Abstract
Argonauts or paper nautiluses are pelagic octopod cephalopods with a cosmopolitan distribution in tropical and subtropical waters around the world. Unlike other species of octopus, these are characterized by the fact that the female has a shell that serves as the breeding chamber for the eggs. Over time, this structure has been used as a taxonomic diagnostic character, causing problems in the systematics of this genus, with around 50 synonymies reported. Only two species, Argonauta argo and A. nouryi, have been reported in the Northern Humboldt Current System; however, there is taxonomic uncertainty regarding these species, which is reflected in the paralarvae (the first stage of life after hatching). In the paralarvae, the chromatophore patterns are considered to be conservative and reliable taxonomic characteristics. The objective of this study is to demonstrate the extensive variability in the chromatophore arrangement of Argonauta paralarvae in the Northern Humboldt Current using DNA barcoding and five species delimitation models. Our results include up to 11 different paralarvae morphotypes according to the pattern of chromatophores (number and arrangement) and 2 shell morphotypes. Species delimitation methods divided the 13 Argonauta morphotypes into two consensus molecular taxonomic units (MOTUs), A. argo and A. nouryi. Additionally, the results revealed an extensive morphological variability in the paralarvae and female shells of A. nouryi, demonstrating the importance of molecular data in studies involving species with different life stages, especially when this extensive morphological variability obscures conventional analyses.
1. Introduction
The Humboldt Current System, a cold-water current, extends along the west coast of South America from southern Chile (42°S) up to the Galapagos Islands near the equator [1]. The Northern Humboldt Current System (NHCS), the northern area facing Peru, represents less than 0.1% of the world’s ocean surface but currently produces about 10% of the world’s fish catch [2]. In this region, the most important fish are Peruvian anchovy Engraulis ringens (Jenyns, 1842), and the jumbo flying squid Dosidicus gigas (d’Orbigny, 1835) [3]. However, there are other species of ecological importance such as the cephalopod genus Argonauta (Linnaeus, 1758), which is an indicator of the approach of warm ocean waters to the coasts of the Peruvian Sea [4], especially during El Niño events.
Argonauts or paper nautiluses (Argonautidae) are a family of pelagic octopuses that inhabit tropical and temperate oceans of the world [5]. These group of octopuses are easily differentiated from benthic octopuses due to the presence of a shell called a paper nautilus, present only in females. This shell is secreted by the first pairs of arms and functions as the breeding chamber for the eggs and their subsequent fertilization [6]. For many years, the morphological characteristics of these shells in adults have been used as a taxonomic characteristic with which to differentiate species [7,8,9,10]. However, this genus presents problems in the organization of its systematics around the world. Web platforms such as WORMS (htpp://www.marinespecies.ogr) (accessed on 23 September 2023), ITIS (htpp://www.itis.gov) (accessed on 23 September 2023), and BoldSystems (htpp://www.boldsystems.org) (accessed on 23 September 2023) show no consensus on the number of species in this molluscan group.
Adults present difficulties in not only taxonomic identification [6,11] but also in their first post-hatching life stages, known as paralarvae [12]. A specialized bibliography on the taxonomic aspects of paralarvae is lacking worldwide, especially for the Pacific Ocean. Some studies suggest that the chromatophore pattern is a conservative and reliable taxonomic characteristic [13,14]. Thus, the number and distribution of each chromatophore, the iridophore pattern, and the arm length formula are considered species-specific [11]. However, the analysis of these characters is problematic, not only because of their low abundance in zooplankton samples (typically between 1 and 5 individuals per net [15]), but also due to difficulties during the fixing of the paralarvae. For example, specimens tend to invert the mantle muscle when dragged in the net during zooplankton sampling collection, different chromatophores are discolored by the fixative, and appendages detach (arms and/or tentacles), hindering determination by conventional morphometric analysis [16]. Despite their small size, paralarvae are capable of evading high-speed sampler equipment [15], which further reduces the possibilities to study them.
For these reasons, the selection of alternative techniques such as DNA barcoding over conventional ones is necessary. This molecular identification tool uses a short and standardized section of the mitochondrial gene Cytochrome Oxidase I (COI) [17] and has been proven efficient in evaluating diversity in the marine ecosystem [18,19]. Recently, molecular data together with species delimitation methods were used to reveal the diversity of cephalopods [20,21,22,23].
Therefore, we used a DNA barcoding approach together with species delimitation methods to identify the Argonauta paralarva diversity in the NHCS, distinguishing molecular operational taxonomic units (MOTUs) and revealing the morphological variability in the number and arrangement of chromatophores. This work contributes to updating the list of species of the genus in the NHCS.
2. Materials and Methods
2.1. Sampling
A total of 15 paralarvae and 1 juvenile of Argonauta were obtained from zooplankton samples collected using different nets (Figure 1, Table 1). The individuals were separated onboard and placed into 2 mL cryovials containing 96° ethanol and stored at 4 °C for one week. Additionally, two female adults with shells were collected via pelagic trawl during research cruises of the Instituto del Mar del Perú (IMARPE) (Table 1). Egg masses from these individuals were stored in 96° ethanol for molecular analysis.
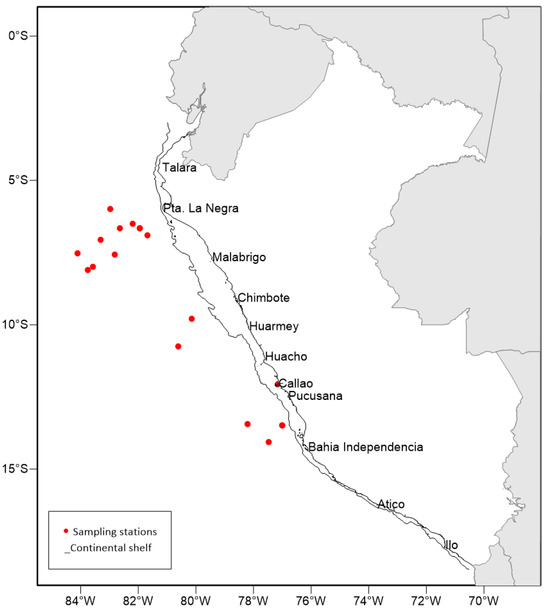
Figure 1.
Sampling area of Argonauta paralarvae in the Northern Humboldt Current System (NHCS). The red dots are the zooplankton sampling stations (different nets), and the line parallel to the coast is the continental shelf.

Table 1.
Paralarvae of Argonauta sampled along the Northern Humboldt Current System (NHCS) between 2017 and 2023.
2.2. Morphological Description
The morphological descriptions of the paralarvae were performed in specimens between 0.8 and 2.5 mm in dorsal mantle length (ML). The morphological determination was performed after consulting a specialized bibliography of cephalopod paralarvae [11,24,25,26]. The shells of females were identified according to Finn [6]. The ML was measured (mm) with a Nikon SMZ 1270 stereomicroscope equipped with a digital camera and NIS Element D v. 4.13 image capture software. The funnel-locking apparatus was reviewed. Specimens were sexed due to sexual dimorphism. Males have a hectocotylized third arm that is wrapped in a bag called a “pouch”. Chromatophore pattern (number, position, and formula) of the DHCP (dorsal head chromatophore pattern), VHCP (ventral head chromatophore pattern), dorsal mantle (DM), ventral mantle (VM), dorsal eye (DE), ventral eye (VE), digestive gland (DG), funnel, arms, and hectocotylus (for males) on each morphotype were described according to De Silva-Dávila [24] (Figure 2). Likewise, diagrams (drawings) were made using a camera lucida suitable for the stereomicroscope. The voucher specimens were deposited in the scientific collection of the Laboratorio de Zooplancton y Producción Secundaria of IMARPE.
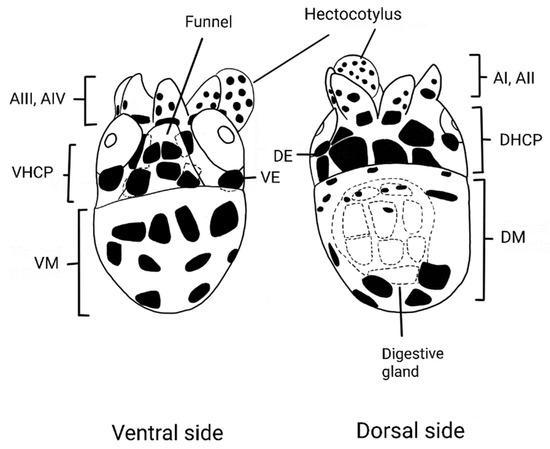
Figure 2.
Argonauta paralarva chromatophore pattern diagram (ML: 1 mm): dorsal head chromatophore pattern (DHCP), ventral head chromatophore pattern (VHCP), dorsal mantle (DM), ventral mantle (VM), dorsal eye (DE), ventral eye (VE), digestive gland (DG), funnel, arms (AI-AIV), and hectocotylus (for males).
2.3. DNA Extraction, Amplification and Sequencing
DNA extraction was performed using the HotSHOT method [27]. Tissues were transferred into 0.2 mL microtubes containing 50 µL of alkaline lysis buffer (25 mM NaOH, 0.2 mM EDTA, pH 8.0) and were ground using a sterile micropipette tip. The samples were incubated at 95 °C for 30 min, followed by 4 °C for 4 min in a thermocycler. Then, 50 µL of neutralizing solution (40 mM Tris-HCl, pH 5.0) was added, followed by a vortex (to homogenize) and a spin in a mini-centrifuge. All samples were stored at 4 °C for use directly in the PCR. For species identification, the cytochrome oxidase subunit I (COI) gene of mitochondrial DNA (mtDNA) was amplified with the primers ZplankF1_t1/ZplankR1_t1 [28]. The PCR was performed using 1X of Taq buffer, 0.25 mM of each dNTP, 0.4 mM of each primer, 2.5 mM of MgCl2, 0.5 U of Vazyme Taq DNA polymerase, and 2 µL of DNA in 15 µL of the final reaction volume. Other PCRs were performed using the HotStartTaq Plus Master Mix kit (QIAGEN). The PCR conditions were an initial denaturation at 95 °C for 5 min, followed by 36 cycles at 95 °C for 40 s, 45 °C for 50 s, and 72 °C for 1 min, with a final extension of 72 °C for 7 min. All reactions were evaluated via electrophoresis in 1% agarose gels, and amplified products with the expected size were then bidirectionally sequenced on a genetic analyzer ABI 3500 (Applied Biosystems) using both a commercial service and at IMARPE. The contigs were assembled using the software CodonCode Aligner v. 6.0.2 (Codon Code Corporation, Dedham, MA, USA). Primer sequences and bad-quality regions were trimmed. The consensus sequences and all specimen data were deposited in the dataset BOLD (https://www.boldsystems.org/, accessed on 6 January 2024) under dataset code DS-ARGNT.
2.4. Species Delimitation
To infer MOTUs using different species delimitation methods, our sequences were combined with thirteen COI sequences from three Argonauta species from different oceans (Accession numbers AF000028, MT216541, MT216542, MK034303, AB191273, ON367817, OP132805, OP132808, OP132809, OP132810, LC596061, KY649285, and OR234732) retrieved from GenBank (www.ncbi.nlm.nih.gov/genbank/, accessed on 6 January 2024).
The thirty-one DNA sequences (627 pb) were aligned in the FASTA format, and five species delimitation methods were performed using pipeline SPdel [29]: ABGD (automatic barcode gap discovery) [30], ASAP (assemble species by automatic partitioning) [31], GMYC (generalized mixed yule coalescent approach) [32], bPTP (Bayesian Poisson tree processes) [33], and mPTP (multi-rate Poisson tree processes) [33]. For bPTP and mPTP methods, an ultrametric tree was generated using BEAST v2.5 [34], with a log-normal relaxed clock model and HKY+G substitution model suggested by the Bayesian Information Criterion (AIC) in jModeltest 2 [35]. The analysis was implemented with 100,000,000 MCMC generations and with a burn-in of 10%. Consensus MOTUs were generated by comparing the five delimitation methods. Values of intragroup and intergroup Kimura 2-parameter (K2P) genetic distances for nominal species and consensus MOTUs were calculated using SPdel.
3. Results
3.1. Morphological Determination
After the morphological revision, 13 different morphotypes were determined, including 11 different chromatophore patterns (n = 16; Argonauta M1 (n = 2), Argonauta M5 (n = 1), Argonauta M6 (n = 1), Argonauta M7 (n = 2), Argonauta M8 (n = 1), Argonauta M10 (n = 1), Argonauta M12 (n = 1), Argonauta M13 (n = 1), Argonauta M15 (n = 1), Argonauta M17 (n = 1), and Argonauta M18 (n = 4)). The two female adults were identified as A. nouryi based on shell morphology. In accordance with the morphotypes reported by Finn [6], the females exhibited two different shell morphotypes A. nouryi shell type I (st_I), (n = 1) and A. nouryi shell type II (st_II), (n = 1) (Figure 3k,i and Table 2),
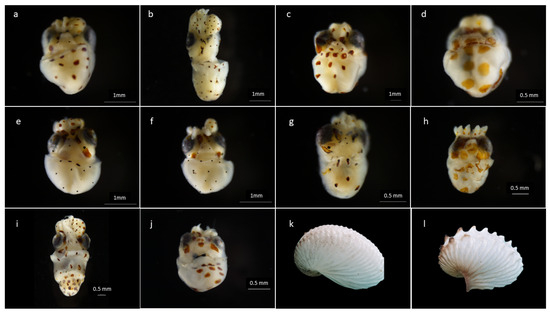
Figure 3.
Argonauta paralarvae, ventral side (a–j) and shell, lateral side (k,l) of an adult female collected in this study. Sample codes: (a) PMZPK063-20 (M5), (b) PMPZK064-20 (M7), (c) PMPZK065-20 (M18), (d) PMPZK066-20 (M6), (e) PMPZK070-20 (M13), (f) PMZPK071-20 (M10), (g) PMPZK072-20 (M12), (h) PMPZK073-20 (M17), (i) PMPZK076-20, (j) PMPZK078-20 (M15), (k) PMPZK086-20 (st_I) and (l) PMPZK085-20 (st_II).

Table 2.
Morphological and meristic characteristics of the Argonauta paralarvae collected in the NHCS. M = male; F = female. The chromatophore pattern is related to the number of chromatophores. (The DHCP, VHCP, dorsal mantle, ventral mantle, and funnel columns are read from top to bottom, each row separated with a “+” sign. In the arms column, the reading extends from the base to the distal part of the appendages. In the dorsal and ventral eye area columns, the number inside the parentheses refers to the number for both eyes. In the digestive gland and hectocotyl columns, the number refers to the total chromatophore count.)
The most variable chromatophore pattern was observed in the funnel, by which individuals were delimited into different morphotypes (Table 2, Figure 4). The chromatophore arrangements that remained consistent in all organisms were the DHCP (2 + 4) and VHCP (2 + 2). The number of chromatophores in the digestive gland ranged from 8 to 11 in all paralarvae, and only one chromatophore was observed at the base of each arm (1b). Due to fixation, the arrangement, number, and disposition of the chromatophores on the dorsal and ventral sides of the mantle and in the hectocotylus could be observed in only a few individuals.
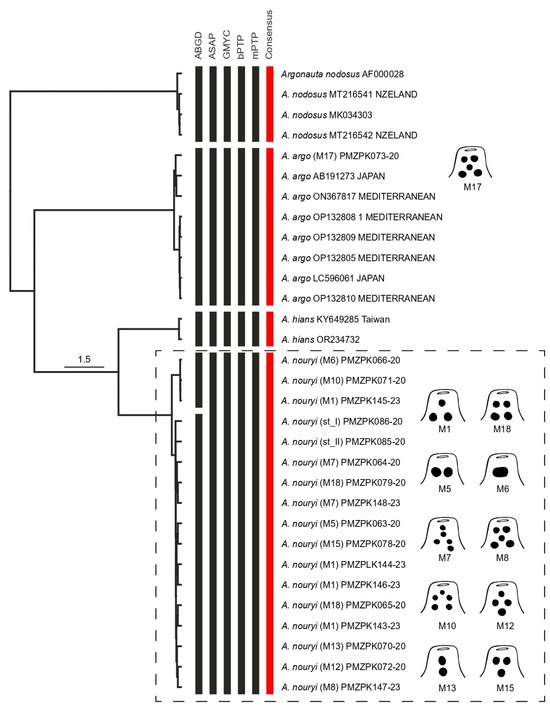
Figure 4.
Bayesian tree showing the clustering of the MOTUs resulting from ABGD, ASAP, bPTP, and mPTP analysis, and consensus MOTUs. The black vertical bars represent the numbers of species delimited by ABGD, ASAP, GMYC, bPTP, and mPTP methods; and the red vertical bars represent consensus MOTUs. The scale bar indicates the nucleotide substitutions per site. The codes in parentheses refer to the described morphotype of Argonauta, and the subsequent code to the barcode accession number. Note the different morphotypes (schemes) corresponding to the A. nouryi MOTU.
For individuals between 0.8 and 2.2 mm, the number of VE chromatophores remained the same (one in each eye). However, the number of chromatophores on the dorsal side (DE) varied. In Argonauta M6 (PMPZK066-20), Argonauta M17 (PMPZK073-20), and Argonauta M18 (PMPZK079-20), only one chromatophore associated with the eye was observed, while the other morphotypes described showed two. For the juvenile (3.5 mm) Argonauta M1 (PMZPK145-23), only the oldest chromatophores (brown) in the funnel were considered (1 + 2) as, in the other parts of the body, it was impossible to establish a pattern due to the number of new chromatophores that appeared (orange).
3.2. DNA Barcode Analysis and Species Delimitation
We obtained 18 sequences from the 13 morphotypes. The final alignment of COI sequences resulted in 627 bp with 102 parsimony-informative sites. The ABGD, ASAP, GYMC, bPTP, and mPTP analyses resulted in four MOTUs (Figure 4). SPdel summarized the different delimitation results in four consensus MOTUs (Figure 4). From the 13 morphotypes, the individual determined as Argonauta M17 was grouped with Argonauta argo from Japan and the Mediterranean Sea in MOTU 01. Meanwhile, the other 12 morphotypes were grouped into MOTU 04, including two female adults of A. nouryi (Figure 4). The other two MOTUs corresponded to Argonauta nodosus and Argonauta hians from New Zealand and Taiwan, respectively. The genetic intra-MOTU and inter-MOTU distances are shown in Table 3. The highest maximum intra-MOTU distance was 0.97% for MOTU 04 (A. nouryi) and the lowest minimum inter-MOTU distance was 4.63% between MOTU 02 (A. hians) and MOTU 04 (A. nouryi).

Table 3.
Genetic K2-P distance of the MOTUs of Argonauta: mean intra-MOTU divergence (Mean), maximum intra-MOTU divergence (Max), nearest neighbor (NN), and distance to the nearest neighbor (DtoNN).
4. Discussion
Our results supported four consensus MOTUs corresponding to the four known nominal species: A. argo, A. hians, A. nodosus, and A. nouryi [6]. From these, two species of the Argonauta genus are considered to be distributed in the Northern Humboldt Current System at the adult level: A. argo and A. nouryi [36]. Previous studies also reported the presence of A. hians [36,37,38]; however, Finn reported A. hians as being restricted to the Atlantic and Indian oceans, and Arabian, Chinese, and Philippine seas after a global review of the systematics and distribution of the genus [6]. At the paralarva level in the NHCS, it had previously only been possible to identify different morphotypes as Argonauta spp. [4]. Our results show the presence of the two species A. argo and A. nouryi in the NHCS, in accordance with Finn [6], the latter being a new public BIN (BOLD:ABA3205) for the BOLDSystem database.
The paralarvae characterized herein were grouped into two consensus MOTUs: MOTU 01 (A. argo) and MOTU 04 (A. nouryi). The MOTU 01 was identified as A. argo using sequences from previous studies [39], including sequences from adult individuals [39]. The MOTU 04 was identified as A. nouryi based on the two female adults herein sequenced. In these MOTUs, paralarvae with different dispositions and numbers of chromatophores in the funnel clustered together, showing high morphological variability. There are many studies that report and analyze the abundance, frequency, and spatial distribution associated with different oceanographic conditions of A. nouryi [40,41]. However, there is no morphological description of its paralarvae, only references to the fact that it was previously named Argonauta boettgeri [24] in the Mexican Pacific Ocean [42], with an arrangement of 1 + 2 in the funnel, 2 + 2 in the VHCP, and 2 + 4 in the DHCP. Our results for A. nouryi show a wide variability in the number and arrangement of chromatophores in the funnel: 2 + 4 in the DHCP, 2 + 2 in the VHCP, 02 chromatophores in each eye (dorsal side), and between 8 and 11 in the digestive gland (Table 2). This highlights the significant challenge of taxonomic determination based solely on morphological characteristics, especially the chromatophore pattern. This complexity increases when we consider that some ancient descriptions did not mention the chromatophore arrangement [43]. However, over the years, some authors have incorporated this information into descriptions, such as that of A. argo (~1 mm) with a 2 + 2 arrangement in the funnel, with 2 chromatophores on the dorsal side of the head [11,25], and 2 + 4 DHCP [24]. In the present work, A. argo is reported with a chromatophore arrangement of 2 + 1 + 2 in the funnel, 2 + 4 in the DHCP, 2 + 2 in the VHCP, 1 chromatophore in each eye (dorsal side), and 9 in the digestive gland, also showing morphological variability. Descriptions of the paralarvae of A. hians (1.0 mm) are scarcer [11,43,44], where some authors describe2 + 2 in the funnel and in the VHCP [24], an arrangement of 4 chromatophores (rectangle shape) on the dorsal side of the head [26], and 4 + 4 in the DHCP [24,40,41,42]. Although the paralarvae of Argonauta nodosus were reported by [45,46], there is no morphological description of them.
Cephalopod chromatophores comprise an elastic saccule containing pigment, to which a set of diagonally striped radial muscles are attached, each with its associated nerves and glial cells [47,48]. As chromatophores are neuronally controlled, an individual can select and display one particular body pattern among many at any time (e.g., at fixation) [49], hindering taxonomic work. In some groups, such as the Loliginidae family, chromatophores are considered good taxonomic characters that allow the identification of larval and juvenile stages [50,51,52]. However, our results show that in Argonauta, the observed chromatophores patterns are more variable than previously reported, including overlapping patterns. Future studies including a large number of individuals of each species are required to determine the variability of chromatophore patterns. This involves seeking constant and repeatable patterns and assessing whether they could be considered to be a taxonomic characteristic of Argonauta.
The shell of the female Argonauta is not a true mollusk shell; it is a secondary structure of calcite secreted by networks at the distal ends of the first pair of (dorsal) arms [53]. Although these shells were used in the past to delimit species of this genus, it is now known that they cannot be used for this purpose since they present a wide plasticity in their shape [6]. In particular, the shells of A. nouryi present a dramatic variation, also observed in this study, where the individuals with the type I shell (PMZPK086-20, Figure 3k), with thin ribs and without ears (the conventional shell of A. nouryi), and the type II shell (PMZPK085-20, Figure 3l), with larger keel tubercles and larger ears (characteristics historically attributed to the synonym Argonauta cornutus) [6], were grouped in the same MOTU (MOTU 04) of A. nouryi.
These results confirm the importance of implementing molecular tools such as DNA barcoding and bioinformatics tools like single-locus species delimitation methods. Furthermore, our results highlight the importance of integrating molecular data into studies involving species with different life stages, especially when extensive morphological variability hinders conventional taxonomic determination. In addition, the application of these results will allow us to practice better ecologic management of these species, especially in the case of Argonauta, whose paralarvae play the role of indicator species for the approach of warm oceanic waters toward the coast in El Niño events in the NHCS [4].
Author Contributions
Conceptualization, X.O., P.A. and J.L.R.; methodology, X.O., P.A., G.S. and J.L.R.; formal analysis, X.O., P.A. and J.L.R.; investigation, X.O. and J.L.R.; Supervision, P.A. and J.L.R.; writing—original draft preparation, X.O.; writing—review and editing, X.O., P.A., G.S. and J.L.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Consejo Nacional de Ciencia, Tecnología e Innovación Tecnológica (CONCYTEC), Peru (Círculo de Investigación 023-2016-FONDECYT), PpR-137 Oceanografia and PpR-Meta 03 Desarrollo Tecnológico, Instituto del Mar del Perú (IMARPE), Ministerio de la Producción.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We are grateful to Manuel Elías Gutierrez and Alma García Morales of the Barcode of Life Laboratory of El Colegio de la Frontera Sur (ECOSUR), Chetumal, México, for the support provided in their facilities for the training of DNA barcoding techniques.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Thiel, M.; Macaya, E.; Acuña, E.; Arntz, W.; Bastias, H.; Brokordt, K.; Camus, P.; Castilla, J.; Castro, L.; Cortés, M.; et al. The Humboldt Current System of Northern and Central Chile. In Oceanography and Marine Biology; CRC Press: Boca Raton, FL, USA, 2007; Volume 45, pp. 195–344. [Google Scholar]
- Chavez, F.P.; Bertrand, A.; Guevara-Carrasco, R.; Soler, P.; Csirke, J. The Northern Humboldt Current System: Brief History, Present Status and a View towards the Future. Prog. Ocean. 2008, 79, 95–105. [Google Scholar] [CrossRef]
- Csirke, J.; Argüelles, J.; Alegre, A.; Ayón, P.; Bouchon, M.; Castillo, G.; Castillo, R.; Cisneros, R.; Guevara-Carrasco, R.; Lau, L.; et al. Biología, Estructura Poblacional y Pesquería de Pota o Calamar Gigante (Dosidicus gigas) En El Perú. Bol. Inst. Mar. Perú 2018, 33, 302–364. [Google Scholar]
- Orosco, X.; Ayón, P. Variación de Paralarvas de Cefalópodos Asociada a Condiciones Oceanográficas En La Región Norte Del Sistema de La Corriente de Humboldt. Bol. Inst. Mar. Perú 2022, 37, 256–270. [Google Scholar] [CrossRef]
- FAO. Cephalopods of the World: An Annotated and Illustrated Catalogue of Cephalopod Species Known to Date; Jereb, P., Roper, C.F.E., Norman, M.D., Finn, J.K., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016; Volume 3, ISBN 9789251079898. [Google Scholar]
- Finn, J.K. Taxonomy and Biology of the Argonauts (Cephalopoda: Argonautidae) with Particular Reference to Australian Material. Molluscan Res. 2013, 33, 143–222. [Google Scholar] [CrossRef]
- Sasaki, M. Report on Cephalopod Collected during 1906 by The United States Bureau of Fisheries Steaivier “Albatross” in the Northwestern Pacific. Proc. U. S. Natl. Mus. 1920, 57, 163–203. [Google Scholar] [CrossRef]
- Robson, G.C. XXXIX.—Notes on the Cephalopoda.—No. 16. On the Variation, Eggs, and Ovipository Habits of Floridan Octopods. Ann. Mag. Nat. Hist. 1932, 10, 368–374. [Google Scholar] [CrossRef]
- Voss, G.L. The Cephalopoda Obtained by: The Harvard-Havana Expedition off the Coast of Cuba in 1938-39. Bull. Mar. Sci. 1955, 5, 81–115. [Google Scholar]
- Nesis, K.N. Cephalopods of the World; T.F.H. Publications: Neptune, NJ, USA, 1987. [Google Scholar]
- Sweeney, M.J.; Roper, C.F.E.; Mangold, K.M.; Clarke, M.R.; Boletzky, S.V. (Eds.) Smithsonian Contributions to Zoology “Larval” and Juvenile Cephalopods: A Manual for Their Identification; Smithsonian Institution Press: Washington, DC, USA, 1992; Volume 513. [Google Scholar]
- Young, R.E.; Harman, R.F. “Larva”, “Paralarva” and “Subadult” in Cephalopod Terminology. Malacologia 1988, 29, 201–207. [Google Scholar]
- Young, R.E.; Harman, R.F.; Hochberg, F.G. Octopodid Paralarvae from Hawaiian Waters. Veliger 1989, 32, 152–165. [Google Scholar]
- Vidal, É.A.G.; Fuentes, L.; da Silva, L.B. Defining Octopus Vulgaris Populations: A Comparative Study of the Morphology and Chromatophore Pattern of Paralarvae from Northeastern and Southwestern Atlantic. Fish. Res. 2010, 106, 199–208. [Google Scholar] [CrossRef]
- Vecchione, M. Juvenile Ecology. In Cephalopod Life Cycles; Boyle, P.R., Ed.; Academic Press: London, UK, 1987; Volume 2, p. 475. [Google Scholar]
- De Silva-Dávila, R.; Hochberg, F.G.; Lindgren, A.R.; del Carmen Franco-Gordo, M. Paralarval Development, Abundance, and Distribution of Pterygioteuthis hoylei (Cephalopoda: Oegopsida: Pyroteuthidae) in the Gulf of California, México. Molluscan Res. 2013, 33, 50–64. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Rosas-Puchuri, U.; Cañedo, R.M.; Alfaro-Shigueto, J.; Ayon, P.; Zelada-Mázmela, E.; Siccha-Ramirez, R.; Velez-Zuazo, X. DNA Barcoding in the Southeast Pacific Marine Realm: Low Coverage and Geographic Representation despite High Diversity. PLoS ONE 2020, 15, e0244323. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Nuñez-Rodriguez, D.; Britzke, R.; Siccha-Ramirez, R.; Ramírez, R. DNA Barcoding for Assessing Biodiversity. In Conservation Genetics in the Neotropics; Pedro, M.G., Jr., Ed.; Springer International Publishing: Sao Carlos, Brazil, 2023; pp. 21–45. [Google Scholar]
- Fernández-Álvarez, F.; Sánchez, P.; Villanueva, R. Morphological and Molecular Assessments of Bobtail Squids (Cephalopoda: Sepiolidae) Reveal a Hidden History of Biodiversity. Front. Mar. Sci. 2021, 7, 632261. [Google Scholar] [CrossRef]
- Ibáñez, C.M.; Fenwick, M.; Ritchie, P.A.; Carrasco, S.A.; Pardo-Gandarillas, M.C. Systematics and Phylogenetic Relationships of New Zealand Benthic Octopuses (Cephalopoda: Octopodoidea). Front. Mar. Sci. 2020, 7, 182. [Google Scholar] [CrossRef]
- Sales, J.B.d.L.; Rodrigues-Filho, L.F.d.S.; Ferreira, Y.d.S.; Carneiro, J.; Asp, N.E.; Shaw, P.W.; Haimovici, M.; Markaida, U.; Ready, J.; Schneider, H.; et al. Divergence of Cryptic Species of Doryteuthis Plei Blainville, 1823 (Loliginidae, Cephalopoda) in the Western Atlantic Ocean Is Associated with the Formation of the Caribbean Sea. Mol. Phylogenet. Evol. 2017, 106, 44–54. [Google Scholar] [CrossRef]
- Xu, R.; Lü, Y.; Tang, Y.; Chen, Z.; Xu, C.; Zhang, X.; Zheng, X. DNA Barcoding Reveals High Hidden Species Diversity of Chinese Waters in the Cephalopoda. Front. Mar. Sci. 2022, 9, 830381. [Google Scholar] [CrossRef]
- De Silva-Dávila, R. Paralarvas de Cefalópodos En El Golfo de California, México; Universidad de Guadalajara-CUCSUR: San Patricio Melaque, México, 2013. [Google Scholar]
- Zaragoza, N.; Quetglas, A.; Moreno, A. Identification Guide for Cephalopod Paralarvae from the Mediterranean Sea; International Council for the Exploration of the Sea: Copenhagen, Denmark, 2015. [Google Scholar]
- Sukhsangchan, C.; Sunthornket, P.; Phuynoi, S. Morphological Characteristics of Paralarvae of Cephalopods Found in Thai Waters. Mar. Biodivers. 2017, 47, 639–645. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Gómez, A.; Muñoz, J. Application of an Inexpensive and High-Throughput Genomic DNA Method for the Molecular Ecology of Zooplanktonic Eggs. Limnol. Ocean. Methods 2008, 6, 218–222. [Google Scholar] [CrossRef]
- Prosser, S.; Martínez-Arce, A.; Elías-Gutiérrez, M. A New Set of Primers for COI Amplification from Freshwater Microcrustaceans. Mol. Ecol. Resour. 2013, 13, 1151–1155. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Valdivia, P.; Rosas-Puchuri, U.; Valdivia, N.L. SPdel: A Pipeline to Compare and Visualize Species Delimitation Methods for Single-Locus Datasets. Mol. Ecol. Resour. 2023, 23, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble Species by Automatic Partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Barraclough, T.G. Delimiting Species Using Single-Locus Data and the Generalized Mixed Yule Coalescent Approach: A Revised Method and Evaluation on Simulated Data Sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Paredes, C.; Huamán, P.; Cardoso, F.; Vivar, R.; Vera, V. Estado Actual Del Conocimiento de Los Moluscos Acuáticos En El Perú. Rev. Peru. Biol. 1999, 6, 5–47. [Google Scholar] [CrossRef]
- Alamo, V.; Valdivieso, V. Lista Sistemática de Moluscos Marinos Del Perú, 2nd ed.; Instituto del Mar del Perú: Callao, Peru, 1997. [Google Scholar]
- Ramírez, R.; Paredes, C.; Arenas, J. Moluscos Del Perú. Rev. Biol. Trop. 2003, 51, 225–284. [Google Scholar]
- Battaglia, P.; Pedà, C.; Rizzo, C.; Stipa, M.G.; Arcadi, E.; Longo, F.; Ammendolia, G.; Cavallaro, M.; Rao, I.; Villari, A.; et al. How Rare Are Argonautoidea Octopuses in the Mediterranean? New Data from Stranding Events, Stomach Contents and Genetics. Biology 2023, 12, 420. [Google Scholar] [CrossRef]
- Alejo-Plata, M.d.C.; León-Guzmán, S.S.; Díaz-Polo, R.; Torres-Huerta, A.M. Diversity, Abundance and Frequency of Argonauts (Cephalopoda: Argonautidae) in the Diet of Pelagic Fishes of Commercial Value in Oaxaca, Mexico. Rev. Biol. Mar. Ocean. 2019, 54, 107–117. [Google Scholar] [CrossRef]
- Martínez-Soler, E.; Gómez-Gutiérrez, J.; De Silva-Dávila, R.; González-Rodríguez, E.; Aburto-Oropeza, O. Cephalopod Paralarval Species Richness, Abundance and Size Structure during the 2014-2017 Anomalous Warm Period in the Southern Gulf of California. J. Plankton Res. 2021, 43, 224–243. [Google Scholar] [CrossRef]
- Martínez, E. Variación Temporal de La Estructura de La Comunidad de Paralarvas de Cefalópodos Del Parque Nacional Cabo Pulmo, B.C.S. En Respuesta a La Variabilidad Ambiental (2014–2017); La Paz, B.C.S.: La Paz, Mexico, 2019. [Google Scholar]
- Nesis, K.N. Cephalopoda. In South Atlantic Zooplankton; Boltovskoy, D., Ed.; Backhuys Publishers: Leiden, The Netherlands, 1999. [Google Scholar]
- Vecchione, M.; Roper, C.F.E.; Sweeney, M.J.; Lu, C.C. Distribution, Relative Abundance and Developmental Morphology of Paralarval Cephalopods in the Western North Atlantic Ocean; US Government Printing Office: Seattle, WA, USA, 2001.
- Vidal, E.A.; Haimovici, M.; Hackbart, V.C. Distribution of Paralarvae and Small Juvenile Cephalopods in Relation to Primary Production in an Upwelling Area off Southern Brazil. ICES J. Mar. Sci. 2010, 67, 1346–1352. [Google Scholar] [CrossRef]
- Franco-Santos, R.M.; Iglesias, J.; Domingues, P.M.; Vidal, E.A.G. Early Beak Development in Argonauta nodosa and Octopus vulgaris (Cephalopoda: Incirrata) Paralarvae Suggests Adaptation to Different Feeding Mechanisms. Hydrobiologia 2014, 725, 69–83. [Google Scholar] [CrossRef]
- Cloney, R.A.; Florey, E. Uhrastructure of Cephalopod Chromatophore Organs. Z. Fiir Zellforsch. 1968, 89, 250–280. [Google Scholar] [CrossRef]
- Reed, C.M. The Ultrastructure and Innervation of Muscles Controlling Chromatophore Expansion in the Squid, Loligo vulgaris. Cell Tissue Res. 1995, 282, 503–512. [Google Scholar] [CrossRef]
- Messenger, J.B. Cephalopod Chromatophores: Neurobiology and Natural History. Biol. Rev. 2001, 76, 473–528. [Google Scholar] [CrossRef]
- Vecchione, M.; Lipiński, M.R. Descriptions of the Paralarvae of Two Loliginid Squids in Southern African Waters. S. Afr. J. Mar. Sci. 1995, 15, 1–7. [Google Scholar] [CrossRef]
- González, Á.F.; Rocha, F.; Guerra, A.; Bückle, L.; Guerra, A.; Rocha, E.; González, A.E.; Bückle, L.E. Embryonic Stages of the Patagonian Squid Loligo Gahi (Mollusca: Cephalopoda). The Veliger 2001, 44, 109–115. [Google Scholar]
- Barón, P.J. The Paralarvae of Two South American Sympatric Squid: Loligo Gahi and Loligo Sanpaulensis. J. Plankton Res. 2003, 25, 1347–1358. [Google Scholar] [CrossRef]
- Troschel, F.H. Bemerkungen Über Die Cephalopoden von Messina. Arch. Naturgeschichte 1857, 23, 41–76. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).