Abundant Species Govern the Altitude Patterns of Bacterial Community in Natural and Disturbed Subalpine Forest Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. Soil Property Measurements
2.3. DNA Extraction, 16S rRNA Gene Amplification and High-Throughput Sequencing
2.4. Bioinformatics Analysis of 16S rRNA Gene Sequencing Data
2.5. Statistical Analysis
3. Results
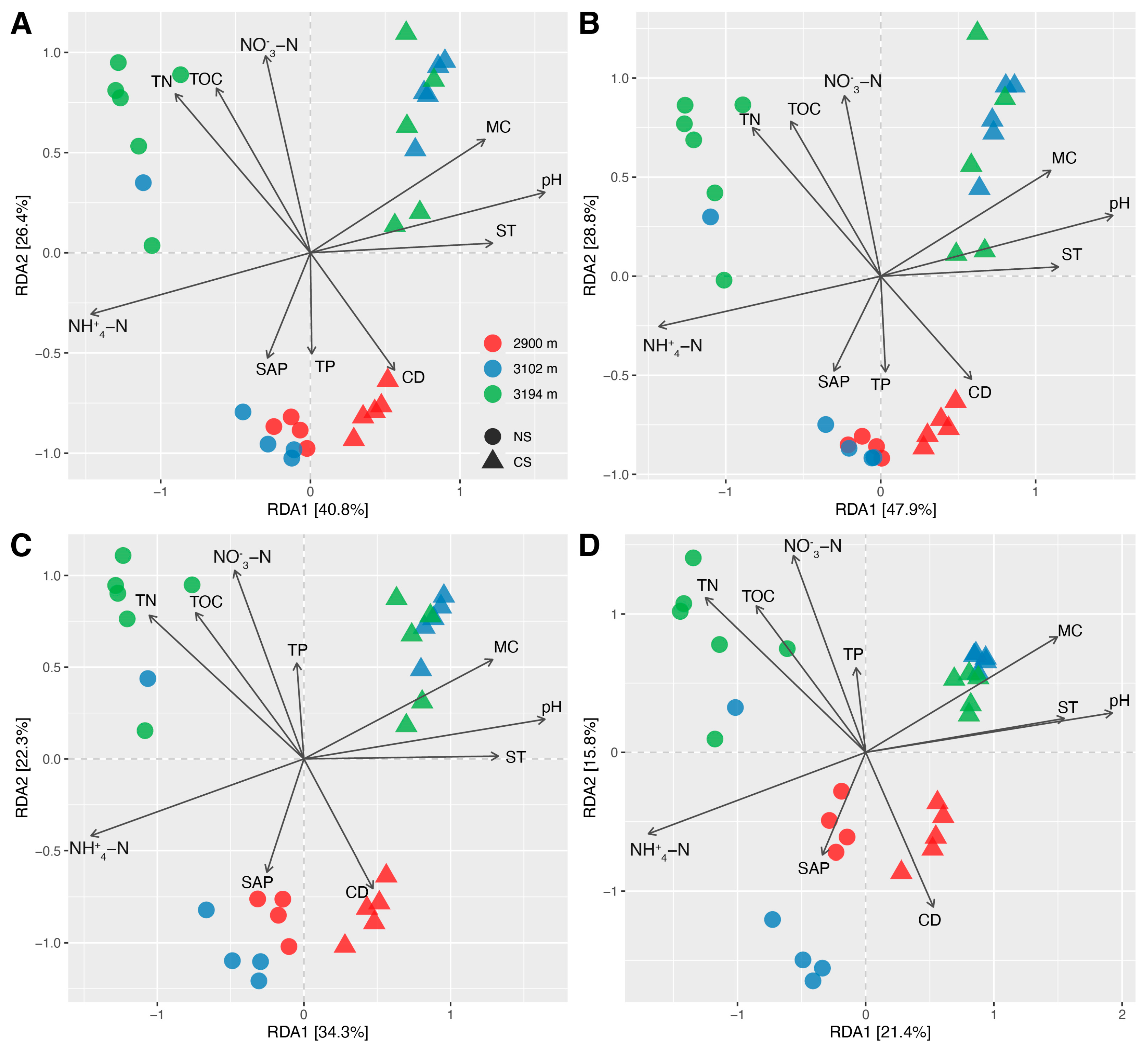
3.1. The Differences in Soil Properties between Natural and Cut Slope Soils, and among Altitudes
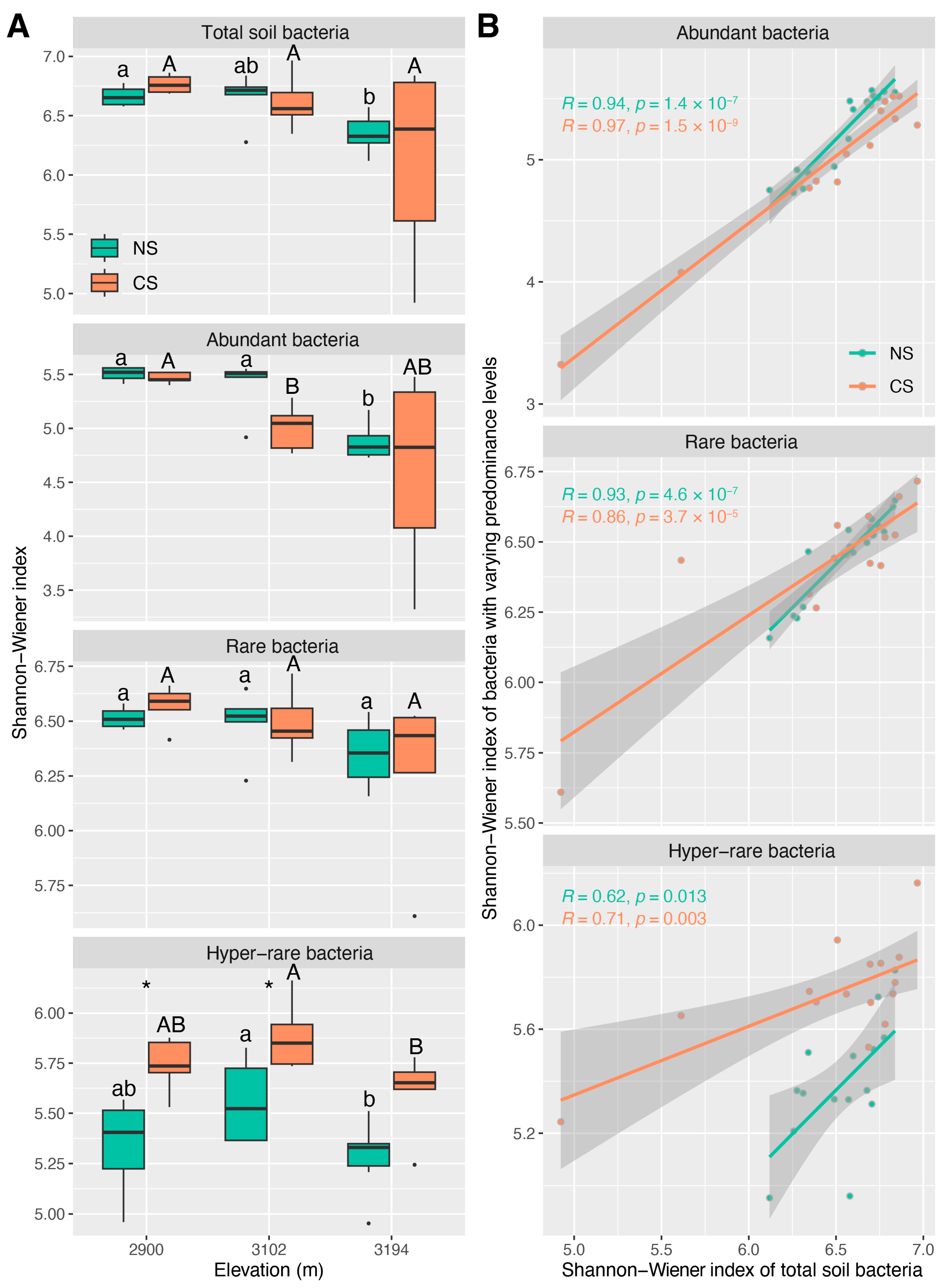
3.2. The Diversity Characteristics of Abundant, Rare and Hyper-Rare Bacteria
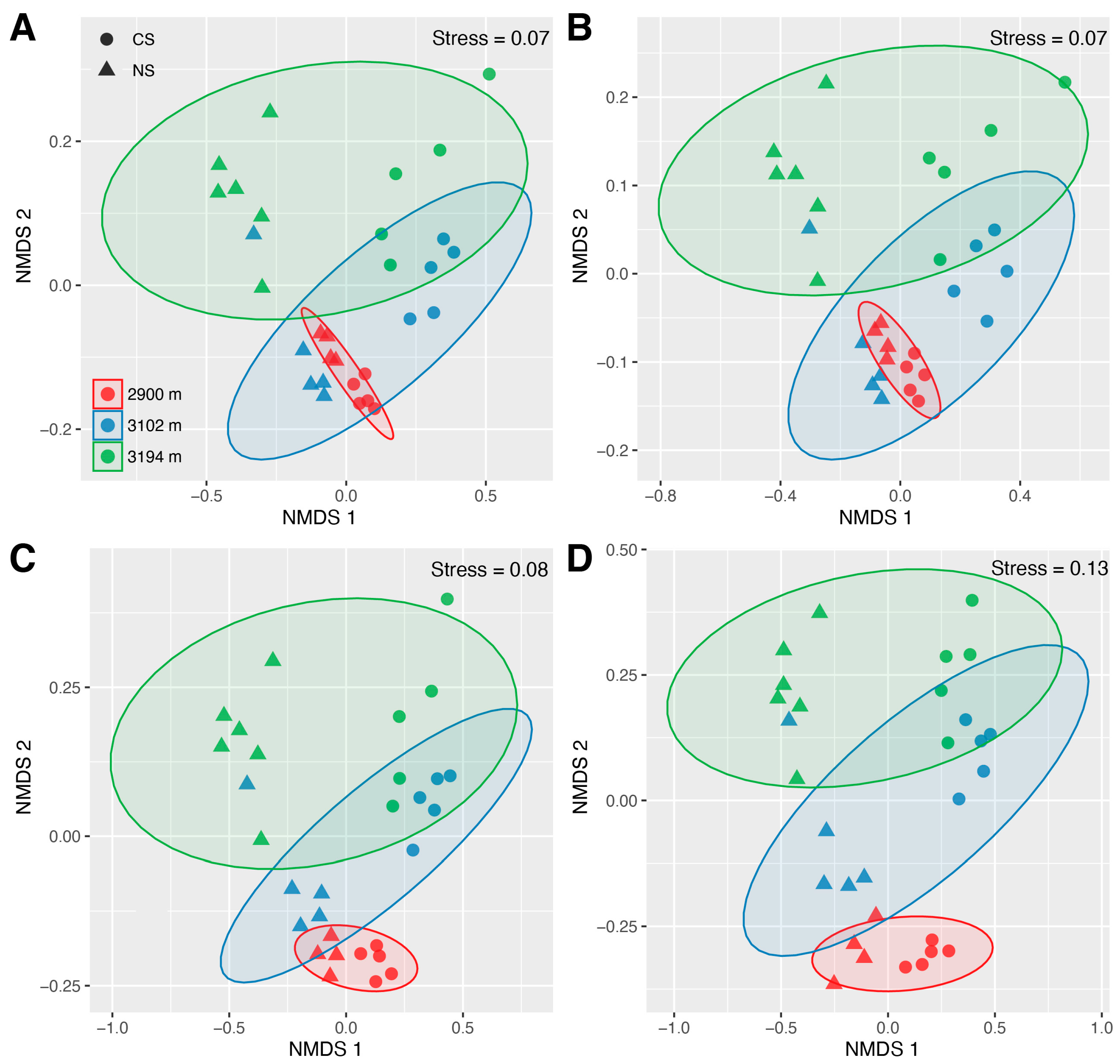
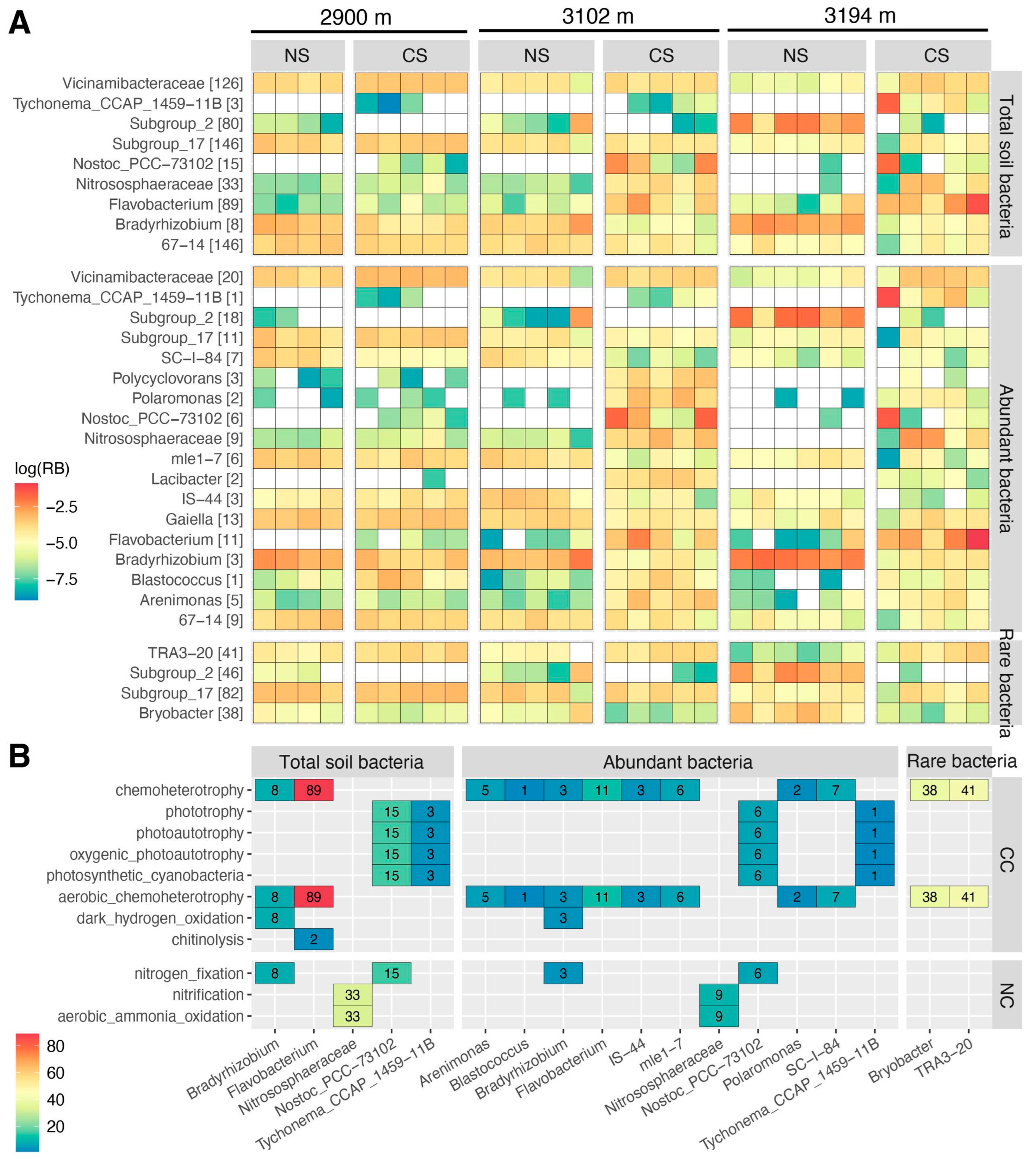
3.3. The Community Characteristics of Abundant, Rare, and Hyper-Rare Bacteria
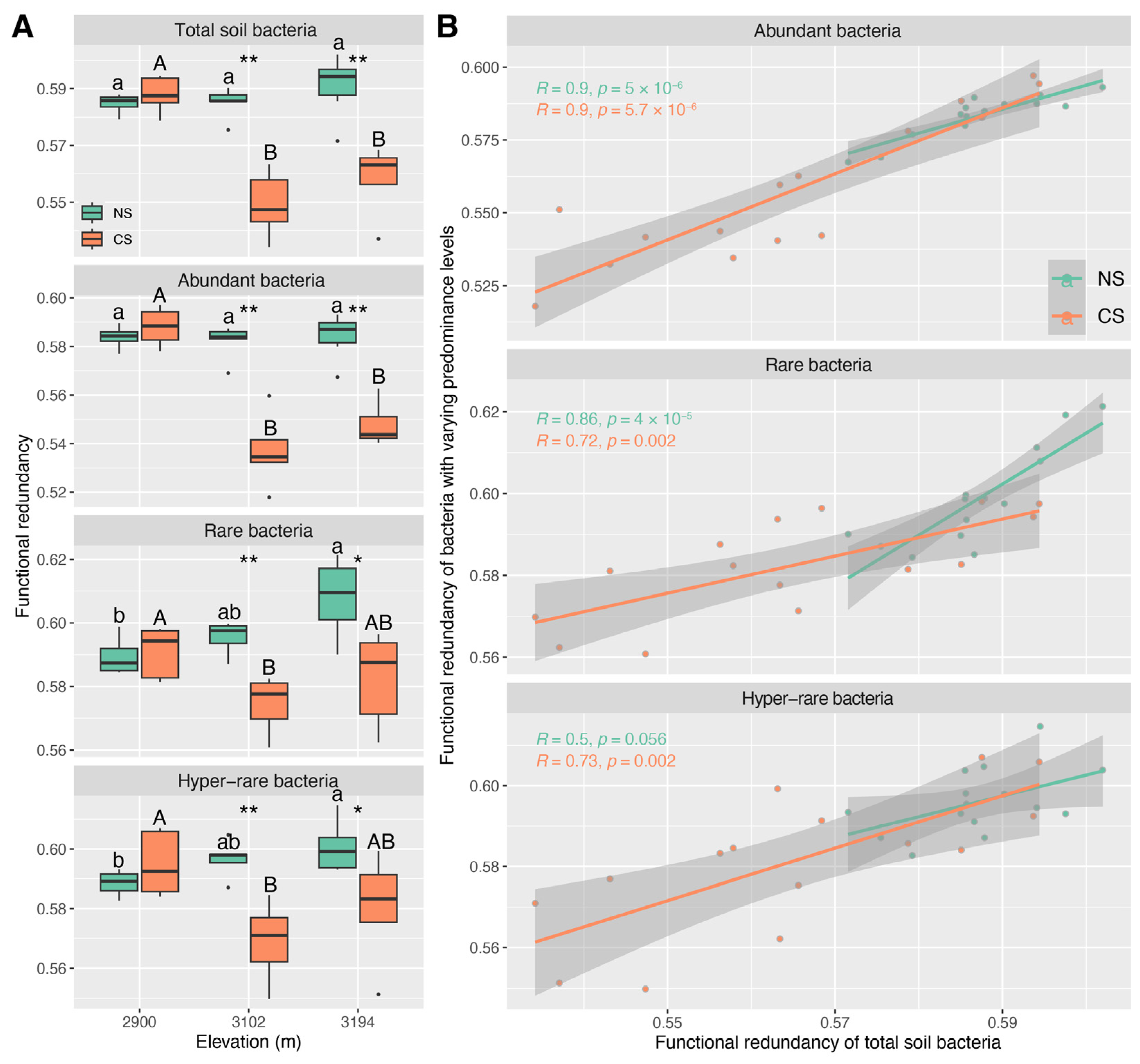
3.4. The Functional Redundancy Characteristics of Abundant, Rare, and Hyper-Rare Bacteria
4. Discussion
4.1. The Influences of Altitude and ESSS on the Communities Consisting of Bacteria with Varying Predominance Levels
4.2. Potential Roles of Bacteria with Varying Predominance Levels in Maintaining the Ecological Characteristics of Total Community across the Altitude Gradient
4.3. Shared Key Driving Factors Result in Similar Ecological Characteristics for the Communities of Total Soil Bacteria and Abundant Ones across the Altitude Gradient
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liao, H.; Sheng, M.; Liu, J.; Ai, X.; Li, C.; Ai, S.; Ai, Y. Soil N availability drives the shifts of enzyme activity and microbial phosphorus limitation in the artificial soil on cut slope in southwestern China. Environ. Sci. Pollut. Res. 2021, 28, 33307–33319. [Google Scholar] [CrossRef] [PubMed]
- Grêt Regamey, A.; Weibel, B. Global assessment of mountain ecosystem services using earth observation data. Ecosyst. Serv. 2020, 46, 101213. [Google Scholar] [CrossRef]
- Beniston, M. Climatic change in mountain regions: A review of possible impacts. Clim. Change 2003, 59, 5–31. [Google Scholar] [CrossRef]
- Fusaro, C.; Sarria Guzmán, Y.; Chávez Romero, Y.A.; Luna Guido, M.; Muñoz Arenas, L.C.; Dendooven, L.; Estrada Torres, A.; Navarro Noya, Y.E. Land use is the main driver of soil organic carbon spatial distribution in a high mountain ecosystem. PeerJ 2019, 7, e7897. [Google Scholar] [CrossRef] [PubMed]
- Forman, R.T.T. Estimate of the area affected ecologically by the road system in the United States. Conserv. Biol. 2000, 14, 31–35. [Google Scholar] [CrossRef]
- Banerjee, P.; Ghose, M.K.; Pradhan, R. Analytic hierarchy process based spatial biodiversity impact assessment model of highway broadening in Sikkim Himalaya. Geocarto Int. 2020, 35, 470–493. [Google Scholar] [CrossRef]
- Van Der Ree, R.; Smith, D.J.; Grilo, C. The ecological effects of linear infrastructure and traffic: Challenges and opportunities of rapid global growth. In Handbook of Road Ecology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- Xu, H.; Li, T.B.; Chen, J.N.; Liu, C.N.; Zhou, X.h.; Xia, L. Characteristics and applications of ecological soil substrate for rocky slope vegetation in cold and high-altitude areas. Sci. Total Environ. 2017, 609, 446–455. [Google Scholar] [CrossRef]
- Ai, S.; Chen, J.; Gao, D.; Ai, Y. Distribution patterns and drivers of artificial soil bacterial community on cut-slopes in alpine mountain area of southwest China. Catena 2020, 194, 104695. [Google Scholar] [CrossRef]
- Fu, D.; Yang, H.; Wang, L.; Yang, S.; Li, R.; Zhang, W.; Ai, X.; Ai, Y. Vegetation and soil nutrient restoration of cut slopes using outside soil spray seeding in the plateau region of southwestern China. J. Environ. Manag. 2018, 228, 47–54. [Google Scholar] [CrossRef]
- Xiao, H.; Huang, J.; Ma, Q.; Wan, J.; Li, L.; Peng, Q.; Rezaeimalek, S. Experimental study on the soil mixture to promote vegetation for slope protection and landslide prevention. Landslides 2017, 14, 287–297. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, J.; Jia, X.; Shangguan, Z.; Wang, R.; Yan, W. Microbial community assembly and metabolic function during wheat straw decomposition under different nitrogen fertilization treatments. Biol. Fertil. Soils 2020, 56, 697–710. [Google Scholar] [CrossRef]
- Thiele Bruhn, S.; Bloem, J.; de Vries, F.T.; Kalbitz, K.; Wagg, C. Linking soil biodiversity and agricultural soil management. Curr. Opin. Environ. Sustain. 2012, 4, 523–528. [Google Scholar] [CrossRef]
- Rocca, J.D.; Simonin, M.; Bernhardt, E.S.; Washburne, A.D.; Wright, J.P. Rare microbial taxa emerge when communities collide: Freshwater and marine microbiome responses to experimental mixing. Ecology 2020, 101, e02956. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xiao, X.; Nuccio, E.E.; Yuan, M.; Zhang, N.; Xue, K.; Cohan, F.M.; Zhou, J.; Sun, B. Differentiation strategies of soil rare and abundant microbial taxa in response to changing climatic regimes. Environ. Microbiol. 2020, 22, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Lu, Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Change Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef] [PubMed]
- Kurm, V.; Geisen, S.; Gera Hol, W.H. A low proportion of rare bacterial taxa responds to abiotic changes compared with dominant taxa. Environ. Microbiol. 2019, 21, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Wang, J.; Wei, G.; Chen, W.; Lu, Y. Dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere 2019, 235, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Pedrós Alió, C. The rare bacterial biosphere. Annu. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef] [PubMed]
- McGill, B.J.; Etienne, R.S.; Gray, J.S.; Alonso, D.; Anderson, M.J.; Benecha, H.K.; Dornelas, M.; Enquist, B.J.; Green, J.L.; He, F.; et al. Species abundance distributions: Moving beyond single prediction theories to integration within an ecological framework. Ecol. Lett. 2007, 10, 995–1015. [Google Scholar] [CrossRef]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Welch, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef]
- Xu, L.; Cao, H.; Li, C.; Wang, C.; He, N.; Hu, S.; Yao, M.; Wang, C.; Wang, J.; Zhou, S. The importance of rare versus abundant phoD-harboring subcommunities in driving soil alkaline phosphatase activity and available P content in Chinese steppe ecosystems. Soil Biol. Biochem. 2022, 164, 108491. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, B.; Li, C.; Zhou, Z.; Yao, M.; Zhou, X.; Wang, J.; Zhang, B.; Li, X. Increasing relative abundance of non-cyanobacterial photosynthetic organisms drives ecosystem multifunctionality during the succession of biological soil crusts. Geoderma 2021, 395, 115052. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, B.; Wang, E.; Zhu, B.; Yao, M.; Li, C.; Li, X. Soil total organic carbon/total nitrogen ratio as a key driver deterministically shapes diazotrophic community assemblages during the succession of biological soil crusts. Soil Ecol. Lett. 2021, 3, 328–341. [Google Scholar] [CrossRef]
- Treplin, M.; Pennings, S.C.; Zimmer, M. Decomposition of leaf litter in a US saltmarsh is driven by dominant species, not species complementarity. Wetlands 2013, 33, 83–89. [Google Scholar] [CrossRef]
- Cottrell, M.T.; David, K.L. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 2003, 48, 168–178. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Dini Andreote, F.; Salles, J.F. Community assembly processes of the microbial rare biosphere. Trends Microbiol. 2018, 26, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; Guo, Y.; Zhang, B.; Zhang, C.; Van Nostrand, J.D.; Lin, Y.; Zhou, J.; Wei, G. Rare prokaryotic sub-communities dominate the complexity of ecological networks and soil multinutrient cycling during long-term secondary succession in China’s Loess Plateau. Sci. Total Environ. 2021, 774, 145737. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; Ding, J.; Zhu, D.; Hu, H.W.; Delgado Baquerizo, M.; Ma, Y.B.; He, J.Z.; Zhu, Y.G. Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol. Biochem. 2020, 141, 107686. [Google Scholar] [CrossRef]
- Zhao, K.; Kong, W.; Wang, F.; Long, X.E.; Guo, C.; Yue, L.; Yao, H.; Dong, X. Desert and steppe soils exhibit lower autotrophic microbial abundance but higher atmospheric CO2 fixation capacity than meadow soils. Soil Biol. Biochem. 2018, 127, 230–238. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, L.; Fu, Y.; Jiang, Z.; Jia, S.; Song, B.; Liu, D.; Zhou, X. Drought-induced changes in rare microbial community promoted contribution of microbial necromass C to SOC in a subtropical forest. Soil Biol. Biochem. 2024, 189, 109252. [Google Scholar] [CrossRef]
- Philippot, L.; Spor, A.; Hénault, C.; Bru, D.; Bizouard, F.; Jones, C.M.; Sarr, A.; Maron, P.-A. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013, 7, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Pester, M.; Bittner, N.; Deevong, P.; Wagner, M.; Loy, A. A ‘rare biosphere’microorganism contributes to sulfate reduction in a peatland. ISME J. 2010, 4, 1591–1602. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, W.; Kou, Y.; Fang, K.; Liu, Y.; He, H.; Liu, Q. The contrasting responses of abundant and rare microbial community structures and co-occurrence networks to secondary forest succession in the subalpine region. Front. Microbiol. 2023, 14, 1177239. [Google Scholar] [CrossRef]
- He, J.; Tan, X.; Nie, Y.; Ma, L.; Liu, J.; Lu, X.; Mo, J.; Leloup, J.; Nunan, N.; Ye, Q. Distinct responses of abundant and rare soil bacteria to nitrogen addition in tropical forest soils. Microbiol. Spectr. 2023, 11, e03003–e03022. [Google Scholar] [CrossRef]
- Liao, H.; Li, C.; Ai, S.; Li, X.; Ai, X.; Ai, Y. A simulated ecological restoration of bare cut slope reveals the dosage and temporal effects of cement on ecosystem multifunctionality in a mountain ecosystem. J. Environ. Manag. 2023, 325, 116672. [Google Scholar] [CrossRef]
- Ai, X.; Wang, L.; Xu, D.; Rong, J.; Ai, S.; Liu, S.; Li, C.; Ai, Y. Stability of artificial soil aggregates for cut slope restoration: A case study from the subalpine zone of southwest China. Soil Tillage Res. 2021, 209, 104934. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, S.; Ai, S.; Ai, X.; Jiang, X.; Chen, J.; Li, R.; Ai, Y. Artificial soil nutrient, aggregate stability and soil quality index of restored cut slopes along altitude gradient in southwest China. Chemosphere 2020, 246, 125687. [Google Scholar] [CrossRef]
- Liao, H.; Li, C.; Ai, Y.; Li, X. Soil bacterial responses to disturbance are enlarged by altitude in a mountain ecosystem. J. Soils Sediments 2023, 23, 3820–3831. [Google Scholar] [CrossRef]
- Zhu, B.; Li, C.; Wang, J.; Li, J.; Li, X. Elevation rather than season determines the assembly and co-occurrence patterns of soil bacterial communities in forest ecosystems of Mount Gongga. Appl. Microbiol. Biotechnol. 2020, 104, 7589–7602. [Google Scholar] [CrossRef]
- Sun, F.; Lü, Y.; Wang, J.; Hu, J.; Fu, B. Soil moisture dynamics of typical ecosystems in response to precipitation: A monitoring-based analysis of hydrological service in the Qilian Mountains. Catena 2015, 129, 63–75. [Google Scholar] [CrossRef]
- Kou, Y.; Liu, Y.; Li, J.; Li, C.; Tu, B.; Yao, M.; Li, X. Patterns and drivers of nirK-type and nirS-type denitrifier community assembly along an elevation gradient. mSystems 2021, 6, e00667-21. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shen, Z.; Li, C.; Kou, Y.; Wang, Y.; Tu, B.; Zhang, S.; Li, X. Stair-step pattern of soil bacterial diversity mainly driven by pH and vegetation types along the elevational gradients of Gongga Mountain, China. Front. Microbiol. 2018, 9, 569. [Google Scholar] [CrossRef]
- Chang, E.H.; Chen, T.H.; Tian, G.; Chiu, C.Y. The effect of altitudinal gradient on soil microbial community activity and structure in moso bamboo plantations. Appl. Soil Ecol. 2016, 98, 213–220. [Google Scholar] [CrossRef]
- Rui, J.; Zhao, Y.; Cong, N.; Wang, F.; Liu, X. Elevational distribution and seasonal dynamics of alpine soil prokaryotic communities. Front. Microbiol. 2023, 14, 1280011. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, Y.; Zhao, W.; He, H.; Li, D.; He, W.; Liu, Q.; Yin, H. Seasonal variations in the soil amino acid pool and flux following the conversion of a natural forest to a pine plantation on the eastern Tibetan Plateau, China. Soil Biol. Biochem. 2017, 105, 1–11. [Google Scholar] [CrossRef]
- Ren, C.; Zhao, F.; Kang, D.; Yang, G.; Han, X.; Tong, X.; Feng, Y.; Ren, G. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. For. Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- Bao, S. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis; Soil Science Society of America, American Society of Agronomy: Madison, WI, USA, 1996; pp. 961–1010. [Google Scholar] [CrossRef]
- Bremner, J.M. Total Nitrogen. In Methods of Soil Analysis: Part 2. Chemical and Micro-Biological Properties; Black, C.A., Evans, D.D., Dinauer, R.C., Eds.; American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1965; pp. 1149–1178. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Kou, Y.; Wang, J.; Tu, B.; Li, H.; Li, X.; Wang, C.; Yao, M. Soil pH is a major driver of soil diazotrophic community assembly in Qinghai-Tibet alpine meadows. Soil Biol. Biochem. 2017, 115, 547–555. [Google Scholar] [CrossRef]
- Kisand, A. Distribution of sediment phosphorus fractions in hypertrophic strongly stratified Lake Verevi. Hydrobiologia 2005, 547, 33–39. [Google Scholar] [CrossRef]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; U.S. Department of Agriculture: Washington, DC, USA, 1954.
- Tamaki, H.; Wright, C.L.; Li, X.; Lin, Q.; Hwang, C.; Wang, S.; Thimmapuram, J.; Kamagata, Y.; Liu, W.-T. Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLoS ONE 2011, 6, e25263. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Amir, A.; McDonald, D.; Navas Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Galand, P.E.; Casamayor, E.O.; Kirchman, D.L.; Lovejoy, C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. USA 2009, 106, 22427–22432. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and customizable approach for metagenome inference. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ricotta, C.; de Bello, F.; Moretti, M.; Caccianiga, M.; Cerabolini, B.E.L.; Pavoine, S. Measuring the functional redundancy of biological communities: A quantitative guide. Methods Ecol. Evol. 2016, 7, 1386–1395. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant graphics for data analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Hines, J.; Maestre, F.T.; Rillig, M.C. Reconsidering functional redundancy in biodiversity research. NPJ Biodivers. 2023, 2, 9. [Google Scholar] [CrossRef]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, e02738-17. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Liu, D.; Shi, Y.; Wu, X.; Dai, Y.; Shang, Y.; Peng, J.; Cui, Z. Broader environmental adaptation of rare rather than abundant bacteria in reforestation succession soil. Sci. Total Environ. 2022, 828, 154364. [Google Scholar] [CrossRef] [PubMed]
- Royalty, T.M.; Steen, A.D. A quantitative measure of functional redundancy in microbial ecosystems. bioRxiv 2020. [Google Scholar] [CrossRef]
- Li, Y.; Ge, Y.; Wang, J.; Shen, C.; Wang, J.; Liu, Y.J. Functional redundancy and specific taxa modulate the contribution of prokaryotic diversity and composition to multifunctionality. Mol. Ecol. 2021, 30, 2915–2930. [Google Scholar] [CrossRef] [PubMed]
- Biggs, C.R.; Yeager, L.A.; Bolser, D.G.; Bonsell, C.; Dichiera, A.M.; Hou, Z.; Keyser, S.R.; Khursigara, A.J.; Lu, K.; Muth, A.F. Does functional redundancy affect ecological stability and resilience? A review and meta-analysis. Ecosphere 2020, 11, e03184. [Google Scholar] [CrossRef]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Chen, W.; Wei, G. Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol. Ecol. 2017, 26, 5305–5317. [Google Scholar] [CrossRef] [PubMed]
- Kearns, P.J.; Holloway, D.; Angell, J.H.; Feinman, S.G.; Bowen, J.L. Effect of short-term, diel changes in environmental conditions on active microbial communities in a salt marsh pond. Aquat. Microb. Ecol. 2017, 80, 29–41. [Google Scholar] [CrossRef]
- Campbell, B.J.; Yu, L.; Heidelberg, J.F.; Kirchman, D.L. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. USA 2011, 108, 12776–12781. [Google Scholar] [CrossRef]
- Zhao, Y.; Lin, H.; Zhang, W. Abundant bacteria shaped by deterministic processes have a high abundance of potential antibiotic resistance genes in a plateau river sediment. Front. Microbiol. 2022, 13, 977037. [Google Scholar] [CrossRef]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Dong, X.M.; Luo, C.; Ma, S.N.; Xu, J.L.; Cui, Y.D. Nitrogen enrichment reduces the diversity of bacteria and alters their nutrient strategies in intertidal zones. Front. Mar. Sci. 2022, 9, 942074. [Google Scholar] [CrossRef]
- Wang, C.; Liu, D.; Bai, E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, Q.; Xia, C.; Zhong, Y.; Sun, G.; Guo, J.; Yuan, T.; Zhou, J.; He, Z. Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. ISME J. 2014, 8, 1932–1944. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Halvorson, J.J.; Bolton, H., Jr. Soil properties and microbial activity across a 500 m elevation gradient in a semi-arid environment. Soil Biol. Biochem. 2002, 34, 1749–1757. [Google Scholar] [CrossRef]
- Cho, H.; Tripathi, B.M.; Moroenyane, I.; Takahashi, K.; Kerfahi, D.; Dong, K.; Adams, J.M. Soil pH rather than elevation determines bacterial phylogenetic community assembly on Mt. Norikura. FEMS Microbiol. Ecol. 2019, 95, fiy216. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, X.; Cai, X.; Gai, J.; Li, X.; Christie, P.; Zhang, J. Soil microbial community structure and activity along a montane elevational gradient on the Tibetan Plateau. Eur. J. Soil Biol. 2014, 64, 6–14. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Liao, H.; Li, D.; Jing, Y. Abundant Species Govern the Altitude Patterns of Bacterial Community in Natural and Disturbed Subalpine Forest Soils. Diversity 2024, 16, 242. https://doi.org/10.3390/d16040242
Li C, Liao H, Li D, Jing Y. Abundant Species Govern the Altitude Patterns of Bacterial Community in Natural and Disturbed Subalpine Forest Soils. Diversity. 2024; 16(4):242. https://doi.org/10.3390/d16040242
Chicago/Turabian StyleLi, Chaonan, Haijun Liao, Dehui Li, and Yanli Jing. 2024. "Abundant Species Govern the Altitude Patterns of Bacterial Community in Natural and Disturbed Subalpine Forest Soils" Diversity 16, no. 4: 242. https://doi.org/10.3390/d16040242
APA StyleLi, C., Liao, H., Li, D., & Jing, Y. (2024). Abundant Species Govern the Altitude Patterns of Bacterial Community in Natural and Disturbed Subalpine Forest Soils. Diversity, 16(4), 242. https://doi.org/10.3390/d16040242




