Changes in Freeze-Thaw Environments in a Cold Lake: Eliciting New Insights into the Activity and Composition of Bacterial Communities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of the Study Area
2.2. Sample Collection
2.3. Analysis of Sample Environmental Factors
2.4. DNA Extraction and PCR Amplification
2.5. Bioinformatics Analysis and Data Processing
2.6. Microbial Co-Occurrence Network Architecture
3. Results
3.1. Physical and Chemical Characteristics of Water and Ice Bodies
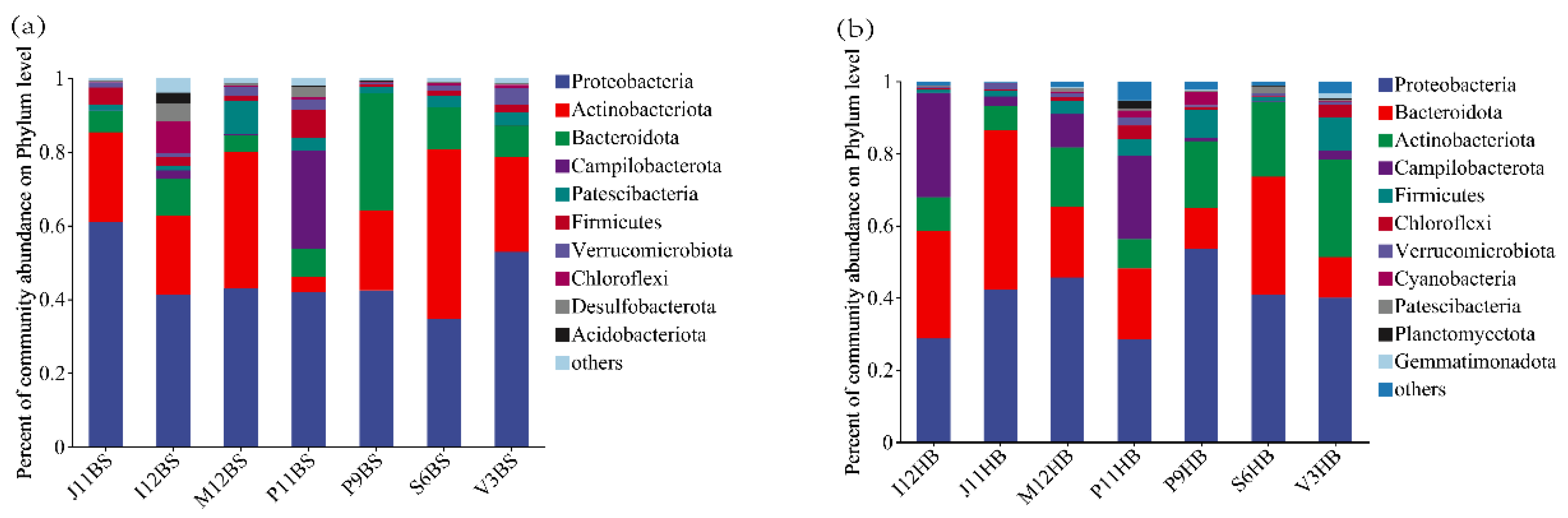
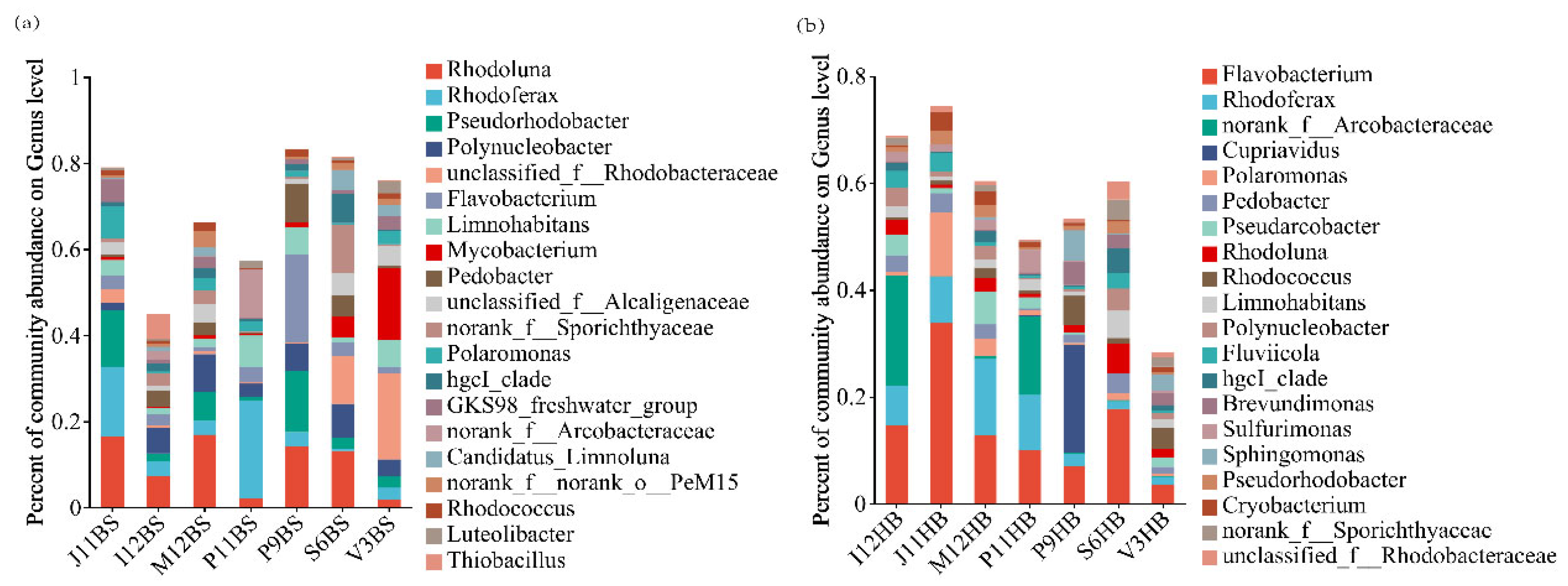
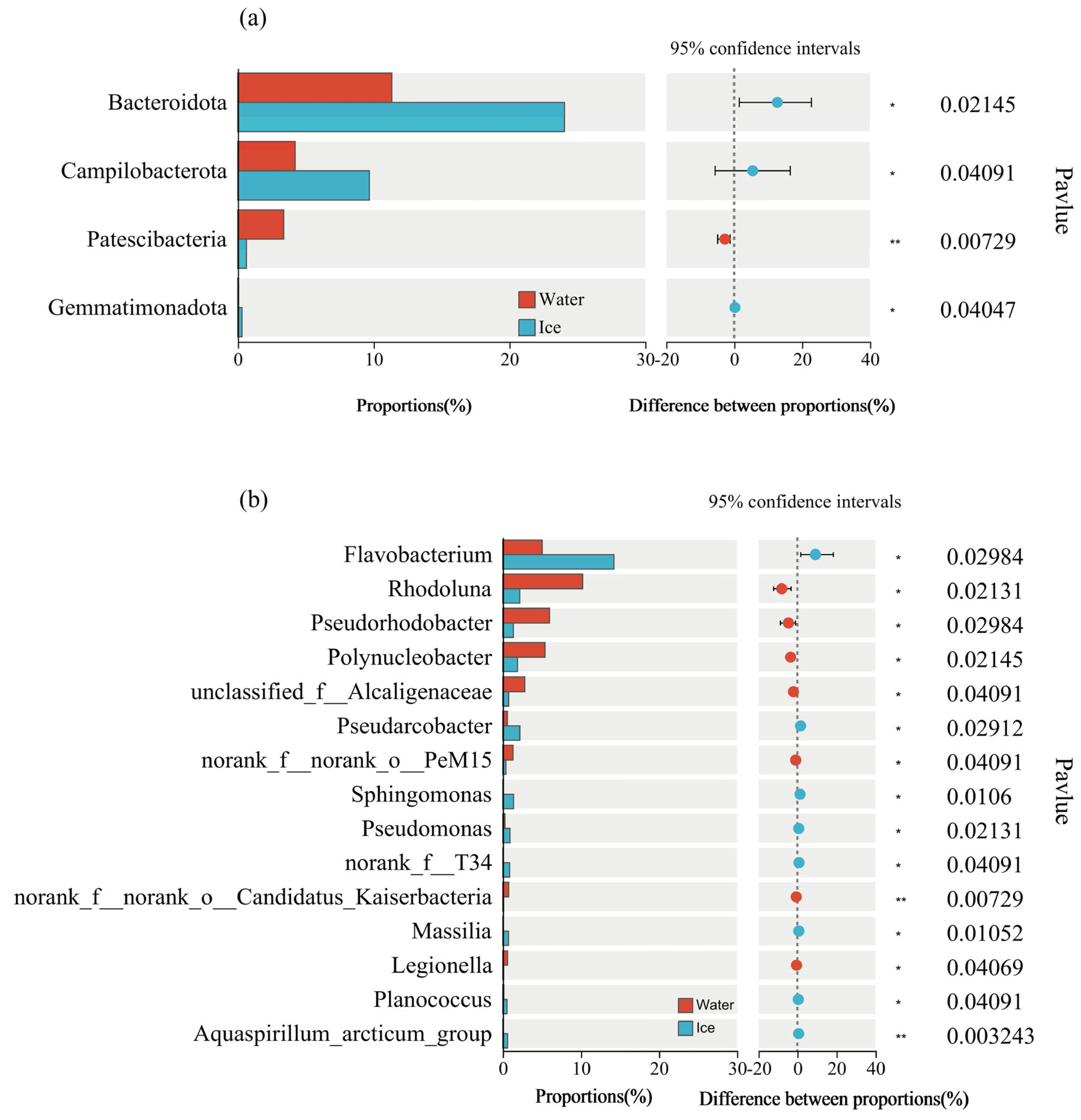
3.2. Bacterial Community Composition
3.3. Structural Diversity of Bacteria
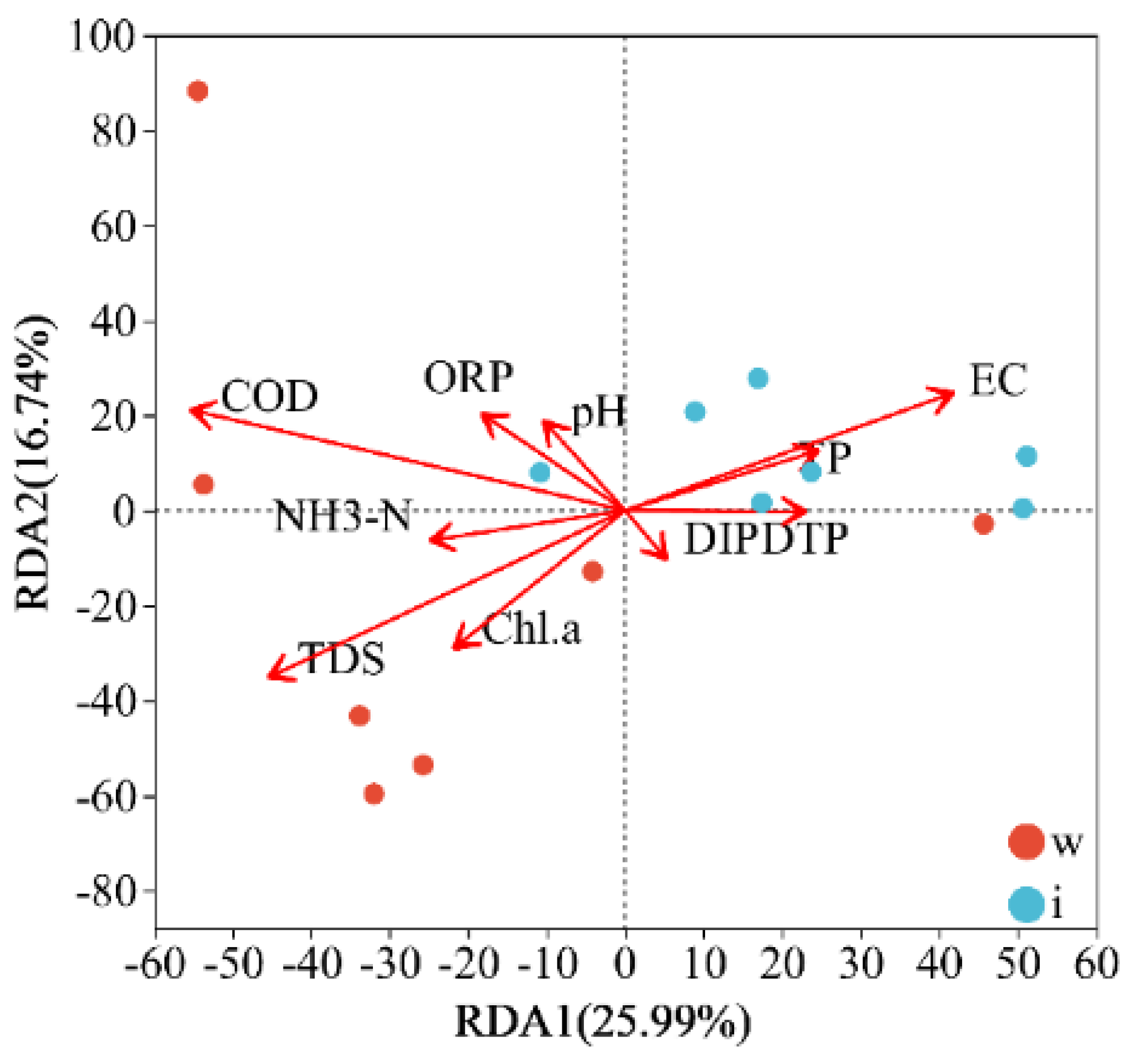
3.4. Environmental Drivers of Bacterial Community Structure
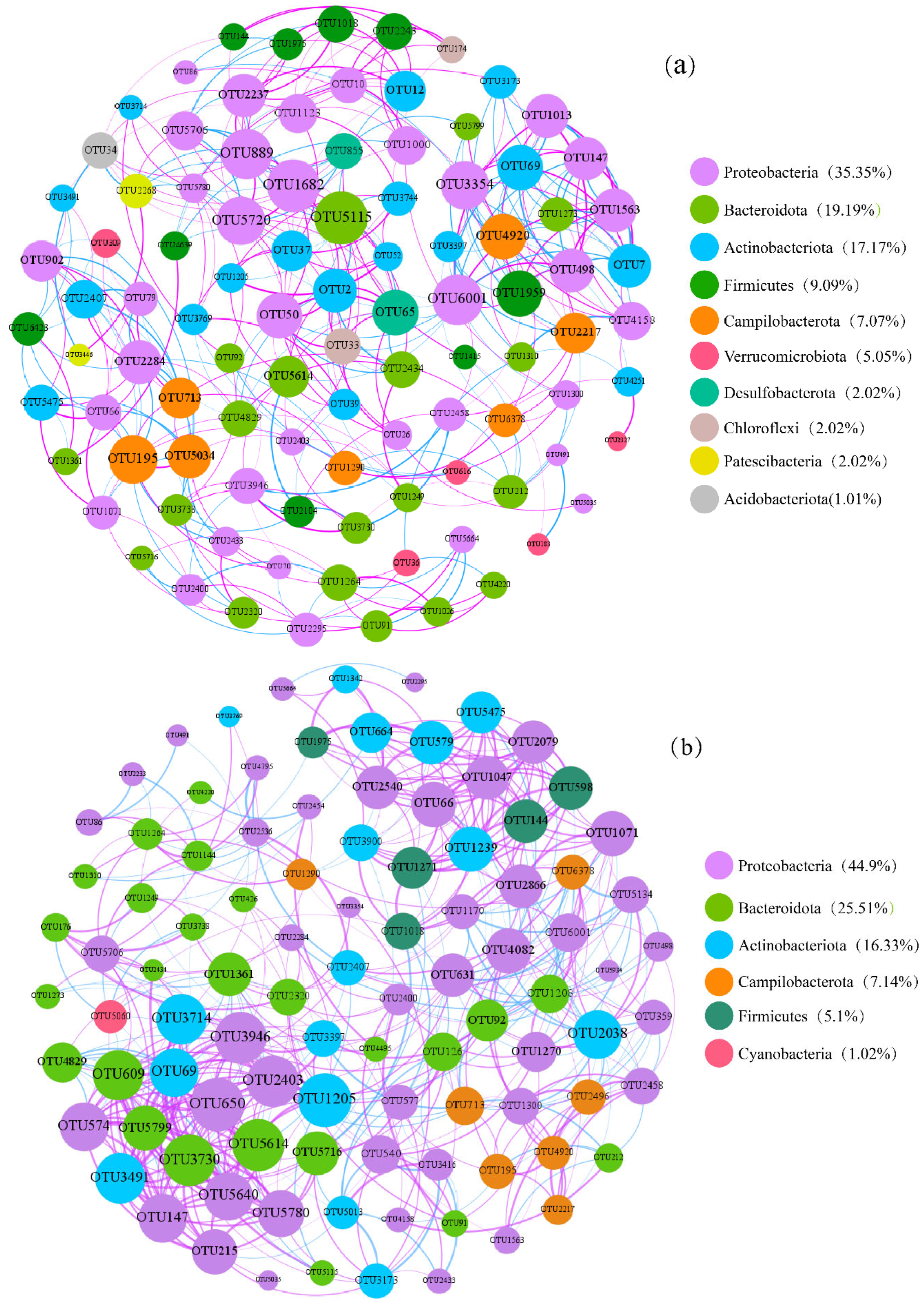
3.5. Co-Occurrence Patterns of Bacteria
4. Discussion
4.1. Habitat Heterogeneity
4.2. Distribution and Composition of Bacterial Communities
4.3. Diversity of Bacterial Communities and Responses to Environmental Factors
4.4. Co-Occurrence Networks of Bacterial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brönmark, C.; Hansson, L.A. Environmental issues in lakes and ponds: Current state and perspectives. Environ. Conserv. 2002, 29, 290–307. [Google Scholar] [CrossRef]
- Davidson, T.A.; Jeppesen, E. The role of palaeolimnology in assessing eutrophication and its impact on lakes. J. Paleolimnol. 2013, 49, 391–410. [Google Scholar] [CrossRef]
- Wang, Y.; Bao, G. Diversity of prokaryotic microorganisms in alkaline saline soil of the Qarhan Salt Lake area in the Qinghai–Tibet Plateau. Sci. Rep. 2022, 12, 3365. [Google Scholar] [CrossRef] [PubMed]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2011, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.; Takemoto, K. Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinform. 2019, 20, 329. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [PubMed]
- Wahdan, S.F.M.; Ji, L.; Schädler, M.; Wu, Y.-T.; Sansupa, C.; Tanunchai, B.; Buscot, F.; Purahong, W. Future climate conditions accelerate wheat straw decomposition alongside altered microbial community composition, assembly patterns, and interaction networks. ISME J. 2022, 17, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hong, W.; Pan, Z.; Fang, W.; Shen, Z.; Cai, H. Wastewater treatment effectiveness is facilitated by crucial bacterial communities in the wetland ecosystem. Sci. Total Environ. 2023, 857, 159375. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Cao, X.; Li, G.; Huang, W.; Leppäranta, M.; Arvola, L.; Huotari, J.; Li, Z. Mass and Heat Balance of a Lake Ice Cover in the Central Asian Arid Climate Zone. Water 2020, 12, 2888. [Google Scholar] [CrossRef]
- Huo, J.; Shi, X.; Zhao, S.; Sun, B.; Li, G.; Yu, H.; Wang, S. Dynamics of water temperature and interfacial heat flux in lake ice and under-ice layers. Hydrol. Curr. Res. 2023, 54, 855–868. [Google Scholar] [CrossRef]
- Shi, X.; Yu, H.; Zhao, S.; Sun, B.; Liu, Y.; Huo, J.; Wang, S.; Wang, J.; Wu, Y.; Wang, Y.; et al. Impacts of environmental factors on Chlorophyll-a in lakes in cold and arid regions: A 10-year study of Wuliangsuhai Lake, China. Ecol. Indic. 2023, 148, 110133. [Google Scholar] [CrossRef]
- Yu, H.; Shi, X.; Zhao, S.; Sun, B.; Liu, Y.; Arvola, L.; Li, G.; Wang, Y.; Pan, X.; Wu, R.; et al. Primary productivity of phytoplankton and its influencing factors in cold and arid regions: A case study of Wuliangsuhai Lake, China. Ecol. Indic. 2022, 144, 109545. [Google Scholar] [CrossRef]
- Shi, R.; Zhao, J.; Shi, W.; Song, S.; Wang, C. Comprehensive Assessment of Water Quality and Pollution Source Apportionment in Wuliangsuhai Lake, Inner Mongolia, China. Int. J. Environ. Res. Public Health 2020, 17, 5054. [Google Scholar] [CrossRef] [PubMed]
- The Editorial Committee of ‘Water and Wastewater Monitoring and Analysis Method’ of the State Environmental Protection 323 Administration. In Water and Wastewater Monitoring and Analysis Method, 4th ed.; China Environmental Science Press: Beijing, China, 2022.
- Zhong, L.; Wu, T.; Ding, J.; Xu, W.; Yuan, F.; Liu, B.F.; Zhao, L.; Li, Y.; Ren, N.Q.; Yang, S.S. Co-composting of faecal sludge and carbon-rich wastes in the earthworm’s synergistic cooperation system: Performance, global warming potential and key microbiome. Sci. Total Environ. 2023, 857, 159311. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, H.; Shen, Y.; Wang, C.; Wang, P.; Zhang, W.; Gao, Y.; Niu, L. Statistical determination of crucial taxa indicative of pollution gradients in sediments of Lake Taihu, China. Environ. Pollut. 2019, 246, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Liu, Y.; Zhang, S.; Shi, X.; Zhao, S.; Lu, J.; Kang, X.; Wang, S.; Wu, Y.; Arvola, L. Characterization of nitrogen and phosphorus at the ice-water-sediment interface and the effect of their migration on overlying water quality in Daihai Lake (China) during the freezing period. Sci. Total Environ. 2023, 893, 164863. [Google Scholar] [CrossRef] [PubMed]
- Pieters, R.; Lawrence, G.A. Effect of salt exclusion from lake ice on seasonal circulation. Limnol. Oceanogr. 2009, 54, 401–412. [Google Scholar] [CrossRef]
- Wu, R.; Zhang, S.; Liu, Y.; Shi, X.; Zhao, S.; Kang, X.; Quan, D.; Sun, B.; Arvola, L.; Li, G. Spatiotemporal variation in water quality and identification and quantification of areas sensitive to water quality in Hulun lake, China. Ecol. Indic. 2023, 149, 110176. [Google Scholar] [CrossRef]
- Rokkan Iversen, K.; Primicerio, R.; Larsen, A.; Egge, J.K.; Peters, F.; Guadayol, O.; Jacobsen, A.; Havskum, H.; Marrase, C. Effects of small-scale turbulence on lower trophic levels under different nutrient conditions. J. Plankton Res. 2009, 32, 197–208. [Google Scholar] [CrossRef]
- Yang, F.; Cen, R.; Feng, W.; Zhu, Q.; Leppäranta, M.; Yang, Y.; Wang, X.; Liao, H. Dynamic simulation of nutrient distribution in lakes during ice cover growth and ablation. Chemosphere 2021, 281, 130781. [Google Scholar] [CrossRef]
- Zhong, M.; Capo, E.; Zhang, H.; Hu, H.; Wang, Z.; Tian, W.; Huang, T.; Bertilsson, S. Homogenisation of water and sediment bacterial communities in a shallow lake (lake Balihe, China). Freshw. Biol. 2022, 68, 155–171. [Google Scholar] [CrossRef]
- Bowman, J.S.; Rasmussen, S.; Blom, N.; Deming, J.W.; Rysgaard, S.; Sicheritz-Ponten, T. Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 2011, 6, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wang, B.C.; Han, J.B.; Yang, J.; Zhang, X.Y.; Guan, X.Y.; Jiang, H.C. Microbial difference and its influencing factors in ice-covered lakes on the three poles. Environ. Res. 2024, 252 Pt 1, 118753. [Google Scholar] [CrossRef]
- Sułowicz, S.; Bondarczuk, K.; Ignatiuk, D.; Jania, J.A.; Piotrowska-Seget, Z. Microbial communities from subglacial water of naled ice bodies in the forefield of Werenskioldbreen, Svalbard. Sci. Total Environ. 2020, 723, 138025. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Keren, R.; Whittaker, M.L.; Farag, I.F.; Doudna, J.A.; Cate, J.H.D.; Banfield, J.F. Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat. Microbiol. 2021, 6, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Cui, Z.; Zhao, M.; Li, J. Seasonal and Spatial Variations of Bacterial Community Structure in the Bailang River Estuary. J. Mar. Sci. Eng. 2023, 11, 825. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Kragelund, C.; Seviour, R.J.; Nielsen, J.L. Identity and ecophysiology of filamentous bacteria in activated sludge. FEMS Microbiol. Rev. 2009, 33, 969–998. [Google Scholar] [CrossRef]
- Kindaichi, T.; Yamaoka, S.; Uehara, R.; Ozaki, N.; Ohashi, A.; Albertsen, M.; Nielsen, P.H.; Nielsen, J.L.; Stams, A. Phylogenetic diversity and ecophysiology of Candidate phylum Saccharibacteria in activated sludge. FEMS Microbiol. Ecol. 2016, 92, fiw078. [Google Scholar] [CrossRef] [PubMed]
- Remmas, N.; Melidis, P.; Zerva, I.; Kristoffersen, J.B.; Nikolaki, S.; Tsiamis, G.; Ntougias, S. Dominance of candidate Saccharibacteria in a membrane bioreactor treating medium age landfill leachate: Effects of organic load on microbial communities, hydrolytic potential and extracellular polymeric substances. Bioresour. Technol. 2017, 238, 48–56. [Google Scholar] [CrossRef]
- Morya, R.; Salvachúa, D.; Thakur, I.S. Burkholderia: An Untapped but Promising Bacterial Genus for the Conversion of Aromatic Compounds. Trends Biotechnol. 2020, 38, 963–975. [Google Scholar] [CrossRef]
- Obbard, R.W.; Sadri, S.; Wong, Y.Q.; Khitun, A.A.; Baker, I.; Thompson, R.C. Global warming releases microplastic legacy frozen in Arctic Sea ice. Earth’s Future 2014, 2, 315–320. [Google Scholar] [CrossRef]
- Moon, K.; Kang, I.; Kim, S.; Kim, S.J.; Cho, J.C. Genomic and ecological study of two distinctive freshwater bacteriophages infecting a Comamonadaceae bacterium. Sci. Rep. 2018, 8, 7989. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A Guide to the Natural History of Freshwater Lake Bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed]
- Mao, R.; Hu, Y.; Zhang, S.; Wu, R.; Guo, X. Microplastics in the surface water of Wuliangsuhai Lake, northern China. Sci. Total Environ. 2020, 723, 137820. [Google Scholar] [CrossRef]
- Taipale, S.J.; Vesamäki, J.; Kautonen, P.; Kukkonen, J.V.K.; Biasi, C.; Nissinen, R.; Tiirola, M. Biodegradation of microplastic in freshwaters: A long-lasting process affected by the lake microbiome. Environ. Microbiol. 2022, 25, 2669–2680. [Google Scholar] [CrossRef]
- Gong, M.; Yang, G.; Zhuang, L.; Zeng, E.Y. Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms. Environ. Pollut. 2019, 252, 94–102. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, W.; Li, X.; Gao, G.; Jiang, J. Linking bacterial community shifts with changes in the dissolved organic matter pool in a eutrophic lake. Sci. Total Environ. 2020, 719, 137387. [Google Scholar] [CrossRef]
- Heylen, K.; Gevers, D.; Vanparys, B.; Wittebolle, L.; Geets, J.; Boon, N.; De Vos, P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ. Microbiol. 2006, 8, 2012–2021. [Google Scholar] [CrossRef]
- He, Z.; Wang, Y.; Bai, X.; Chu, M.; Yi, Y.; Zhu, J.; Gu, M.; Jiang, L.; Zhang, Z. Bacterial Community Composition and Isolation of Actinobacteria from the Soil of Flaming Mountain in Xinjiang, China. Microorganisms 2023, 11, 489. [Google Scholar] [CrossRef]
- Essoussi, I.; Ghodhbane-Gtari, F.; Amairi, H.; Sghaier, H.; Jaouani, A.; Brusetti, L.; Daffonchio, D.; Boudabous, A.; Gtari, M. Esterase as an enzymatic signature of Geodermatophilaceaeadaptability to Sahara desert stones and monuments. J. Appl. Microbiol. 2010, 108, 1723–1732. [Google Scholar] [CrossRef]
- Dastrup, D.B.; Carling, G.T.; Collins, S.A.; Nelson, S.T.; Fernandez, D.P.; Tingey, D.G.; Hahnenberger, M.; Aanderud, Z.T. Aeolian dust chemistry and bacterial communities in snow are unique to airshed locations across northern Utah, USA. Atmos. Environ. 2018, 193, 251–261. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Zaichikov, E.; Belkova, N.; Denissova, L.; Pernthaler, J.; Pernthaler, A.; Amann, R. Comparative 16S rRNA Analysis of Lake Bacterioplankton Reveals Globally Distributed Phylogenetic Clusters Including an Abundant Group of Actinobacteria. Appl. Environ. Microbiol. 2000, 66, 5053–5065. [Google Scholar] [CrossRef] [PubMed]
- Michaud, L.; Caruso, C.; Mangano, S.; Interdonato, F.; Bruni, V.; Lo Giudice, A. Predominance of Flavobacterium, Pseudomonas, andPolaromonaswithin the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol. Ecol. 2012, 82, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Brinkmeyer, R.; Knittel, K.; Jürgens, J.; Weyland, H.; Amann, R.; Helmke, E. Diversity and Structure of Bacterial Communitiesin Arctic versus Antarctic Pack Ice. Appl. Environ. Microbiol. 2003, 69, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Selivanova, E.A.; Poshvina, D.V.; Khlopko, Y.A.; Gogoleva, N.E.; Plotnikov, A.O. Diversity of Prokaryotes in Planktonic Communities of Saline Sol-Iletsk lakes (Orenburg Oblast, Russia). Microbiology 2018, 87, 569–582. [Google Scholar] [CrossRef]
- Li, W.B.; Yang, X.; Tian, Y.N.; Du, L. Differences of plankton community structure in ice-water in Dari Lake under frozen condition. Environ. Sci. 2021, 42, 2276–2286. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Changyou, L.; Leppäranta, M.; Xiaonghong, S.; Shengnan, Z.; Chengfu, Z. Notable increases in nutrient concentrations in a shallow lake during seasonal ice growth. Water Sci. Technol. 2016, 74, 2773–2783. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, J.; Nawaz, M.Z.; Mahboob, S.; Al-Ghanim, K.A.; Khan, I.A.; Lu, Z.; Chen, T. Seasonal succession and spatial distribution of bacterial community structure in a eutrophic freshwater Lake, Lake Taihu. Sci. Total Environ. 2019, 669, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Xie, G.; Zhang, Y.; Yu, B.; Shao, K.; Gao, G.; Tang, X. Similar assembly mechanisms but distinct co-occurrence patterns of free-living vs. particle-attached bacterial communities across different habitats and seasons in shallow, eutrophic Lake Taihu. Environ. Pollut. 2022, 314, 120305. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Chen, S.; Zhao, Z.; Feng, J.; Zhang, Z.; Lu, K.; Jia, J. Water Bacterial and Fungal Community Compositions Associated with Urban Lakes, Xi’an, China. Int. J. Environ. Res. Public Health 2018, 15, 469. [Google Scholar] [CrossRef]
- Shao, K.; Yao, X.; Wu, Z.; Jiang, X.; Hu, Y.; Tang, X.; Xu, Q.; Gao, G. The bacterial community composition and its environmental drivers in the rivers around eutrophic Chaohu Lake, China. BMC Microbiol. 2021, 21, 179. [Google Scholar] [CrossRef]
- Yuan, W.; Su, X.; Cui, G.; Wang, H. Microbial community structure in hypolentic zones of a brine lake in a desert plateau, China. Environ. Earth Sci. 2016, 75, 1132. [Google Scholar] [CrossRef]
- Wan, F.; Zhang, F.; Zheng, X.; Xiao, L. Study on Ecological Water Demand and Ecological Water Supplement in Wuliangsuhai Lake. Water 2022, 14, 1262. [Google Scholar] [CrossRef]
- Newman, M.E.J. The structure and function of complex networks. Siam Rev. 2003, 45, 167–256. [Google Scholar] [CrossRef]
- Shen, Z.; Yu, B.; Shao, K.; Gao, G.; Tang, X. Warming reduces microeukaryotic diversity, network complexity and stability. Environ. Res. 2023, 238, 117235. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Allesina, S.; Bodini, A.; Pascual, M. Functional links and robustness in food webs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1701–1709. [Google Scholar] [CrossRef]
- Bernardet, J.F.; Bowman, J.P. The Genus Flavobacterium; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Collado, L.; Inza, I.; Guarro, J.; Figueras, M.J. Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 2008, 10, 1635–1640. [Google Scholar] [CrossRef]
- Morales Medina, W.R.; Eramo, A.; Fahrenfeld, N.L. Metabolically Active Prokaryotes and Actively Transcribed Antibiotic Resistance Genes in Sewer Systems: Implications for Public Health and Microbially Induced Corrosion. Microb. Ecol. 2021, 83, 583–595. [Google Scholar] [CrossRef]
- Di Pippo, F.; Venezia, C.; Sighicelli, M.; Pietrelli, L.; Di Vito, S.; Nuglio, S.; Rossetti, S. Microplastic-associated biofilms in lentic Italian ecosystems. Water Res. 2020, 187, 116429. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, C.; Lu, J.; Jia, Y.; Tian, Z.; Zhang, Z.; Hu, Y.; Liu, Y. Changes in Freeze-Thaw Environments in a Cold Lake: Eliciting New Insights into the Activity and Composition of Bacterial Communities. Diversity 2024, 16, 311. https://doi.org/10.3390/d16060311
Feng C, Lu J, Jia Y, Tian Z, Zhang Z, Hu Y, Liu Y. Changes in Freeze-Thaw Environments in a Cold Lake: Eliciting New Insights into the Activity and Composition of Bacterial Communities. Diversity. 2024; 16(6):311. https://doi.org/10.3390/d16060311
Chicago/Turabian StyleFeng, Chen, Junping Lu, Yongqin Jia, Zhiqiang Tian, Zixuan Zhang, Yaxin Hu, and Yinghui Liu. 2024. "Changes in Freeze-Thaw Environments in a Cold Lake: Eliciting New Insights into the Activity and Composition of Bacterial Communities" Diversity 16, no. 6: 311. https://doi.org/10.3390/d16060311
APA StyleFeng, C., Lu, J., Jia, Y., Tian, Z., Zhang, Z., Hu, Y., & Liu, Y. (2024). Changes in Freeze-Thaw Environments in a Cold Lake: Eliciting New Insights into the Activity and Composition of Bacterial Communities. Diversity, 16(6), 311. https://doi.org/10.3390/d16060311




