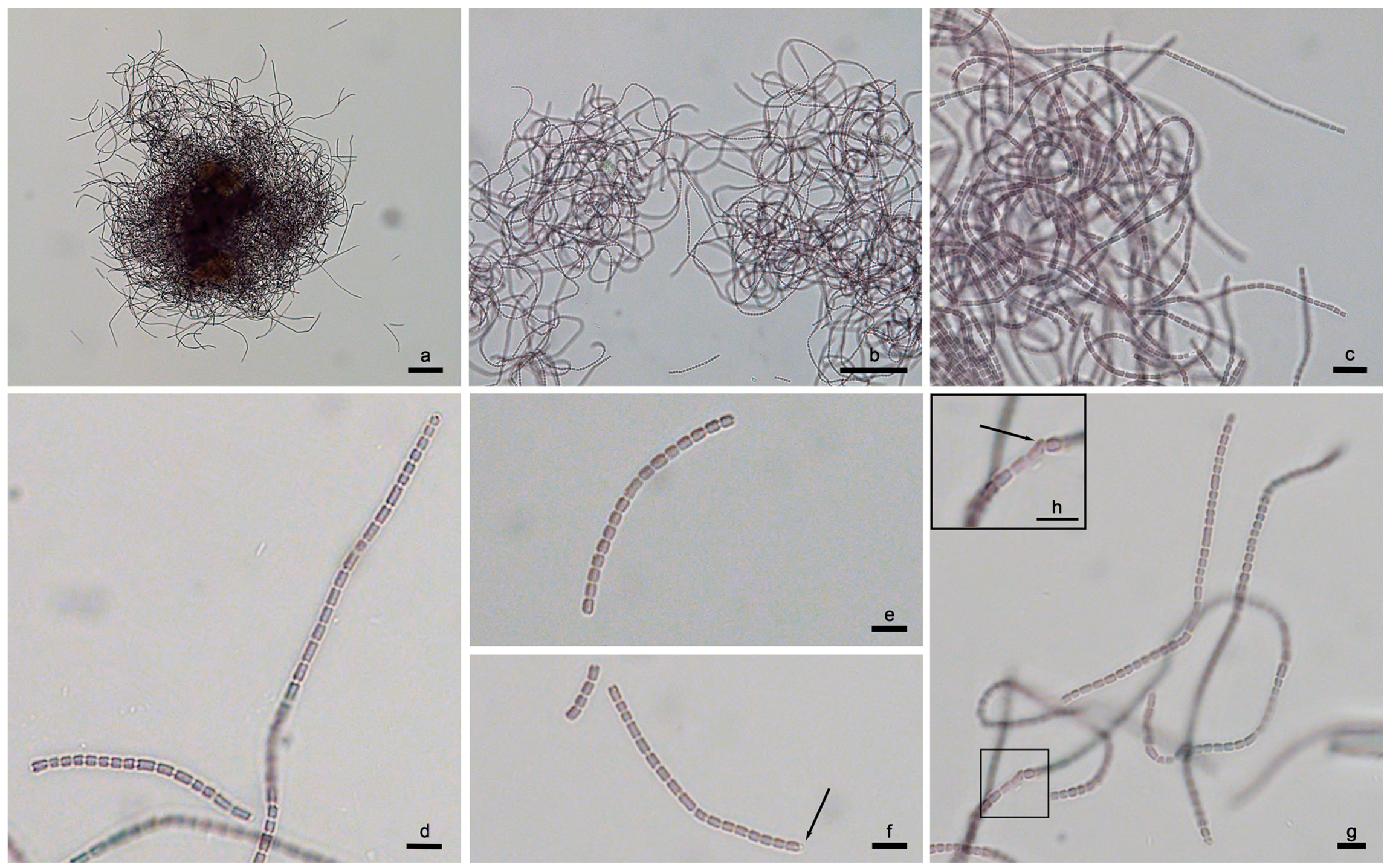
3.1. Strain Morphology and Habitat
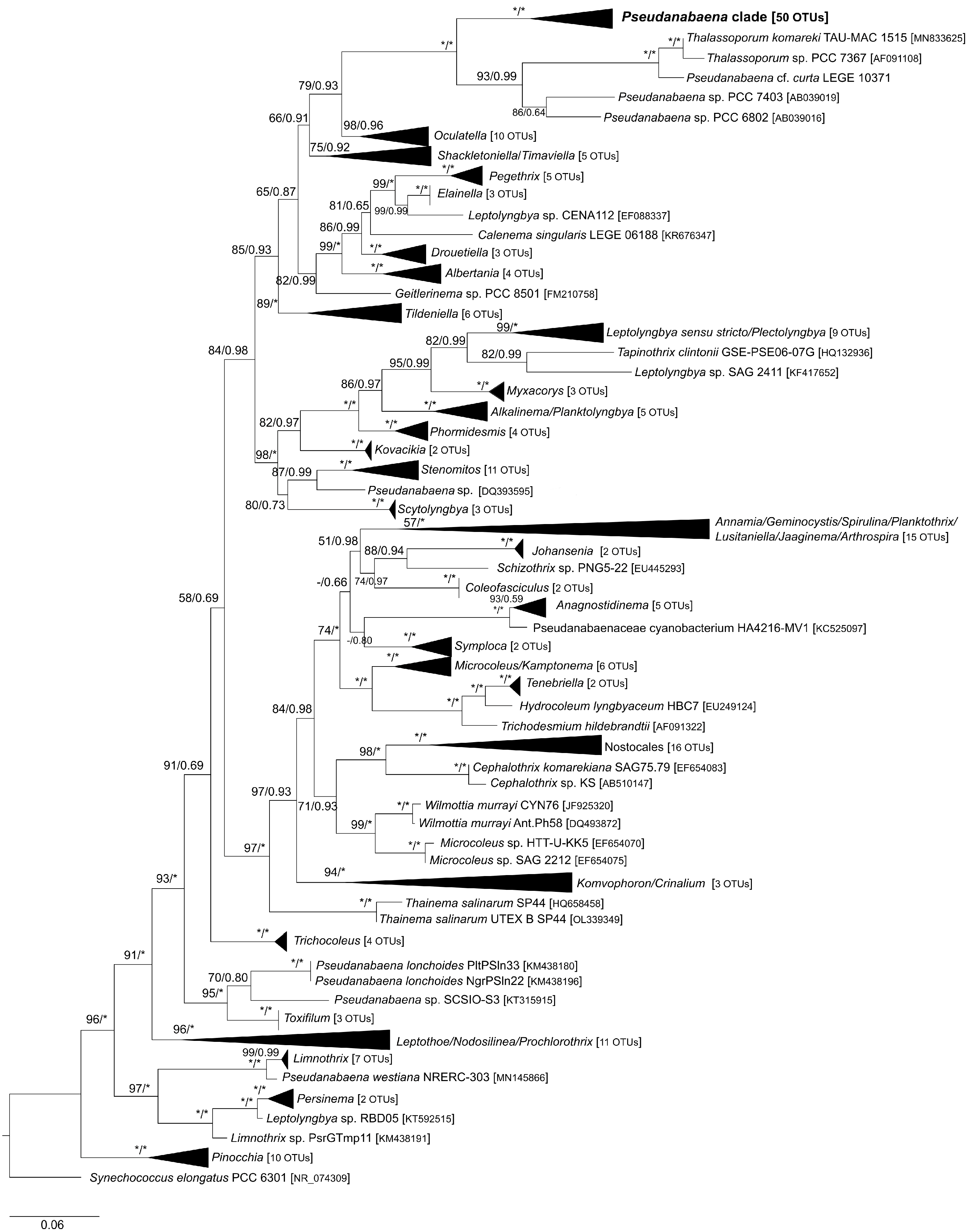
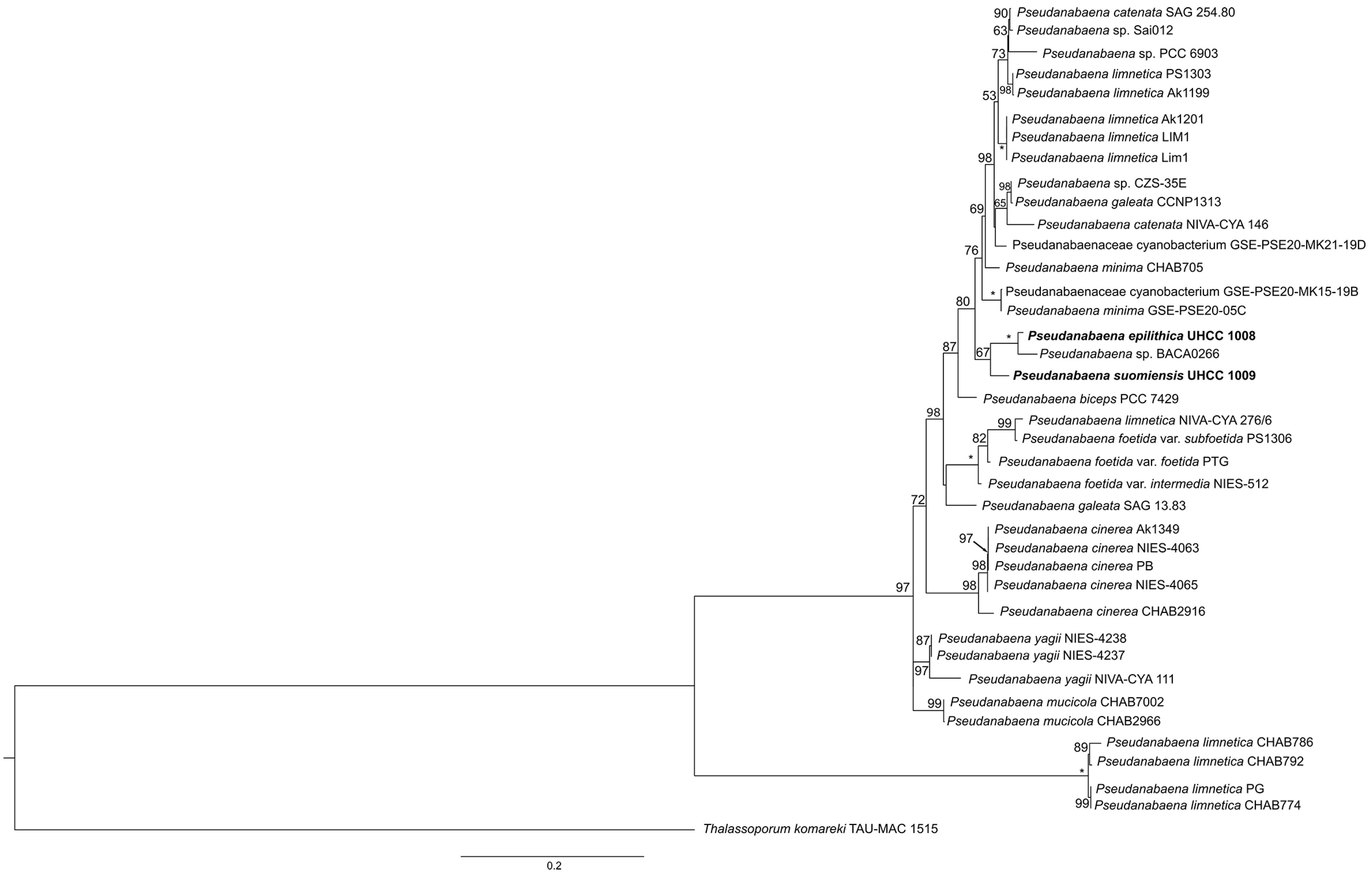
3.2. Phylogenetic Analysis of 16S rRNA Gene and Phylogeny Based on Concatenate Alignment

3.3. Study of 16S–23S ITS
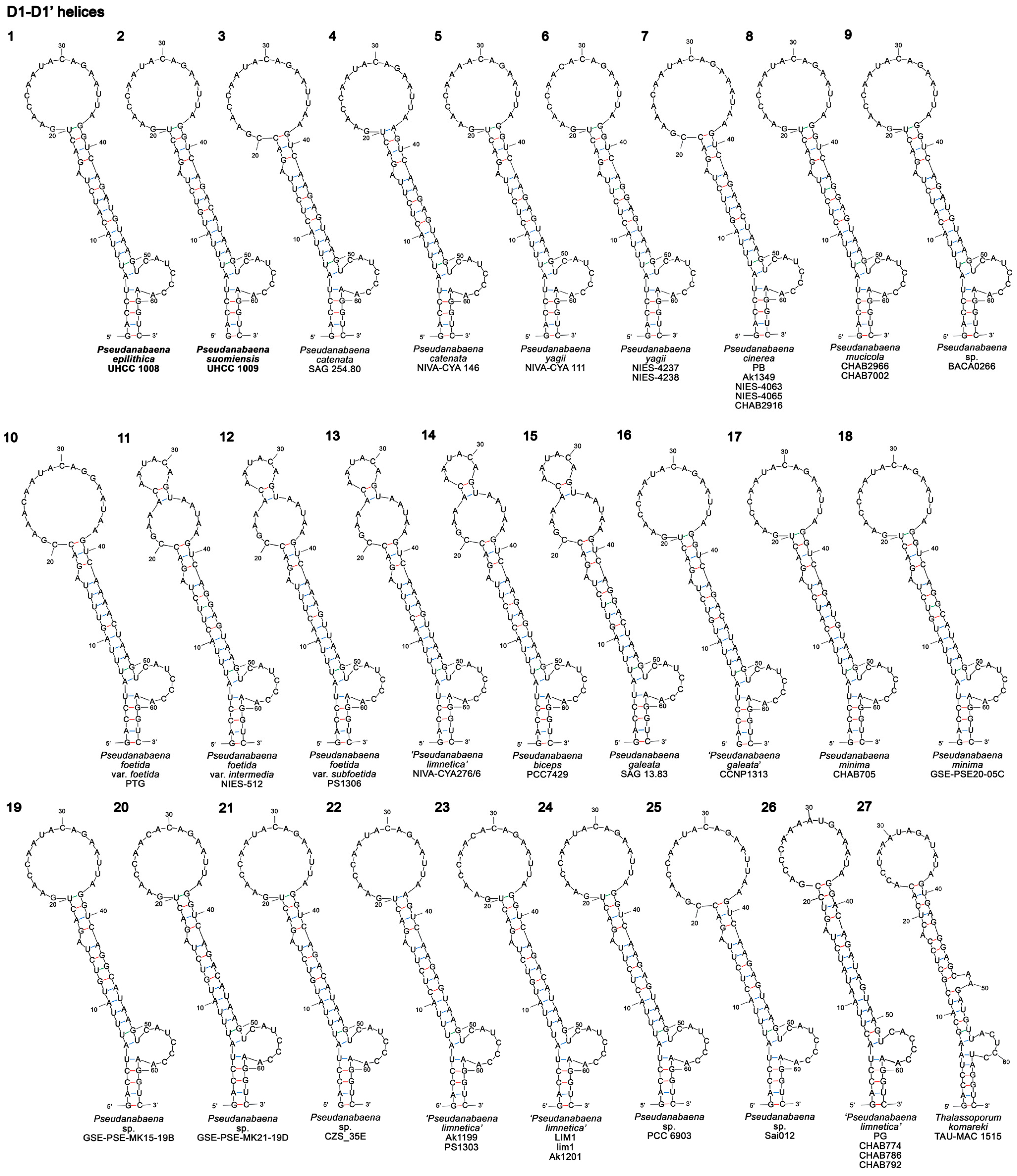
3.3.1. D1–D1′ Helix
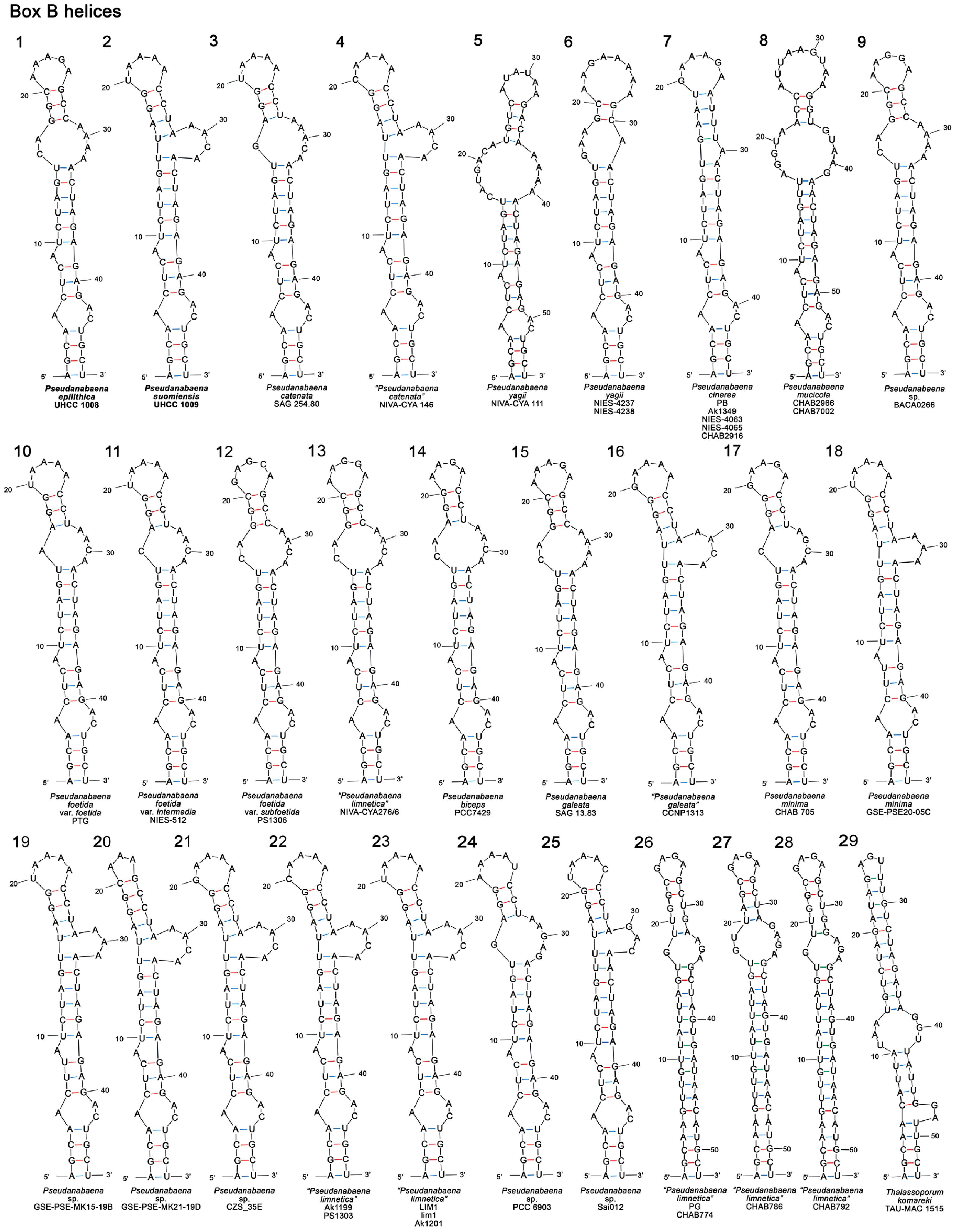
3.3.2. Box B Secondary Structure
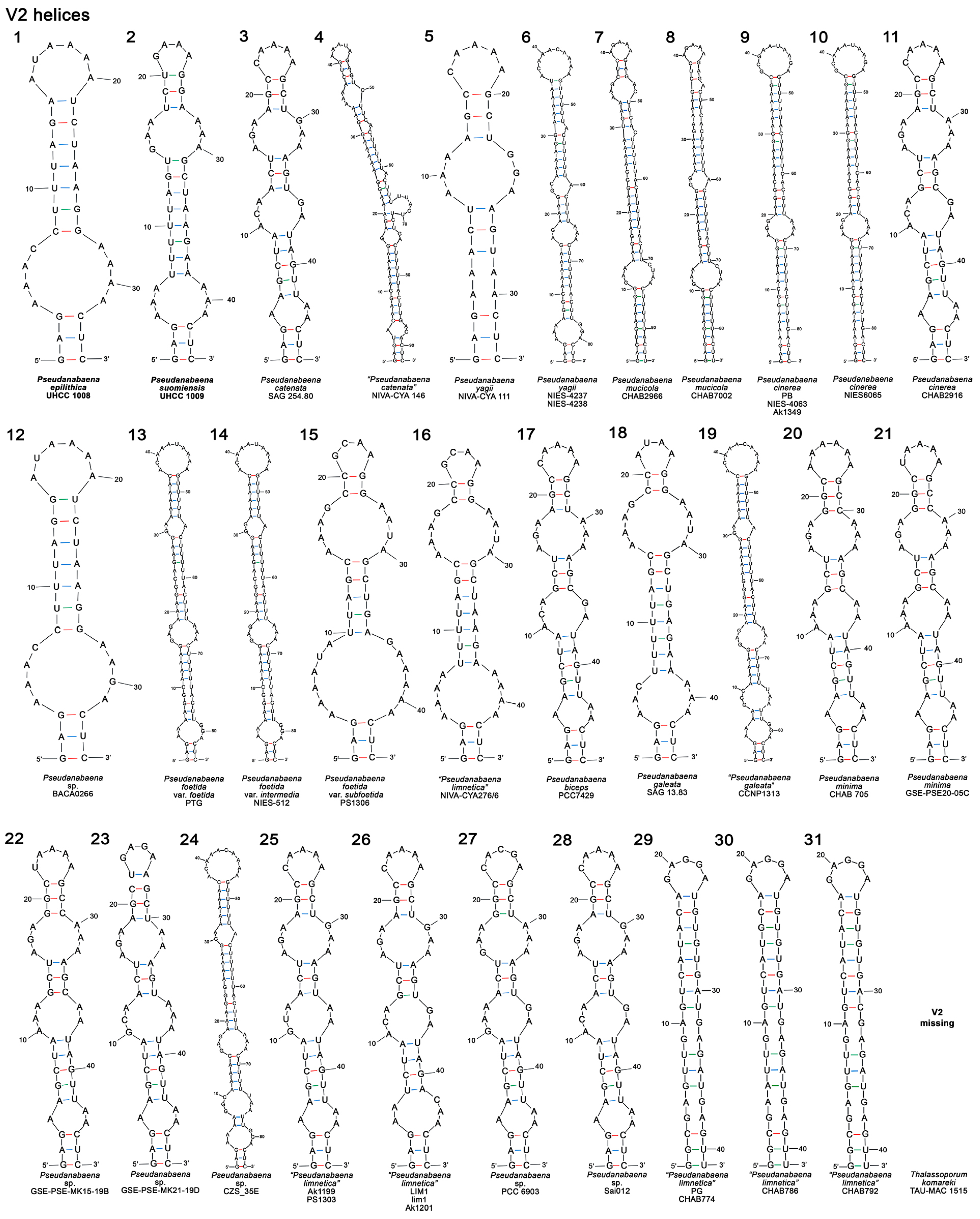
3.3.3. V2 and V3 Helices
3.4. Biochemical Analysis
3.4.1. Antimicrobial Susceptibility Tests and Bioactive Metabolites Screening
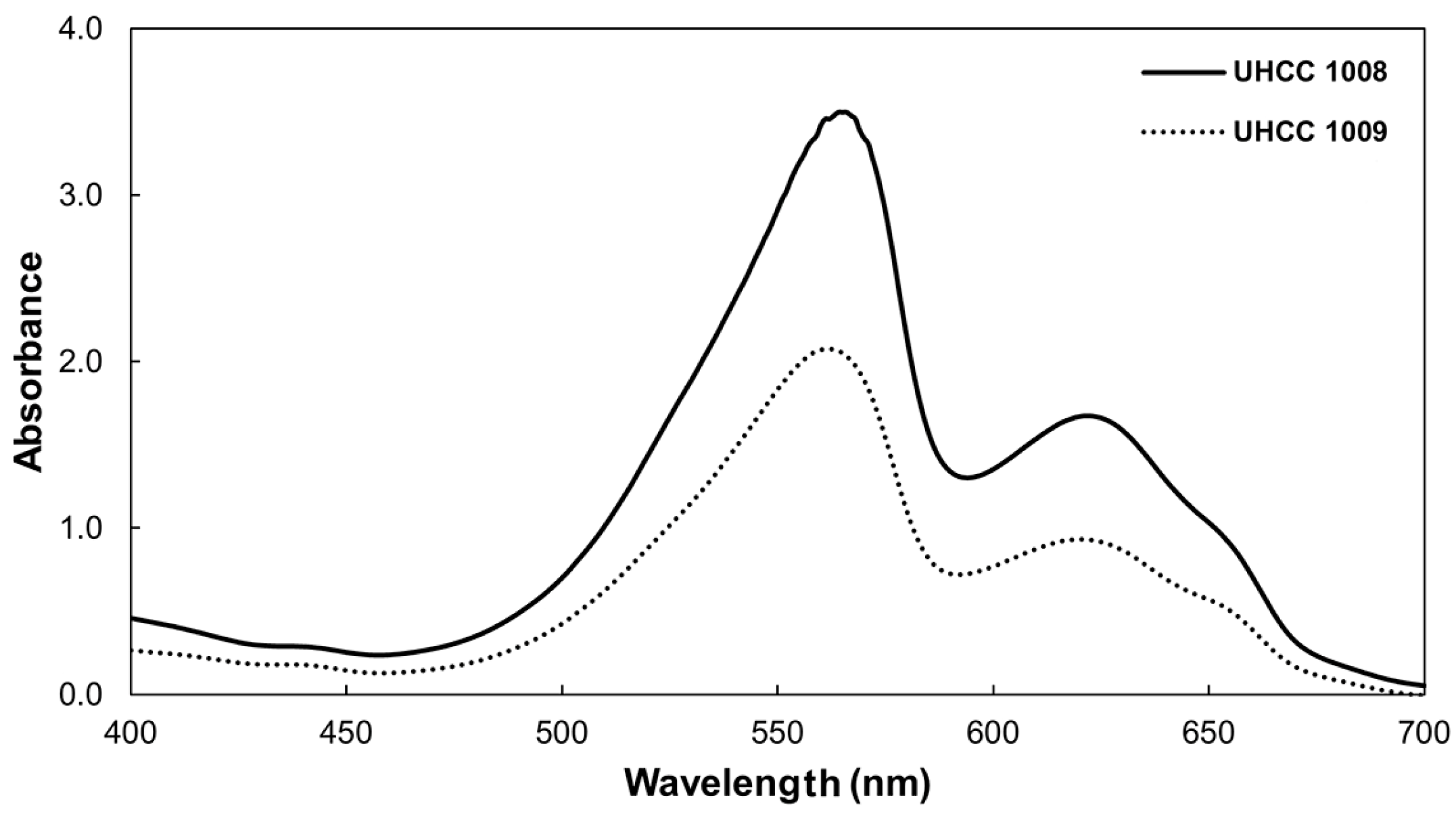
3.4.2. Phycobiliproteins
3.5. Taxonomic Assessments
4. Discussion
4.1. Morphology and Ecology
4.2. 16S rRNA and Combined Phylogenetic Analyses
4.3. The Use of 16S–23S ITS in Polyphasic Approach Studies
4.4. A Polyphasic Approach Workflow Is Necessary for Accurate Taxonomic Assignments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Whitton, B.A.; Potts, M. Introduction to the Cyanobacteria. In Ecology of Cyanobacteria II; Whitton, B.A., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–13. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota 2. Teil/2nd Part: Oscillatoriales. In Süsswasserflora von Mitteleuropa 19/2; Büdel, B., Krienitz, L., Gärtner, G., Schagerl, M., Eds.; Elsevier/Spektrum: Heidelberg, Germany, 2005; p. 759. [Google Scholar]
- Engene, N.; Choi, H.; Esquenazi, E.; Rottacker, E.C.; Ellisman, M.H.; Dorrestein, P.C.; Gerwick, W.H. Underestimated biodiversity as a major explanation for the perceived rich secondary metabolite capacity of the cyanobacterial genus Lyngbya. Environ. Microbiol. 2011, 13, 1601–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, L.; Komárek, J.; Kaštovský, J. System of cyanoprokaryotes (cyanobacteria)—State in 2004. Arch. Hydrobiol. Suppl. Algol. Stud. 2005, 117, 95–115. [Google Scholar] [CrossRef]
- Komárek, J. A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. Eur. J. Phycol. 2016, 51, 346–353. [Google Scholar] [CrossRef] [Green Version]
- Boyer, S.L.; Flechtner, V.R.; Johansen, J.R. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol. Biol. Evol. 2001, 18, 1057–1069. [Google Scholar] [CrossRef] [Green Version]
- Johansen, J.R.; Casamatta, D.A. Recognizing cyanobacterial diversity through adoption of a new species paradigm. Arch. Hydrobiol. Suppl. Algol. Stud. 2005, 117, 71–93. [Google Scholar] [CrossRef]
- Osorio-Santos, K.; Pietrasiak, N.; Bohunická, M.; Miscoe, L.H.; Kovácik, L.; Martin, M.P.; Johansen, J.R. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): Taxonomically recognizing cryptic diversification. Eur. J. Phycol. 2014, 49, 450–470. [Google Scholar] [CrossRef] [Green Version]
- Bohunická, M.; Pietrasiak, N.; Johansen, J.R.; Gómez, E.B.; Hauer, T.; Gaysina, L.A.; Lukešová, A. Roholtiella, gen. nov. (Nostocales, Cyanobacteria)—A tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa 2015, 197, 84–103. [Google Scholar] [CrossRef] [Green Version]
- Jung, P.; Sommer, V.; Karsten, U.; Lakatos, M. Salty twins: Salt-tolerance of terrestrial Cyanocohniella strains (Cyanobacteria) and description of C. rudolphia sp. nov. point towards a marine origin of the genus and terrestrial long distance dispersal patterns. Microorganisms 2022, 10, 968. [Google Scholar] [CrossRef]
- Baldarelli, L.M.; Pietrasiak, N.; Osorio-Santos, K.; Johansen, J.R. Mojavia aguilerae and M. dolomitestris—Two new Nostocaceae (Cyanobacteria) species from the Americas. J. Phycol. 2022, 58, 502–516. [Google Scholar] [CrossRef]
- Pietrasiak, N.; Osorio-Santos, K.; Shalygin, S.; Martin, M.P.; Johansen, J.R. When is a lineage a species? A case study in Myxacorys gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. J. Phycol. 2019, 55, 976–996. [Google Scholar] [CrossRef]
- Mai, T.; Johansen, J.R.; Pietrasiak, N.; Bohunická, M.; Martin, M.P. Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 2018, 365, 1–59. [Google Scholar] [CrossRef] [Green Version]
- Erwin, P.M.; Thacker, R.W. Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Mol. Ecol. 2008, 17, 2937–2947. [Google Scholar] [CrossRef] [PubMed]
- Mareš, J.; Johansen, J.R.; Hauer, T.; Zima, J.; Ventura, S.; Cuzman, O.; Tiribilli, B.; Kaštovský, J. Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea, and Zehria. J. Phycol. 2019, 55, 578–610. [Google Scholar] [CrossRef] [PubMed]
- González-Resendiz, L.; Johansen, J.R.; León-Tejera, H.; Sánchez, L.; Segal-Kischinevzky, C.; Escobar-Sánchez, V.; Morales, M. A bridge too far in naming species: A total evidence approach does not support recognition of four species in Desertifilum (Cyanobacteria). J. Phycol. 2019, 55, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Zammit, G.; Billi, D.; Albertano, P. The subaerophytic cyanobacterium Oculatella subterranea (Oscillatoriales, Cyanophyceae) gen. et sp. nov.: A cytomorphological and molecular description. Eur. J. Phycol. 2012, 47, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Dvořák, P.; Jahodářová, E.; Hašler, P.; Gusev, E.; Poulíčková, A. A new tropical cyanobacterium Pinocchia polymorpha gen et sp nov derived from the genus Pseudanabaena. Fottea 2015, 15, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Miscoe, L.H.; Johansen, J.R.; Kociolek, J.P.; Lowe, R.L.; Vaccarino, M.A.; Pietrasiak, N.; Sherwood, A.R. Novel Cyanobacteria from Caves on Kauai, Hawaii. In The Diatom Flora and Cyanobacteria from Caves on Kauai, Hawaii; Miscoe, L.H., Johansen, J.R., Kociolek, J.P., Lowe, R.L., Vaccarino, M.A., Pietrasiak, N., Sherwood, A.R., Eds.; J. Cramer: Stuttgart, Germany, 2016; pp. 75–152. ISBN 9783443600471. [Google Scholar]
- Perkerson, R.B.; Johansen, J.R.; Kovácik, L.; Brand, J.; Kaštovský, J.; Casamatta, D.A. A unique pseudanabaenalean (cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. J. Phycol. 2011, 47, 1397–1412. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J. Phycol. 2023, 59, 12–51. [Google Scholar] [CrossRef]
- Turicchia, S.; Ventura, S.; Komárková, J.; Komárek, J. Taxonomic evaluation of cyanobacterial microflora from alkaline marshes of northern Belize. 2. Diversity of oscillatorialean genera. Nov. Hedwig. 2009, 89, 165–200. [Google Scholar] [CrossRef]
- Skuja, H. Some Algae and Protists. In Vegetation of Amchitka Island, Aleutian Islands, Alaska; Shacklette, H.T., Durrell, L.W., Erdman, J.A., Keith, J.R., Klein, W.M., Krog, H., Persson, H., Skuja, H., Weber, W.A., Eds.; United States Government Printing Office: Washington, DC, USA, 1969. [Google Scholar]
- Konstantinou, D.; Voultsiadou, E.; Panteris, E.; Gkelis, S. Revealing new sponge-associated cyanobacterial diversity: Novel genera and species. Mol. Phylogenet. Evol. 2021, 155, 106991. [Google Scholar] [CrossRef]
- Niiyama, Y.; Tuji, A.; Takemoto, K.; Ichise, S. Pseudanabaena foetida sp. nov. and P. subfoetida sp. nov. (cyanophyta/cyanobacteria) producing 2–methylisoborneol from Japan. Fottea 2016, 16, 1–11. [Google Scholar] [CrossRef]
- Tuji, A.; Niiyama, Y. The Identity and phylogeny of Pseudanabaena strain, NIES-512, producing 2-methylisoborneol (2-MIB). Bull. Natl. Mus. Nat. Sci. Ser. B 2016, 42, 83–89. [Google Scholar]
- Tuji, A.; Niiyama, Y. Two new Pseudanabaena (Cyanobacteria, Synechococcales) species from Japan, Pseudanabaena cinerea and Pseudanabaena yagii, which produce 2-methylisoborneol. Phycol. Res. 2018, 66, 291–299. [Google Scholar] [CrossRef]
- Chorus, I.; Walker, M. Cyanobacterial Toxins. In Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring and Management; Chorus, I., Walker, M., Eds.; CRC Press, on behalf of the World Health Organization: Geneva, Switzerland, 2021; pp. 15–162. [Google Scholar]
- Olvera-Ramírez, R.; Centeno-Ramos, C.; Martínez-Jerónimo, F. Toxic effects of Pseudanabaena tenuis (cyanobacteria) on the cladocerans Daphnia magna and Ceriodaphnia dubia. Hidrobiologica 2010, 20, 203–212. [Google Scholar]
- Humisto, A.; Herfindal, L.; Jokela, J.; Karkman, A.; Bjørnstad, R.; Choudhury, R.R.; Sivonen, K. Cyanobacteria as a source for novel anti-leukemic compounds. Curr. Pharm. Biotechnol. 2016, 17, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Felczykowska, A.; Pawlik, A.; Mazur-Marzec, H.; Toruńska-Sitarz, A.; Narajczyk, M.; Richert, M.; Wegrzyn, G.; Herman-Antosiewicz, A. Selective inhibition of cancer cells’ proliferation by compounds included in extracts from Baltic Sea cyanobacteria. Toxicon 2015, 108, 1–10. [Google Scholar] [CrossRef]
- Cano-Europa, E.; Ortiz-Butrón, R.; Gallardo-Casas, C.A.; Blas-Valdivia, V.; Pineda-Reynoso, M.; Olvera-Ramírez, R.; Franco-Colin, M. Phycobiliproteins from Pseudanabaena tenuis rich in c-phycoerythrin protect against HgCl2-caused oxidative stress and cellular damage in the kidney. J. Appl. Phycol. 2010, 22, 495–501. [Google Scholar] [CrossRef]
- Cegłowska, M.; Szubert, K.; Grygier, B.; Lenart, M.; Plewka, J.; Milewska, A.; Lis, K.; Szczepański, A.; Chykunova, Y.; Barreto-Duran, E.; et al. Pseudanabaena galeata CCNP1313—Biological activity and peptides production. Toxins 2022, 14, 330. [Google Scholar] [CrossRef]
- Turland, N.; Wiersema, J.; Barrie, F.; Greuter, W.; Hawksworth, D.; Herendeen, P.; Knapp, S.; Kusber, W.-H.; Li, D.-Z.; Marhold, K.; et al. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017; Turland, N., Wiersema, J., Barrie, F., Greuter, W., Hawksworth, D., Herendeen, P., Knapp, S., Kusber, W.-H., Li, D.-Z., Marhold, K., et al., Eds.; Regnum Vegetabile; Koeltz Botanical Books: Glashütten, Germany, 2018; Volume 159, ISBN 9783946583165. [Google Scholar]
- Kotai, J. Instructions for preparation of modified nutrient solution Z8 for algae. Nor. Inst. Water Res. 1972, 11, 1–5. [Google Scholar]
- Rippka, R.; Waterbury, J.B.; Stanier, R.Y. Isolation and purification of cyanobacteria: Some general principles. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin/Heidelberg, Germany, 1981; pp. 212–220. [Google Scholar]
- Christodoulou, M.; Jokela, J.; Wahlsten, M.; Saari, L.; Economou-Amilli, A.; de Fiore, M.F.; Sivonen, K. Description of Aliinostoc alkaliphilum sp. nov. (Nostocales, Cyanobacteria), a new bioactive metabolite-producing strain from Salina Verde (Pantanal, Brazil) and taxonomic distribution of bioactive metabolites in Nostoc and Nostoc-like genera. Water 2022, 14, 2470. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase; World-Wide Electronic Publication, National University of Ireland: Galway, Ireland, 2022. [Google Scholar]
- Kling, H.J.; Dail Laughinghouse, I.; Šmarda, J.; Komárek, J.; Acreman, J.; Bruun, K.; Watson, S.B.; Chen, F. A new red colonial Pseudanabaena (Cyanoprokaryota, Oscillatoriales) from North American large lakes. Fottea 2012, 12, 327–339. [Google Scholar] [CrossRef] [Green Version]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef] [PubMed]
- Taton, A.; Grubisic, S.; Brambilla, E.; De Wit, R.; Wilmotte, A. Cyanobacterial diversity in natural and artificial microbial mats of lake Fryxell (McMurdo Dry Valleys, Antarctica): A morphological and molecular approach. Appl. Environ. Microbiol. 2003, 69, 5157–5169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrouzek, P.; Ventura, S.; Lukešová, A.; Mugnai, M.-A.; Turicchia, S.; Komárek, J. Diversity of soil Nostoc strains: Phylogenetic and phenotypic variability. Arch. Hydrobiol. Suppl. Algol. Stud. 2005, 117, 251–264. [Google Scholar] [CrossRef]
- Wilmotte, A.; Van der Auwera, G.; De Wachter, R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Iteman, I.; Rippka, R.; De Marsac, N.T.; Herdman, M. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 2000, 146, 1275–1286. [Google Scholar] [CrossRef] [Green Version]
- Boyer, S.L.; Johansen, J.R.; Flechtner, V.R.; Howard, G.L. Phylogeny and genetic variance in terrestrial Microcoleus (cyanophyceae) species based on sequence analysis of the 16s rRNA gene and associated 16S–23S ITS region. J. Phycol. 2002, 38, 1222–1235. [Google Scholar] [CrossRef]
- Wright, E.S.; Yilmaz, L.S.; Noguera, D.R. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 2012, 78, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Vences, M.; Patmanidis, S.; Kharchev, V.; Renner, S.S. Concatenator, a user-friendly program to concatenate DNA sequences, implementing graphical user interfaces for MAFFT and FastTree. Bioinform. Adv. 2022, 2, vbac050. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 20 April 2022).
- The Inkscape team. The Inkscape Project. Available online: https://inkscape.org (accessed on 10 May 2018).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Bohunická, M.; Johansen, J.R.; Fučíková, K. Tapinothrix clintonii sp. nov. (Pseudanabaenaceae, Cyanobacteria), a new species at the nexus of five genera. Fottea 2011, 11, 127–140. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Zavřel, T.; Chmelík, D.; Sinetova, M.A.; Červený, J. Spectrophotometric determination of phycobiliprotein content in cyanobacterium Synechocystis. J. Vis. Exp. 2018, 2018, 58076. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a comprehensive public database of secondary metabolites from cyanobacteria. Water Res. 2021, 196, 117017. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Tuji, A.; Niiyama, Y. The taxonomy of Pseudanabaena (Cyanobacteria) and characters for judging the production of odor compounds. Japanese J. Water Treat. Biol. 2018, 54, 115–120. [Google Scholar] [CrossRef]
- Yu, G.; Zhu, M.; Chen, Y.; Pan, Q.; Chai, W.; Li, R. Polyphasic characterization of four species of Pseudanabaena (Oscillatoriales, Cyanobacteria) from China and insights into polyphyletic divergence within the Pseudanabaena genus. Phytotaxa 2015, 192, 1–12. [Google Scholar] [CrossRef]
- Cegłowska, M.; Toruńska-Sitarz, A.; Stoń-Egiert, J.; Mazur-Marzec, H.; Kosakowska, A. Characteristics of cyanobacterium Pseudanabaena galeata CCNP1313 from the Baltic Sea. Algal Res. 2020, 47, 101861. [Google Scholar] [CrossRef]
- Albrecht, M.; Pröschold, T.; Schumann, R. Identification of cyanobacteria in a eutrophic coastal lagoon on the Southern Baltic Coast. Front. Microbiol. 2017, 8, 923. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.T.; Yusoff, F.M.; Khaw, Y.S.; Noor Mazli, N.A.I.; Nazarudin, M.F.; Shaharuddin, N.A.; Katayama, T.; Ahmad, S.A. A review on a hidden gem: Phycoerythrin from blue-green algae. Mar. Drugs 2023, 21, 28. [Google Scholar] [CrossRef]
- Dagnino-Leone, J.; Figueroa, C.P.; Castañeda, M.L.; Youlton, A.D.; Vallejos-Almirall, A.; Agurto-Muñoz, A.; Pavón Pérez, J.; Agurto-Muñoz, C. Phycobiliproteins: Structural aspects, functional characteristics, and biotechnological perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 1506–1527. [Google Scholar] [CrossRef]
- Patel, S.N.; Sonani, R.R.; Jakharia, K.; Bhastana, B.; Patel, H.M.; Chaubey, M.G.; Singh, N.K.; Madamwar, D. Antioxidant activity and associated structural attributes of Halomicronema phycoerythrin. Int. J. Biol. Macromol. 2018, 111, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Blas-Valdivia, V.; Rojas-Franco, P.; Serrano-Contreras, J.I.; Sfriso, A.A.; Garcia-Hernandez, C.; Franco-Colín, M.; Cano-Europa, E. C-phycoerythrin from Phormidium persicinum prevents acute kidney injury by attenuating oxidative and endoplasmic reticulum stress. Mar. Drugs 2021, 19, 589. [Google Scholar] [CrossRef] [PubMed]
- Soni, B.; Visavadiya, N.P.; Madamwar, D. Ameliorative action of cyanobacterial phycoerythrin on CCl4-induced toxicity in rats. Toxicology 2008, 248, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Soni, B.; Visavadiya, N.P.; Madamwar, D. Attenuation of diabetic complications by C-phycoerythrin in rats: Antioxidant activity of C-phycoerythrin including copper-induced lipoprotein and serum oxidation. Br. J. Nutr. 2009, 102, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Komárek, J. Several problems of the polyphasic approach in the modern cyanobacterial system. Hydrobiologia 2018, 811, 7–17. [Google Scholar] [CrossRef]
- Dvořák, P.; Jahodářová, E.; Casamatta, D.A.; Hašler, P.; Poulíčková, A. Difference without distinction? Gaps in cyanobacterial systematics; when more is just too much. Fottea 2018, 18, 130–136. [Google Scholar] [CrossRef] [Green Version]
- Komárek, J. Quo vadis, taxonomy of cyanobacteria (2019). Fottea 2020, 20, 104–110. [Google Scholar] [CrossRef] [Green Version]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Jonas, E. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 8, 152–155. [Google Scholar]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Řeháková, K.; Mareš, J.; Lukešová, A.; Zapomělová, E.; Bernardová, K.; Hrouzek, P. Nodularia (Cyanobacteria, Nostocaceae): A phylogenetically uniform genus with variable phenotypes. Phytotaxa 2014, 172, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Martins, M.D.; Rigonato, J.; Taboga, S.R.; Branco, L.H.Z. Proposal of Ancylothrix gen. nov., a new genus of Phormidiaceae (Cyanobacteria, Oscillatoriales) based on a polyphasic approach. Int. J. Syst. Evol. Microbiol. 2016, 66, 2396–2405. [Google Scholar] [CrossRef]
- Buch, B.; Martins, M.D.; Branco, L.H.Z. A widespread cyanobacterium supported by polyphasic approach: Proposition of Koinonema pervagatum gen. & sp. nov. (Oscillatoriales). J. Phycol. 2017, 53, 1097–1105. [Google Scholar] [CrossRef]
- Martins, M.D.; Branco, L.H.Z. Potamolinea gen. nov. (Oscillatoriales, Cyanobacteria): A phylogenetically and ecologically coherent cyanobacterial genus. Int. J. Syst. Evol. Microbiol. 2016, 66, 3632–3641. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A Place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef] [Green Version]


| Strain Name | Vegetative Cell Color, Shape and Size (μm) | Sheath | Constrictions | Cell Content | Apical Cell | Motility | Ecology | Ref. |
|---|---|---|---|---|---|---|---|---|
| P. epilithica UHCC 1008T | purple to purple-brown, cylindrical to barrel-shaped W: (1.5) 1.7–1.9 (2.1) L: up to 2.5–3.0 | − | + | one to two granules and/or one to two refractive granules | rounded, ring-like or cupola-like large refractive granules at the tip | + | epilithic | This study |
| P. suomiensis UHCC 1009T | brownish, cylindrical with rounded ends W: (1.5) 1.6–1.9 (2.1) L: up to 3.5 | − | + | with scarce but prominent granules and/or one to two refractive granules | rounded, with two small (or one large) ring-like or cupola-like refractive granules at the tip | + | epilithic | This study |
| ‘P. catenata’ | ||||||||
| SAG 254.80 | N/A | N/A | N/A | N/A | N/A | N/A | freshwater | SAG online catalogue 3 |
| PCC 7408 1 | greenish, cylindrical, W: 2.0–2.5 L: longer than wide | − | + | homogeneous | N/A | + | freshwater | [64] |
| ‘P. limnetica’ | ||||||||
| PS1303 | blue-green to bright blue-green, cylindrical, W: 1.1–1.5 L: 1.5–5.0 | + | + | homogeneous | long and thin cylindrical with rounded ends | + | freshwater | [26] |
| Ak1199 | blue-green to bright blue-green, cylindrical, W: 1.1–1.5 L: 1.5–5.0 | N/A | + | homogeneous | long and thin cylindrical with rounded ends | N/A | freshwater | [65] |
| Ak1201 | grayish green to pale brown, cylindrical, W: 0.9–1.4 L: 1.4–7.1 | N/A | N/A | homogeneous | N/A | N/A | freshwater | [65] |
| Lim1 2 | grayish green to pale brown, cylindrical, W: 0.9–1.4 L: 1.4–7.1 | N/A | N/A | homogeneous | N/A | N/A | freshwater | [65] |
| P. minima | ||||||||
| CHAB705 | few-celled, blue green or pale blue green, cylindrical W: 1.7–2.5 L: 2.0–4.2 | − | + | homogeneous | cylindrical with rounded ends | N/A | freshwater | [66] |
| GSE-PSE20-05C | N/A | N/A | N/A | N/A | N/A | N/A | N/A | − |
| P. biceps | ||||||||
| PCC 7429 | >10 cells in each trichome W: 2.0–2.5 L: 2.0–4.2 | N/A | + | N/A | N/A | + | sphagnum bog | [64] |
| ‘P. galeata’ | ||||||||
| CCNP1313 | up to 10 cells, greenish, barrel-shaped, W: 1.0–1.4 L: 1.0–3.8 | − | + | granular, polar gas vesicles present | rounded | N/A | Baltic Sea | [67] |
| SAG 13.83 | N/A | N/A | N/A | N/A | N/A | N/A | freshwater | SAG online catalogue 3 |
| Pseudanabaena sp. | ||||||||
| Sai012 | N/A | N/A | N/A | N/A | N/A | N/A | freshwater, sediment | NCBI |
| CZS 35E | N/A | N/A | N/A | N/A | N/A | N/A | eutrophic, shallow lagoon | [68] |
| GSE-PSE-MK15-19B | N/A | N/A | N/A | N/A | N/A | N/A | − | NCBI |
| GSE-PSE-MK21-19D | N/A | N/A | N/A | N/A | N/A | N/A | seep wall | NCBI |
| PCC 6903 | three to six cells W: 2.5–3.0 | N/A | N/A | polar gas vesicles | N/A | + | brackish pool | [64] |
| PCC 7402 | three to six cells W: 2.0–3.5 | N/A | N/A | polar gas vesicles | + | eutrophic pond | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christodoulou, M.; Wahlsten, M.; Sivonen, K. Morphological and Molecular Evaluation of Pseudanabaena epilithica sp. nov. and P. suomiensis sp. nov. (Pseudanabaenaceae, Cyanobacteria) from Finland. Diversity 2023, 15, 909. https://doi.org/10.3390/d15080909
Christodoulou M, Wahlsten M, Sivonen K. Morphological and Molecular Evaluation of Pseudanabaena epilithica sp. nov. and P. suomiensis sp. nov. (Pseudanabaenaceae, Cyanobacteria) from Finland. Diversity. 2023; 15(8):909. https://doi.org/10.3390/d15080909
Chicago/Turabian StyleChristodoulou, Maria, Matti Wahlsten, and Kaarina Sivonen. 2023. "Morphological and Molecular Evaluation of Pseudanabaena epilithica sp. nov. and P. suomiensis sp. nov. (Pseudanabaenaceae, Cyanobacteria) from Finland" Diversity 15, no. 8: 909. https://doi.org/10.3390/d15080909