Abstract
Dendrobium officinale Kimura & Migo in the genus Dendrobium of Orchidaceae is an important medicinal plant that produces various bibenzyl and phenanthrene derivatives. In some orchids, these derivatives have been reported to increase with fungal infection. Bibenzyl biosynthesis is regulated by bibenzyl synthase (BBS). Although six genes of the BBS family have been registered from D. officinale, their gene regulation mechanisms are unclear. The infection of Dendrobium with mycorrhizal fungi also reportedly increases the expression of genes involved in biosynthesis; however, the effect of mycorrhizal fungi on bibenzyl production is unknown. The present study examined the effects of three mycorrhizal fungi isolated from D. officinale on BBS gene expression and bibenzyl production over time. One of the Tulasnellaceae operational taxonomic units induced BBS gene expression and increased two representative bibenzyls, gigantol and dendrophenol, at specific time points. Furthermore, 19 BBS sequences were cloned from 12 Dendrobium species, and a phylogenetic analysis was performed. The results indicated that repeated BBS gene duplication occurred during the evolution of the genus, and further duplication occurred after speciation. These results suggest that it is possible to optimize metabolite production by selecting suitable symbiotic fungi.
1. Introduction
Symbiotic relationships between plants and fungi play a crucial role in more than 80% of terrestrial plants. A previous study showed that plant secondary metabolites facilitate beneficial symbiotic relationships with fungi by promoting mycorrhizal formation or suppressing fungal growth [1]. For instance, Vierheiling et al. demonstrated that flavonoids were involved in the regulation of symbiosis between plants and arbuscular mycorrhizal fungi (AMF) [2]. Moreover, some plants form a unique symbiosis with specific mycorrhizal fungi. For example, the symbiosis between orchids and mycorrhizal fungi is essential for their nutrient acquisition during germination [3].
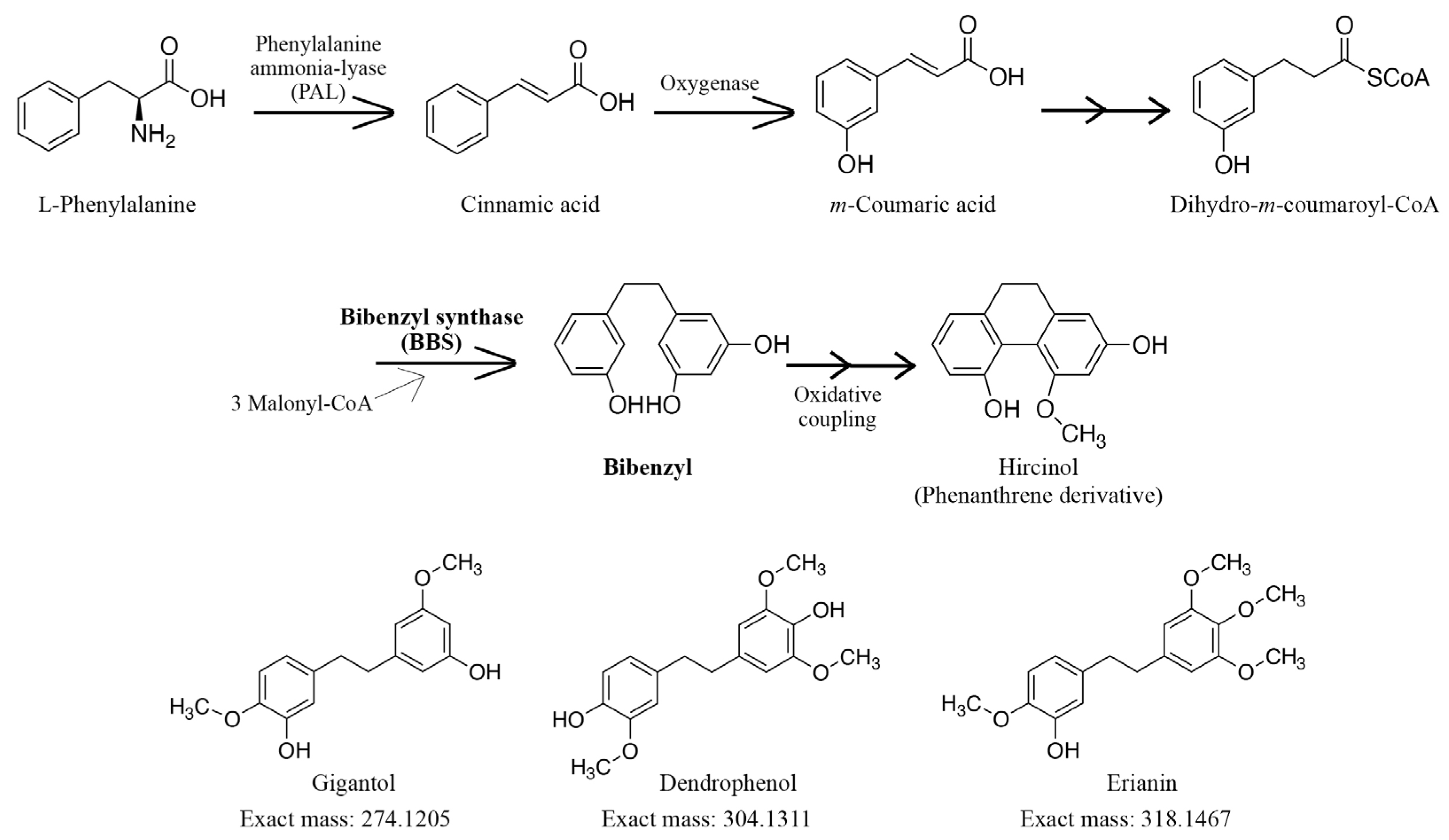
Bibenzyl and phenanthrene derivatives are secondary metabolites that have garnered attention in recent years due to their structural diversity and beneficial biological activities, such as antitumor, anti-inflammatory, and antimicrobial activities [4,5,6]. Several studies have shown that the production of these derivatives is increased after fungal infection, indicating their roles in protection against plant pathogens and inhibition of excessive fungal growth [7,8]. However, despite their biological significance, these derivatives have been isolated from a limited number of plants, including members of Orchidaceae, Ricciaceae, Dioscoreaceae, and Cannabaceae [4,9,10,11,12]. As shown in Figure 1, the biosynthesis of bibenzyl is regulated by bibenzyl synthase (BBS), which catalyzes condensation reactions with dihydro-m-coumaroyl-CoA and three malonyl-CoA as substrates, followed by cyclization reactions [13,14]. Subsequent oxidative radical coupling of bibenzyl leads to the formation of phenanthrene derivatives [13,14]. Although BBS is a type III polyketide synthase, only a few genes encoding BBS have been identified [14,15,16,17].

Figure 1.
Putative bibenzyl biosynthetic pathway. Bibenzyl synthase (BBS) catalyzes the biosynthesis of bibenzyl using dihydro-m-coumaroyl-CoA and three malonyl-CoA molecules. The subsequent oxidative coupling leads to the formation of phenanthrene derivatives.
The genus Dendrobium (Orchidaceae) comprises approximately 1450 species [18] and is divided into two major clades (Asian and Australasian) [19,20]. Various bibenzyl and phenanthrene derivatives have been isolated from Dendrobium [5,6]. The members of this genus exhibit medicinal properties [21]. Among these, D. officinale is an important medicinal plant recorded in the Pharmacopoeia of the People’s Republic of China (2020 edition), and its draft genome sequence was determined in 2016 [22]. Moreover, the metabolites and biosynthetic pathways in D. officinale have been well studied [23,24]. It has recently been reported that nitrogen uptake and NH4+ assimilation in D. officinale are affected by mycorrhizal fungi [25].
The quantity of useful bibenzyl and phenanthrene derivatives produced by Dendrobium is insufficient for their multifaceted use [26]. BBS had only been identified from D. officinale and D. sinense in the genus [17,22]. The regulation of bibenzyl biosynthesis, particularly the expression and genetic diversity of BBS genes, remains poorly understood. Therefore, it is desirable to understand the diversity of BBS and the regulation of bibenzyl biosynthesis to increase the production of beneficial compounds.
The aim of the present study was to investigate the effect of three mycorrhizal fungi isolated from D. officinale in Japan on BBS gene expression and bibenzyl production. Additionally, we cloned BBS genes from 12 Dendrobium species, presenting a genetically diverse panel, and performed phylogenetic analysis to understand the similarity and duplication of the BBS genes in this genus, and the phylogenetic relationships among the six genes of the BBS family in D. officinale.
2. Materials and Methods
2.1. Biological Materials and Culture Conditions
Seeds of D. officinale (voucher: KNY91 in Tsukuba Botanical Garden) were treated with 1% (v/v) sodium hypochlorite and sown in culture bottles containing H medium (comprising 3% (w/v) sucrose, 0.3% (w/v) Hyponex® (Hyponex Japan Corp., Osaka, Japan), 0.2% (w/v) tryptone, and 1.3% (w/v) agar, pH 6). The culture bottles were then incubated (BioTRON LH-220S) under 16 h light/8 h dark photoperiod conditions at 25 °C and an illumination intensity of 3000 lx for four months.
The Tulasnellaceae operational taxonomic units (OTUs) TU22 and TU27 and the Serendipitaceae OTU SE1B, isolated from D. officinale in Japan [27], were used for symbiotic cultures. The OTUs were defined at 97% sequence similarity for the internal transcribed spacer region. The mycorrhizal fungi were precultured in potato dextrose agar (PDA) medium comprising 2.4% (w/v) potato dextrose broth and 1.5% (w/v) agar for one week. Next, the PDA medium was hollowed out with sterilized straws and placed on oatmeal and agar medium (OMA medium: 0.25% (w/v) oatmeal and 1.5% (w/v) agar) in culture bottles. Subsequently, the four-month-old D. officinale seedlings were transferred to culture bottles containing OMA medium and grown for twelve weeks under the same conditions described above. Young seedlings grown in the absence of mycorrhizal fungi were used as the control.
2.2. RNA Extraction
RNA was extracted from young seedlings (0.2 g) grown on OMA medium for 2, 4, 8, and 12 weeks using a modified method of Liu et al. [28]. Briefly, the frozen young seedlings were added to 300 μL of 8 M guanidine hydrochloride and crushed. The RNA in the solution was precipitated by adding a 1.5-fold volume of high-salt solution (0.4 M sodium citrate and 0.6 M NaCl), then gently mixed with a 2-fold volume of isopropanol, followed by centrifugation. Next, the RNA was treated with DNase I using an RNase-free DNase Set (Qiagen, Frankfurt, Germany) according to the manufacturer’s instructions. The RNA was purified using phenol–chloroform, and the integrity of RNA was checked using a 1.0% (w/v) agarose gel. The purity and quantity of the RNA were evaluated using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at A260/230 and A260/280.
2.3. Analysis of Gene Expression by Reverse Transcription Quantitative PCR (RT-qPCR)
Reverse transcription was performed on DNase-treated RNA using a ReverTra AceTM qPCR RT Kit (TOYOBO, Osaka, Japan) according to the manufacturer’s instructions. Briefly, the RNA (500 ng) was incubated at 65 °C for 5 min and then immediately placed on ice. An RT mixture containing buffer, reverse transcriptase, and primer mix (random primer and oligo(dT) primer) was then added to it and incubated first at 37 °C for 15 min, and then at 98 °C for 5 min. Next, RT-qPCR was performed using GoTaq® qPCR Master Mix (Promega, Madison, WI, USA), according to the manufacturer’s instructions, on an AriaMx Real-Time PCR system (Agilent Technologies, Santa Clara, CA, USA). The cycling conditions were as follows: hot start at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing and extension at 60 °C for 30 s, denaturation by 1 cycle at 95 °C for 30 s, next at 65 °C for 30 s, and finally at 95 °C for 30 s. The mRNA levels of the target genes were normalized to those of Actin 1 [29] and calculated using the 2−ΔΔCt method. The primers used in this study are listed in Table 1.

Table 1.
Primers used for reverse transcription quantitative PCR (RT-qPCR).
2.4. Quantitative Analysis of Bibenzyl Using Liquid Chromatography–Mass Spectrometry (LC-MS)
The amounts of the bibenzyls gigantol, dendrophenol (moscatilin), and erianin, which have been isolated from D. officinale and reported to have antitumor activity, in the young seedlings cultured for 6 and 12 weeks were quantified. Firstly, the young seedlings were freeze-dried and then ground to powder. An amount of 5 milligrams of powder was extracted with 1.0 mL of 80% (v/v) aqueous methanol at 50 °C for 30 min under ultrasonication. After centrifugation, the supernatant was collected. The residue was again extracted with 1.0 mL of 80% (v/v) aq. methanol, as mentioned above. After centrifugation, the supernatant was collected and combined with the supernatant described above. The supernatant was passed through DISMIC®-13HP (ADVANTEC, Tokyo, Japan) and evaporated in vacuo. The dried extract was suspended in methanol and water, and the final methanol concentration was 5% (v/v). Next, the solution was purified using MonoSpin® (GL Sciences, Tokyo, Japan). The methanol fraction from MonoSpin® was evaporated and then suspended in methanol at a final concentration of 1 mg/mL (extract solution). The same volume of internal standard (0.1 mg/L ethyl 4-aminophenylacetate) was added to the extract solution and the mixture was subjected to LC-MS.
Acquity UPLC I-Class coupled with Xevo G2-S Q-TOF (Waters Co., Milford, MA, USA) was used for the LC-MS analysis. MassLynxTM 4.1 software was used for LC-MS system control and to collect spectral data (m/z 100–1000). Chromatographic separation was carried out using a gradient elution of solvent A (water containing 0.1% (v/v) formic acid) and solvent B (acetonitrile containing 0.1% (v/v) formic acid) at a flow rate of 0.25 mL/min, as follows: solvent A 95% (0→0.5 min), solvent A 95→5% (0.5→20.0 min), solvent A 5% (20.0→25.0 min), solvent A 5→95% (25.0→26.0 min), solvent A 95% (26.0→30.0 min). An ACQUITY UPLC Peptide BEH C18 Column (1.7 μm, 2.1 × 150 mm; Waters Co.) was heated to 40 °C in a column oven. The MS conditions were as follows: desolvation gas flow of 800 L/h, desolvation temperature of 450 °C, cone gas flow of 50 L/h, source temperature of 120 °C, and capillary and cone voltages of 1.5 kV and 40 V, respectively. The TOF mass spectrometer was calibrated routinely in positive electrospray ionization (ESI+) mode using a sodium formate solution. The data were centralized during acquisition using independent reference lock mass ions via the LockSpray interface to ensure mass accuracy and reproducibility. A leucine enkephalin solution (Waters Co.) was used as the lock mass, with an [M+H]+ ion of m/z 556.2771. The accurate mass and composition of the precursor and fragment ions were calculated using MassLynx 4.1 (Waters Co.) incorporated in the instrument. The amounts of gigantol (exact mass: 274.1205, m/z 275.1300), dendrophenol (exact mass: 304.1311, m/z 305.1391), and erianin (exact mass: 318.1467, m/z 319.1541) (Figure 1) in the extract were quantified using an internal standard method. Calibration curves were prepared at eight points in the range of 0.84 ng/mL to 0.13 μg/mL for gigantol, 1.25 ng/mL to 0.20 μg/mL for dendrophenol, and 0.41 ng/mL to 0.065 μg/mL for erianin.
2.5. DNA Extraction and BBS Gene Cloning
In this study, we selected 12 important medicinal plants or representative horticultural species based on phylogenetic analysis [30,31,32], morphological classification of the genus Dendrobium [18,33], and the availability of plant specimens. Genomic DNA was extracted from 0.1 g of fresh leaves using a DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions, using a template for PCR amplification. The BBS gene regions were amplified using the primer sets BBS-F (GAACARAGCRYYTWYCCDGA) and BBS-R (CRCTYGACATATTACCRTAYTCT), which were developed in the present study. The forward primer BBS-F was designed for exon 1 and the reverse primer BBS-R was designed for exon 2. These primers were designed with reference to the BBS gene sequences from D. officinale, Phalaenopsis equestris, and a Cymbidium hybrid cultivar. The amplification reactions were performed using 2 × Ampdirect® Plus and BIOTAQTM HS DNA polymerase (Shimadzu, Kyoto, Japan). The PCR profile consisted of an initial 10 min denaturation at 95 °C, 40 cycles of 30 s at 94 °C (denaturation), 1.5 min at 55 °C (annealing), and 1 min at 72 °C (extension), followed by a final extension at 72 °C for 7 min. The PCR products were confirmed by agarose gel electrophoresis, followed by TA cloning using TArget CloneTM (TOYOBO), according to the manufacturer’s instructions. DNA sequencing was performed using the T7-F (TAATACGACTCACTATAGGG) and T3-R (ATTAACCCTCACTAAAGGGAA) primers. The plant voucher information and DDBJ accession numbers (LC771154–LC771172) for all the sequences are shown in Table 2.

Table 2.
List of Dendrobium spp. used for phylogenetic analysis.
2.6. Phylogenetic Analysis of BBS Sequences
A total of 31 sequences were aligned using ClustalW in MEGA-X [34]. This dataset included 19 sequences obtained in this study, 6 genes registered as BBS or BBS-like in the D. officinale draft genome sequence [22], 4 BBS or BBS-like genes in the Phalaenopsis equestris draft genome sequence [35], and 2 BBS cDNA sequences in D. sinense (OP887149 and OP887150). The 10 DNA sequences of D. officinale and P. equestris and the 2 cDNA sequences of D. sinense were obtained from GenBank. Maximum likelihood analysis (1000 bootstrap replicates) was performed using RAxML-NG ver. 1.1.0 [36], implemented in raxmlGUI 2.0.10 [37]. The best-fitting model (GTR + G4) was selected by ModelTest-NG v0.1.7 [38], according to the Akaike information criterion.
2.7. Statistical Analysis
Statistical analysis of the RT-qPCR data was performed using IBM SPSS Statistics 29 (IBM Corp., Armonk, NY, USA). Dunnett’s multiple comparisons test was used to analyze the differences between the control (no inoculation) and the plants inoculated with each fungus, as well as the time course of gene expression after inoculation.
3. Results
3.1. Effects of Mycorrhizal Fungi on Plant Growth
The fresh weight of the young seedlings inoculated with each fungal strain (TU22, TU27, and SE1B) after 12 weeks showed an increasing trend compared to that of the seedlings without fungal inoculation (control). Among the three fungal strains, the cultivation with SE1B showed the greatest increase in fresh plant weight (Supplementary Table S1 and Figure 2).

Figure 2.
Seedlings of Dendrobium officinale at 12 weeks after inoculation with mycorrhizal fungi.
3.2. Effects of Mycorrhizal Fungi on BBS Gene Expression
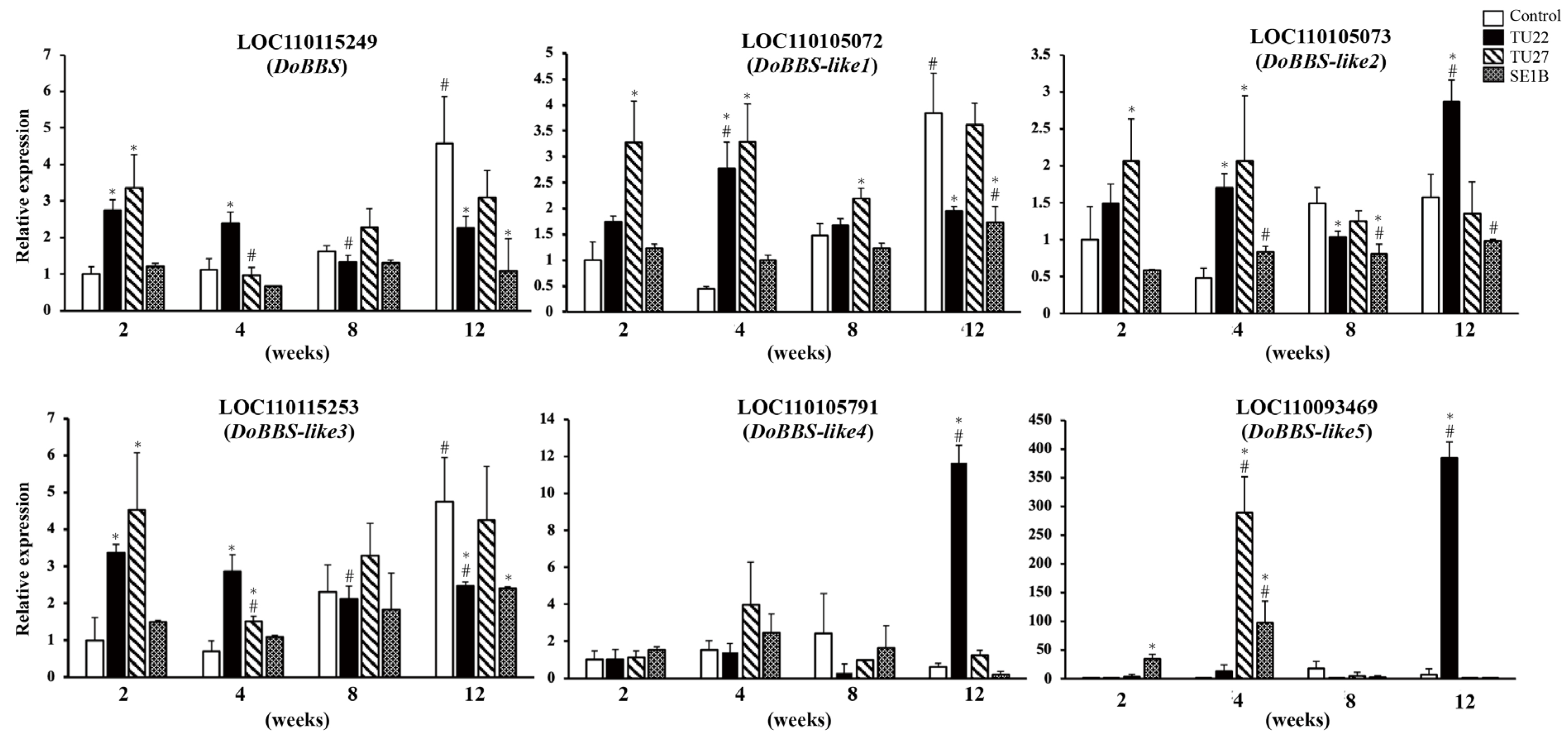
There was one BBS gene and five BBS-like genes in the D. officinale genome. To clarify the effect of the three fungal strains on the gene expression of the BBS family in D. officinale, we analyzed the expression of the six genes, namely, LOC110115249 (DoBBS), LOC110105072 (DoBBS-like1), LOC110105073 (DoBBS-like2), LOC110115253 (DoBBS-like3), LOC110105791 (DoBBS-like4), and LOC110093469 (DoBBS-like5), using RT-qPCR. The results showed that the variation in gene expression differed between the control and symbiotic seedlings (Figure 3). Additionally, the effect of symbiosis on their gene expression was dependent on the fungal strain.
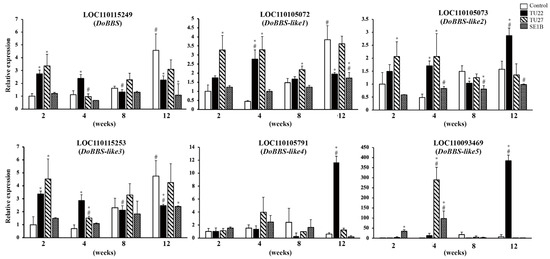
Figure 3.
Reverse transcription quantitative PCR (RT-qPCR) analysis of bibenzyl synthase (BBS) genes. The y-axis values are presented as the mean ± SD of three independent experiments, and the x-axis labels represent the duration of the symbiotic cultures (in weeks). Statistical significance was evaluated using Dunnett’s multiple comparisons test; * p < 0.05 vs. control in the same weeks, # p < 0.05 vs. 2-week sample with the same treatment.
In the control, the gene expression of DoBBS, DoBBS-like1, and DoBBS-like3 increased with plant growth, peaking at 12 weeks. In contrast, the expression of DoBBS-like2, DoBBS-like4, and DoBBS-like5 experienced no significant change. Compared to the gene expression in the control, those of DoBBS, DoBBS-like1, DoBBS-like2, and DoBBS-like3 were upregulated at two or four weeks, wherein those of DoBBS-like4 and DoBBS-like5 were significantly upregulated at twelve weeks in the TU22-inoculated seedlings. TU27 inoculation also increased the gene expression of DoBBS, DoBBS-like1, DoBBS-like2, and DoBBS-like3 in the young seedlings two or four weeks after inoculation compared to the control. Furthermore, in contrast to the effects of TU22, the expression of DoBBS-like4 showed no significant change in the TU27-inoculated seedlings. Conversely, the expression of DoBBS-like5 was upregulated transiently at four weeks in the TU27-inoculated seedlings. SE1B had a different effect on the BBS gene expression compared to TU22 and TU27. The DoBBS, DoBBS-like3, and DoBBS-like4 genes had a constant expression level in the SE1B-inoculated seedlings, whereas the expression of DoBBS-like1 and DoBBS-like2 was increased slightly at 12 weeks compared to at 2 weeks. In addition, the gene expression of DoBBS-like5 was upregulated two and four weeks after SE1B inoculation in comparison to the control.
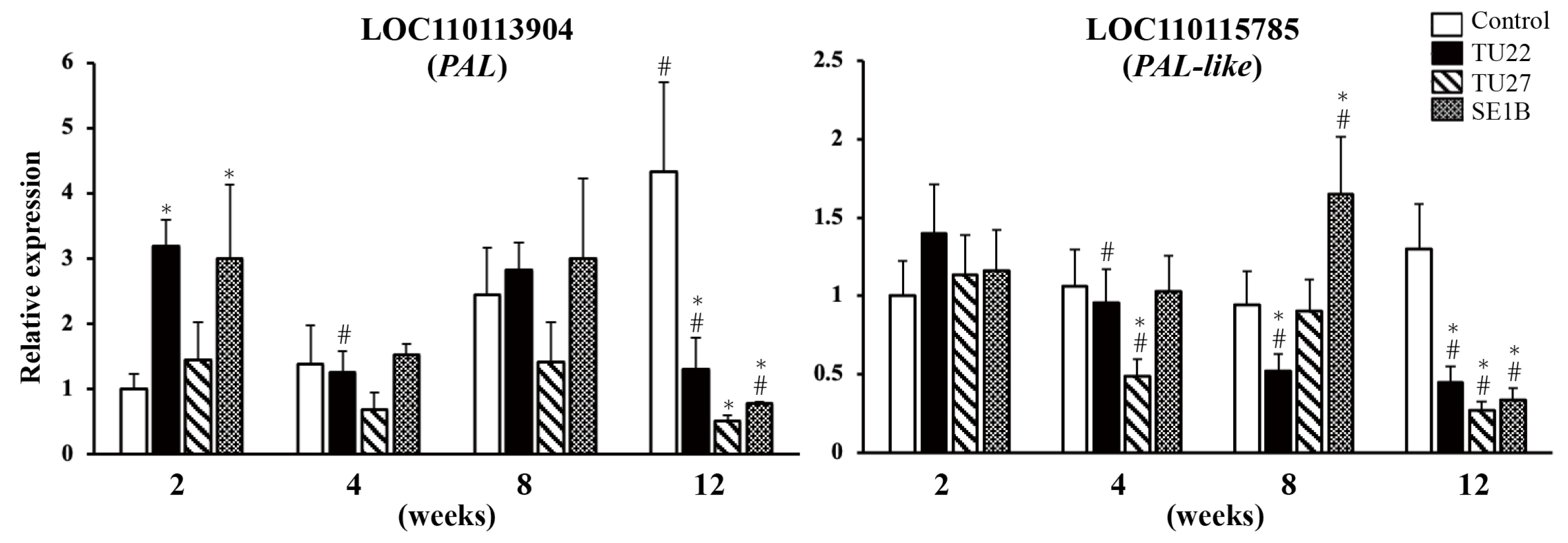
3.3. Effects of Mycorrhizal Fungi on PAL Gene Expression
Phenylalanine ammonia-lyase (PAL) is the first key enzyme in the phenylpropanoid pathway responsible for the biosynthesis of many secondary metabolites such as bibenzyl and phenanthrene (Figure 1). To clarify the effects of the three fungal strains on the gene expression of the PAL family in D. officinale, we analyzed the expression of two genes, namely, LOC110113904 (PAL) and LOC110115785 (PAL-like). In the control, the expression of PAL increased with growth (Figure 4). Symbioses with TU22 increased PAL expression at 2 weeks, which decreased at 12 weeks, compared to that in the control. In contrast, the PAL expression in the TU27-inoculated seedlings did not show significant changes at two, four, or eight weeks, and it decreased at twelve weeks, compared to that in the control. In SE1B-inoculated seedlings, PAL expression was significantly increased at 2 weeks and significantly reduced at 12 weeks compared to that in the control. The PAL-like expression did not change significantly in the control, while its expression in the TU22-inoculated seedlings decreased with growth. The PAL-like expression in the TU27-inoculated seedlings was not consistent, that is, its expression decreased at 4 and 12 weeks compared to that in the control. In the SE1B-inoculated seedlings, the PAL-like expression was increased at 8 weeks and significantly reduced at 12 weeks. These results show that the expression of PAL and PAL-like was reduced at 12 weeks in young seedlings inoculated with TU22, TU27, and SE1B.
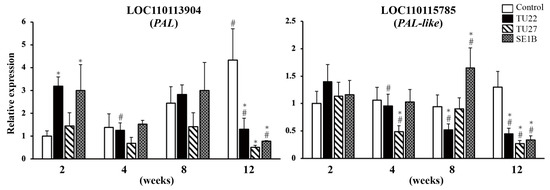
Figure 4.
RT-qPCR analysis of phenylalanine ammonia-lyase (PAL) genes. The y-axis values are presented as the mean ± SD of three independent experiments. * p < 0.05 vs. control in the same weeks, # p < 0.05 vs. 2-week sample with the same treatment.
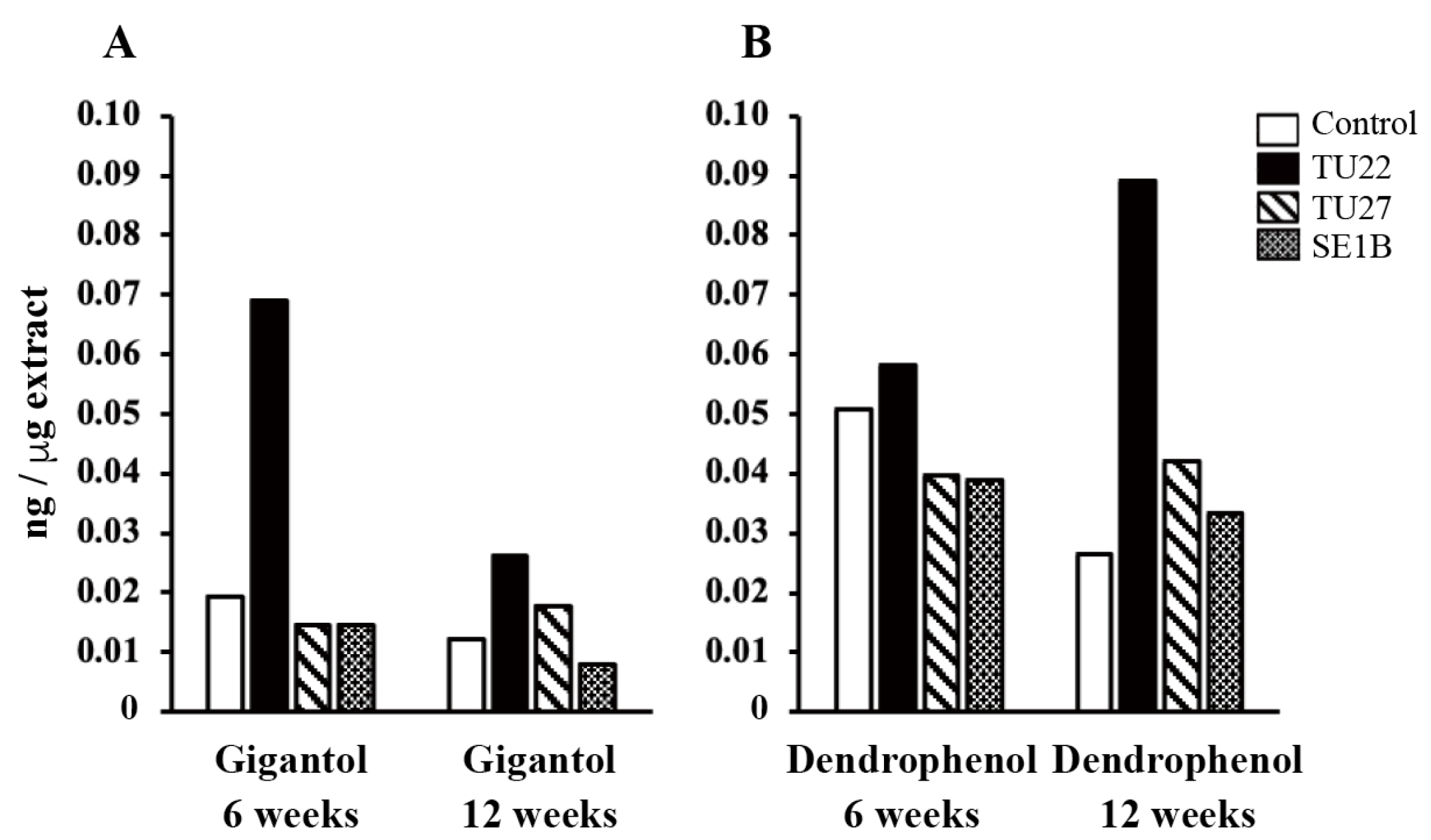
3.4. Effects of Mycorrhizal Fungi on Bibenzyl Production
The contents of gigantol, dendrophenol, and erianin (Figure 1) in the extracts from young seedlings were analyzed (Figure 5). Erianin was not detected in any sample. The TU22-inoculated seedlings had the highest gigantol content at six weeks, 3.6-fold higher than that in the control (Figure 5A). As for TU22 inoculation, the 12-week seedlings had a lower gigantol content than the 6-week seedlings. The dendrophenol content was the highest in the TU22-inoculated seedlings 12 weeks after inoculation, 3.3-fold higher than that in the control (Figure 5B). Meanwhile, no significant changes in the gigantol and dendrophenol contents were observed in the young seedlings inoculated with TU27 or SE1B (Figure 5).
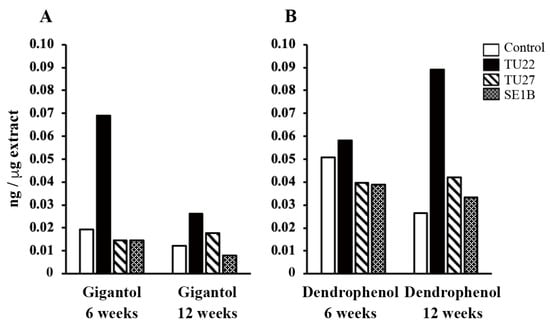
Figure 5.
Comparison of the bibenzyl contents. The y-axis represents the amounts of gigantol (A) and dendrophenol (B). The values are presented as the mean of two independent experiments.
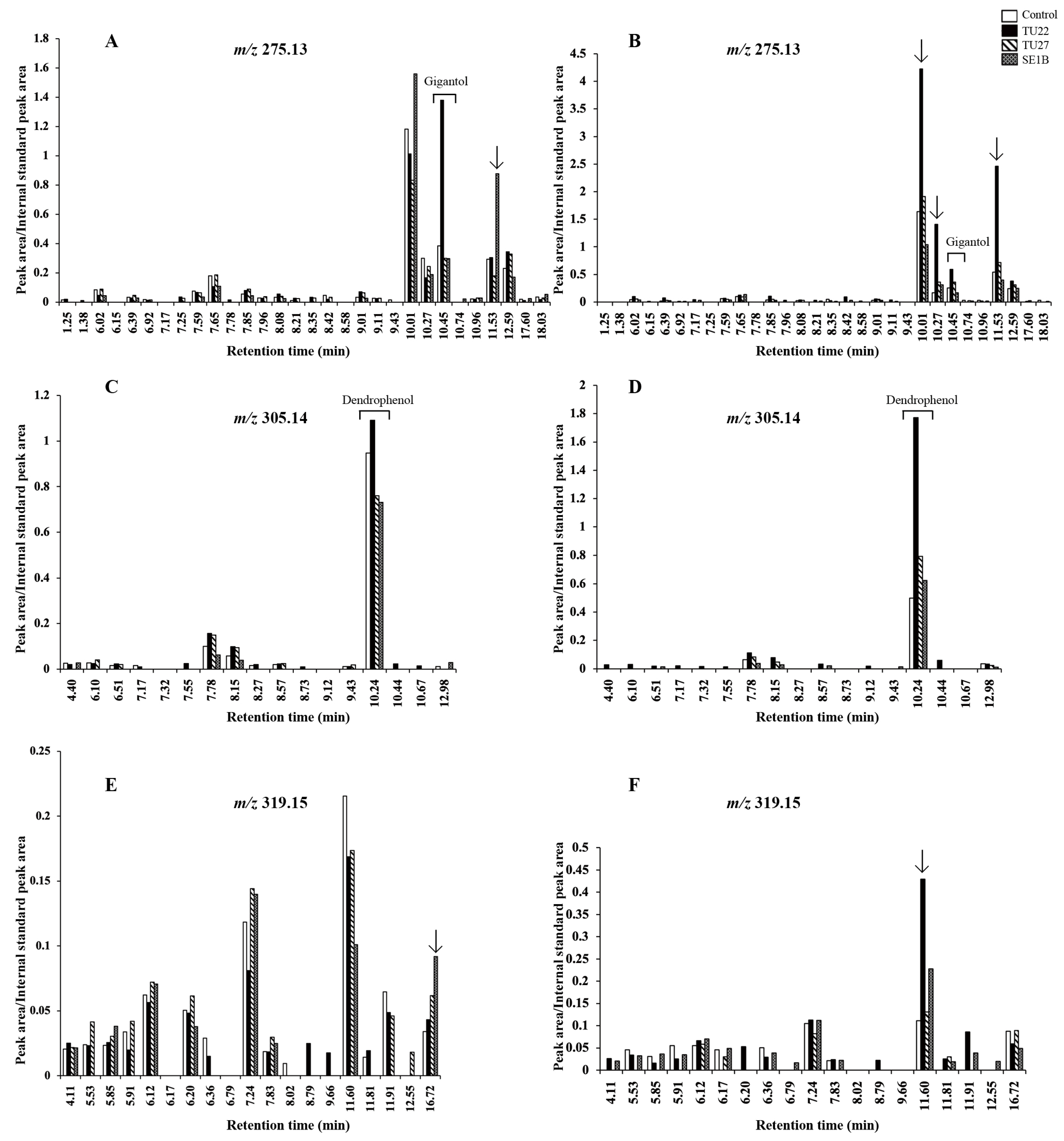
Several other metabolites, which had the same molecular weight as gigantol, dendrophenol, and erianin, were detected in the LC-MS chromatograms (Figure 6). Therefore, the chromatograms were compared to determine whether there were characteristic metabolites in the young seedlings inoculated with TU27 and SE1B. The SE1B-inoculated seedlings had higher signals at a retention time (Rt) of 11.53 min (Figure 6A) and Rt of 16.72 min (Figure 6E). Conversely, no significant metabolites specific to the TU27-inoculated seedlings were detected. In addition to gigantol and dendrophenol, several signals in the TU22-inoculated seedlings were higher than those in the other young seedlings (Rt of 10.01, 10.27, and 11.53 min in Figure 6B and Rt of 11.60 min in Figure 6F).
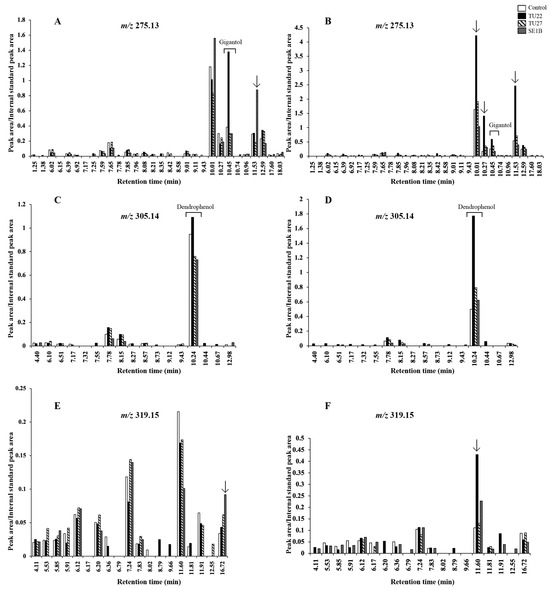
Figure 6.
Comparison of metabolite peak areas in the extracted ion chromatogram. Panels (A,B) show m/z 275.13 ± 0.02 metabolite peaks in 6- and 12-week young seedlings, respectively. The retention time (Rt) of gigantol was 10.45 min. Panels (C,D) show m/z 305.14 ± 0.02 metabolite peaks in 6- and 12-week young seedlings, respectively. The Rt of dendrophenol was 10.24 min. Panels (E,F) show m/z 319.15 ± 0.02 metabolite peaks in 6- and 12-week young seedlings, respectively. Erianin was not detected. Arrows denote values greater than 2.5-fold compared to the control.
3.5. Phylogenetic Analysis of the BBS Gene in the Genus Dendrobium
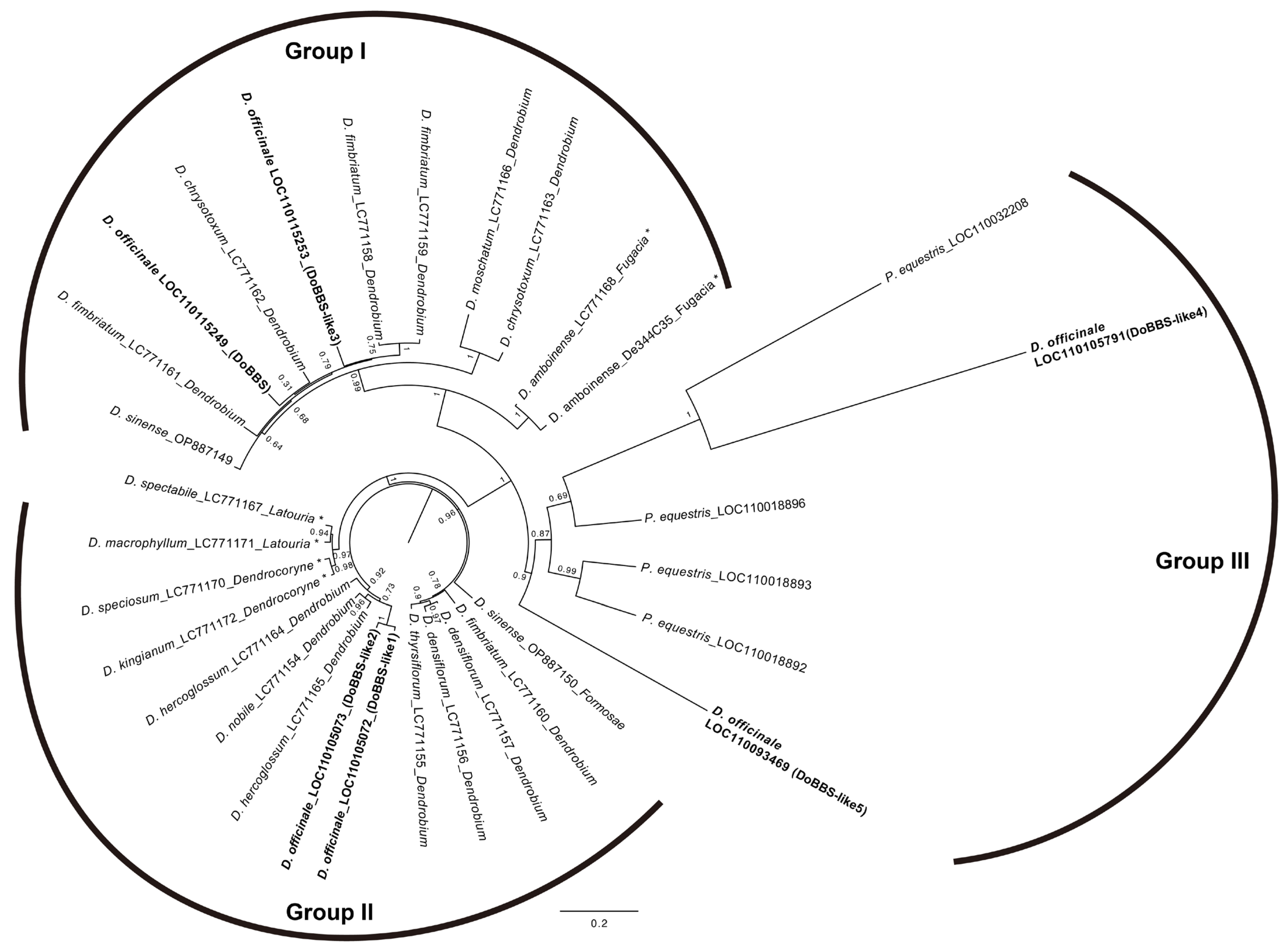
The above results indicate that the gene expression of the BBS family is affected by mycorrhizal fungi and participates in plant–fungus interactions. However, only six genes from D. officinale and two cDNA from D. sinense had been registered in the genus Dendrobium. To clarify the diversity of the BBS genes in this genus, we cloned DNA fragments containing first exon, intron and second exon from 12 Dendrobium spp. using BBS gene-specific primers. Nineteen DNA sequences homologous to the BBS gene family in D. officinale were identified (Table 2). Several homologous sequences were identified from the same individuals of D. amboinese, D. chrysotoxum, D. densiflorum, D. fimbriatum, and D. hercoglossum, respectively. For example, four homologous sequences were identified from D. fimbriatum.
Phylogenetic analysis was performed using these 19 sequences, along with 6 genes from D. officinale, 4 genes (LOC110018892, LOC110018893, LOC110018896, and LOC110032208) from Phalaenopsis equestris, and 2 cDNA sequences (OP887149 and OP887150) from D. sinense (Figure 7). The phylogenetic tree of 31 sequences showed that the BBS gene family diverged into three major groups (Groups I, II, and III). Group I included two sequences from D. amboinese in the Australasian clade, eight sequences from four species (D. chrysotoxum, D. fimbriatum, D. moschatum, and D. officinale), and one cDNA sequence from D. sinense (OP887149) in the Asian clade. Group II included four sequences from four species (D. kingianum, D. speciosum, D. macrophyllum, and D. spectabile) in the Australasian clade, nine sequences from six species (D. densiflorum, D. fimbriatum, D. hercoglossum, D. nobile, D. officinale, and D. thyrsiflorum), and one cDNA sequence (OP887150) from D. sinense in the Asian clade. The six genes of D. officinale were divided into Groups I, II, and III (bold font in Figure 7). The sequences of D. fimbriatum and D. sinense were also divided into Groups I and II.
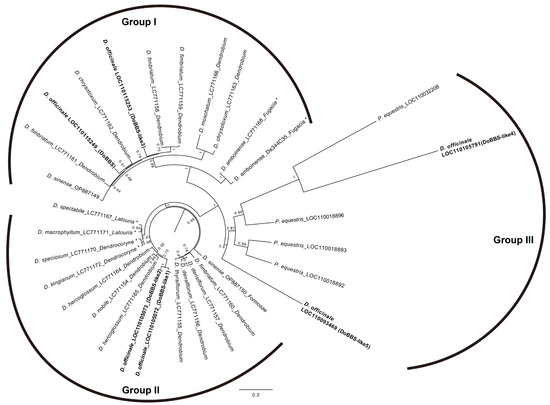
Figure 7.
Phylogenetic analysis of the bibenzyl synthase (BBS) gene family in Dendrobium spp. A phylogenetic tree was constructed using the maximum likelihood method bootstrapped with 1000 replications. Asterisks indicate species belonging to the Australasian clade.
4. Discussion
The present study focused on the symbiotic relationship between mycorrhizal fungi and D. officinale, exploring the effects of three fungi on the gene expression of the BBS family during plant growth. Our results demonstrated that each fungus had distinct effects on the BBS gene expression. Moreover, the amounts of metabolites in the young seedlings differed depending on the mycorrhizal fungus. The results indicated that the fungi selected in this study increased specific secondary metabolites through interaction with the plant. This highlights the importance of selecting suitable symbiotic fungi for optimizing metabolite production. Furthermore, phylogenetic analysis of BBS genes in Dendrobium shed light on their diversity.
Zhang et al. performed symbiotic cultures of three Dendrobium species and nine fungal strains, including the three strains (TU22, TU27, and SE1B) used in this study, to investigate the effect of fungi on plant germination and growth [39]. Consistent with their study, our results demonstrated that D. officinale seedlings inoculated with SE1B had the highest fresh and dry weights. In contrast, their study showed that the fresh and dry weights of D. moniliforme and D. okinawense increased when inoculated with other fungi [39]. Together, these results suggest that the effect of SE1B on plant growth might be species-specific.
Previous studies have investigated the effects of fungal infection on the activity of BBS. For example, the treatment of sliced rhizomes of Epipactis palustris with Rhizoctonia fungal mycelium increased BBS activity [40]. Moreover, infection with Rhizoctonia sp. and Botrytis cinerea increased the amount and enzyme activity of BBS in young sterile Phalaenopsis plants [41]. BBS gene expression was upregulated transiently when Botrytis cinerea infected Phalaenopsis sp. [14]. In this study, we investigated the variation in BBS gene expression during the growth of D. officinale inoculated with mycorrhizal fungi and clarified the following: (1) each fungus had a different effect on the BBS gene expression and (2) the variation pattern of gene expression differed for each gene.
The effects of TU22 and TU27 on the BBS gene expression were similar. However, the effects of SE1B were different. Chen et al. performed a comparative transcriptome analysis for D. officinale inoculated with members of Tulasnellaceae and Serendipitaceae and showed that the genes related to plant hormone signal transduction and phenylpropanoid biosynthesis were upregulated in plants inoculated with Tulasnellaceae fungi [42].
DoBBS, DoBBS-like1, DoBBS-like2, and DoBBS-like3 exhibited similar expression patterns in the young seedlings inoculated with TU22 and TU27, i.e., the BBS expression increased two or four weeks after inoculation compared to that in the control. In contrast, DoBBS-like4 and DoBBS-like5 expression was increased at a specific time in the TU22- or TU27-inoculated seedlings. A previous study showed that methyl jasmonate treatment increases the expression of DoBBS, DoBBS-like1, and DoBBS-like2 in D. officinale [26]. Their and our findings suggest that there is a common gene regulation mechanism for DoBBS, DoBBS-like1, and DoBBS-like2.
In bibenzyl biosynthesis, PAL works upstream of BBS (Figure 1). The gene expression of PAL, DoBBS, DoBBS-like1, and DoBBS-like3 increased with growth in the control. However, we did not find any commonality between PAL and BBS gene expression in the young seedlings inoculated with mycorrhizal fungi. One common feature in the young seedlings inoculated with each fungus is that the expression of PAL and PAL-like decreased 12 weeks after inoculation. Similar trends of reduced PAL mRNA levels and PAL activities have been demonstrated in Medicago sativa L. inoculated with AMF [43,44]. Moreover, a transient increase in PAL activity has been reported in Piper nigrum L. ‘Bragantina’ upon inoculation with AMF [45]. Overall, the findings indicate that infection with mycorrhizal fungi might have a common effect on PAL expression in a wide range of plant species. Additionally, our observations demonstrated that BBS and PAL expression varied with growth in young seedlings of D. officinale. Consistent with the results, Li et al. reported the upregulation of several genes at specific times from one to nine weeks of symbiotic culture while analyzing the expression of genes associated with the mevalonate pathway in D. nobile inoculated with Mycena sp. [46]. Collectively, a time course analysis of plant growth is warranted to reveal the effects of fungal symbiosis on gene expression in plants.
To reveal the effect of each fungus on the bibenzyl production in D. officinale, the contents of gigantol and dendrophenol were compared in young seedlings cultured for 6 and 12 weeks. The results showed that the gigantol and dendrophenol contents were higher in the TU22-inoculated seedlings than in the other seedlings. In particular, the gigantol and dendrophenol contents were the highest in the TU22-inoculated seedlings 6 and 12 weeks after inoculation, respectively. Bibenzyl production may change over time. DoBBS and DoBBS-like3 exhibited high expression in the TU22-inoculated seedlings two and four weeks after inoculation. Meanwhile, the expression of DoBBS-like4 and DoBBS-like5 was increased in the TU22-inoculated seedlings 12 weeks after inoculation. Chen et al. showed that BBS gene expression was positively related to bibenzyl contents [17]. It is necessary to determine whether such variations in BBS expression cause changes in bibenzyl production. To this end, a wide range of metabolite analyses should be performed to determine the fungus-specific effects on plants. In addition to BBS, the effects of mycorrhizal fungi on enzymes that add substituents such as hydroxyl and methyl groups to bibenzyl need to be clarified. As for erianin, we could not detect it in any samples. The production of erianin in D. officinale may require different environmental conditions to those of gigantol and dendrophenol.
Although BBS duplication and its diversity were previously unknown in Dendrobium species other than D. officinale, we isolated multiple BBS genes from D. amboinese, D. chrysotoxum, D. densiflorum, D. fimbriatum, and D. hercoglossum. Notably, D. amboinese, D. densiflorum, and D. fimbriatum had two highly similar sequences. This suggests that duplication of BBS genes occurred after speciation. In addition, six genes from D. officinale were divided into Groups I, II, and III, while four genes from D. fimbriatum were divided into Groups I and II. Therefore, it is presumed that the BBS genes of Dendrobium diverged into several lineages prior to speciation. The next step is to investigate whether Dendrobium species with BBS genes belonging to the same lineages have common metabolites and whether the same fungus has similar effects on the gene expression of BBS in the same lineages. This will provide a clue to clarify the relationship among BBS gene evolution, metabolite diversification, and mycorrhizal fungi in Dendrobium.
Considering previous studies and our results, mycorrhizal fungi with symbiosis capacity have potentially specific effects on the expression of genes involved in plant metabolite biosynthesis. Therefore, screening for suitable symbiotic fungi is essential to increase the production of target metabolites through Dendrobium–fungus interactions. In the future, a comprehensive analysis of BBS genes, clarification of the catalytic function and products of BBS, and further elucidation of downstream biosynthetic pathways are required.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16060337/s1, Table S1: fungal strains used for symbiotic culture and its effect on seedling growth.
Author Contributions
Conceptualization, T.T., A.M., Y.O.-T. and T.Y. (Tomohisa Yukawa); design of this study and writing the manuscript, H.I. and K.M.; data analysis and revision of the manuscript, M.S., M.O. and L.Z.; cultivation, K.Y., E.O., Y.T., Y.S., M.W. and T.Y. (Tadahiro Yahagi); gene expression analysis, metabolome analysis, and phylogenetic analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI grant numbers 21K06635 to T.T., 23K05914 to T. Yukawa, and 21K06306 to Y.O.-T.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Sequence data have been deposited in DDBJ under accession numbers (LC771154–LC771172).
Acknowledgments
We thank Taira Wada (Nihon University) and Dai Hirose (Nihon University) for their valuable insights into this study and Kazuhiro Suzuki (National Museum of Nature and Science) and Kentarou Komura (Nihon University) for plant cultivation.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Elhamouly, N.A.; Hewedy, O.A.; Zaitoon, A.; Miraples, A.; Elshorbagy, O.T.; Hussien, S.; El-Tahan, A.; Peng, D. The hidden power of secondary metabolites in plant-fungi interactions and sustainable phytoremediation. Front. Plant Sci. 2022, 13, 1044896. [Google Scholar] [CrossRef]
- Vierheiling, H.; Bago, B.; Albrecht, C.; Poulin, M.J.; Piché, Y. Flavonoids and arbuscular-mycorrhizal fungi. Adv. Exp. Med. Biol. 1998, 439, 9–33. [Google Scholar]
- Rasmussen, H.N.; Rasmussen, F.N. Orchid mycorrhiza: Implications of a mycophagous lifestyle. Oikos 2009, 118, 334–345. [Google Scholar] [CrossRef]
- Kovács, A.; Vasas, A.; Hohmann, J. Natural phenanthrenes and their biological activity. Phytochemistry 2008, 69, 1084–1110. [Google Scholar] [CrossRef]
- Zhai, D.; Lv, X.; Chen, J.; Peng, M.; Cai, J. Recent research progress on natural stilbenes in Dendrobium species. Molecules 2022, 27, 7233. [Google Scholar] [CrossRef]
- He, L.; Su, Q.; Bai, L.; Li, M.; Liu, J.; Liu, X.; Zhang, C.; Jiang, Z.; He, J.; Shi, J.; et al. Recent research progress on natural small molecule bibenzyls and its derivatives in Dendrobium species. Eur. J. Med. Chem. 2020, 204, 112530. [Google Scholar] [CrossRef]
- Stoessl, A.; Arditti, J. Orchid phytoalexins. In Orchid Biology. Reviews and Perspectives; Arditti, J., Ed.; Cornell University Press: New York, NY, USA, 1984; Volume 3, pp. 151–175. [Google Scholar]
- Shimura, H.; Matsuura, M.; Takada, N.; Koda, Y. An antifungal compound involved in symbiotic germination of Cypripedium macranthos var. rebunense (Orchidaceae). Phytochemistry 2007, 68, 1442–1447. [Google Scholar]
- Hashimoto, T.; Hasegawa, K.; Yamaguchi, H.; Saito, M.; Ishimoto, S. Structure and synthesis of batatasins, dormancy-inducing substances of yam bulbils. Phytochemistry 1974, 13, 2849–2852. [Google Scholar] [CrossRef]
- Kunz, S.; Becker, H. Bibenzyl derivatives from the liverwort Ricciocarpos natans. Phytochemistry 1994, 36, 675–677. [Google Scholar] [CrossRef]
- Fritzemeier, K.H.; Kindl, H.; Schlösser, E. Two different pathways leading to phenanthrenes and 9,10-dihydrophenanthrenes of the Genus Dioscorea. Z. Naturforsch 1984, 39c, 217–221. [Google Scholar]
- Crombie, L.; Mary, W.; Crombie, L. Dihydrostilbenes of Thailand Cannabis. Tetrahedron Lett. 1978, 19, 4711–4714. [Google Scholar] [CrossRef]
- Fritzemeier, K.H.; Kindl, H. 9,10-Dihydrophenanthrenes as phytoalexins of Orchidaceae. Biosynthetic studies in vitro and in vivo proving the route from L-phenylalanine to dihydro-m-coumaric acid, dihydrostilbene and dihydrophenanthrenes. Eur. J. Biochem. 1983, 133, 545–550. [Google Scholar] [CrossRef]
- Preisig-Müller, R.; Gnau, P.; Kindl, H. The inducible 9, 10-dihydrophenanthrene pathway: Characterization and expression of bibenzyl synthase and S-adenosylhomocysteine hydrolase. Arch. Biochem. Biophys. 1995, 317, 201–207. [Google Scholar] [CrossRef]
- Wang, L.; Albert, N.W.; Zhang, H.; Arathoon, S.; Boase, M.R.; Ngo, H.; Schwinn, K.E.; Davies, K.M.; Lewis, D.H. Temporal and spatial regulation of anthocyanin biosynthesis provide diverse flower colour intensities and patterning in Cymbidium orchid. Planta 2014, 240, 983–1002. [Google Scholar] [CrossRef]
- Boddington, K.F.; Soubeyrand, E.; Van Gelder, K.; Casaretto, J.A.; Perrin, C.; Forrester, T.J.B.; Parry, C.; Al-Abdul-Wahid, M.S.; Jentsch, N.G.; Magolan, J.; et al. Bibenzyl synthesis in Cannabis sativa L. Plant J. 2022, 109, 693–707. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Liang, C.; Liu, L.; Song, X.; Zhao, Y.; Wang, J.; Niu, J. Characterization of the key bibenzyl synthase in Dendrobium sinense. Int. J. Mol. Sci. 2022, 23, 6780. [Google Scholar] [CrossRef]
- Schuiteman, A.; Adams, P.B. Dendrobium. In Genera Orchidacearum; Pridgeon, A.M., Cribb, P.J., Chase, M.W., Rasmussen, F.N., Eds.; Oxford University Press: Oxford, UK, 2014; Volume 6, Epidendroideae (Part three); pp. 62–73. [Google Scholar]
- Clements, M.A. Molecular phylogenetic systematics in the Dendrobiinae (Orchidaceae), with emphasis on Dendrobium section Pedilonum. Telopea 2003, 10, 247–298. [Google Scholar] [CrossRef]
- Yukawa, T.; Kurita, S.; Nishida, M.; Hasebe, M. Phylogenetic implications of chloroplast DNA restriction site variation in subtribe Dendrobiinae (Orchidacaeae). Lindleyana 1993, 8, 211–221. [Google Scholar]
- Lawler, L.J. Ethnobotany of the Orchidaceae. In Orchid Biology. Reviews and Perspectives; Arditti, J., Ed.; Cornell University Press: New York, NY, USA, 1984; Volume 3, pp. 27–149. [Google Scholar]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Yu, Z.; Zhao, C.; Zhang, G.; Teixeira da Silva, J.A.; Duan, J. Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int. J. Mol. Sci. 2020, 30, 5419. [Google Scholar] [CrossRef]
- Si, C.; Teixeira da Silva, J.A.; He, C.; Yu, Z.; Zhao, C.; Wang, H.; Zhang, M.; Duan, J. DoRWA3 from Dendrobium officinale plays an essential role in acetylation of polysaccharides. Int. J. Mol. Sci. 2020, 28, 6250. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Zhou, L.; Li, B.; Chen, X.; Guo, S.; Wang, A.; Tian, L.; Liu, J. The plant growth-promoting fungus MF23 (Mycena sp.) increases production of Dendrobium officinale (Orchidaceae) by affecting nitrogen uptake and NH4+ assimilation. Front. Plant Sci. 2021, 12, 693561. [Google Scholar] [CrossRef] [PubMed]
- Adejobi, O.I.; Guan, J.; Yang, L.; Hu, J.M.; Yu, A.; Muraguri, S.; Liu, A. Transcriptomic analyses shed light on critical genes associated with bibenzyl biosynthesis in Dendrobium officinal. Plants 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rammitsu, K.; Tetsuka, K.; Yukawa, T.; Ogura-Tsujita, Y. Dominant Dendrobium officinale mycorrhizal partners vary among habitats and strongly induce seed germination in vitro. Front. Ecol. Evol. 2022, 10, 994641. [Google Scholar] [CrossRef]
- Liu, L.; Han, R.; Yu, N.; Zhang, W.; Xing, L.; Xie, D.; Peng, D. A method for extracting high-quality total RNA from plant rich in polysaccharides and polyphenols using Dendrobium huoshanense. PLoS ONE 2018, 13, e0196592. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Zhu, Q.; Pei, W.; Fan, J.; Liang, Y.; Cui, Y.; Lv, N.; Wang, W. Whole-transcriptome selection and evaluation of internal reference genes for expression analysis in protocorm development of Dendrobium officinale Kimura et Migo. PLoS ONE 2016, 11, e0163478. [Google Scholar] [CrossRef] [PubMed]
- Takamiya, T.; Wongsawad, P.; Sathapattayanon, A.; Tajima, N.; Suzuki, S.; Kitamura, S.; Shioda, N.; Handa, T.; Kitanaka, S.; Iijima, H.; et al. Molecular phylogenetics and character evolution of morphologically diverse groups, Dendrobium section Dendrobium and allies. AoB Plants 2014, 6, plu045. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, W.; Yang, J.; Hou, Z.; Li, C.; Niu, Z.; Zhang, B.; Xue, Q.; Liu, W.; Ding, X. Mitochondrial genome comparison and phylogenetic analysis of Dendrobium (Orchidaceae) based on whole mitogenomes. BMC Plant Biol. 2023, 23, 586. [Google Scholar] [CrossRef] [PubMed]
- Burzacka-Hinz, A.; Dudek, M.; Szlachetko, D.L. Potential use of low-copy nuclear gene Xdh at lower taxonomic levels based on phylogenetic analysis of the nominal section of Dendrobium. Acta Soc. Bot. Pol. 2024, 93, 1–13. [Google Scholar] [CrossRef]
- Wood, H.P. The Dendrobiums. Schettler, R., Ed.; A.R.G. Gantner Verlag Ruggell: Liechtenstein, Germany, 2006. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.C.; Liu, K.W.; Chen, L.J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2020, 12, 373–377. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rammitsu, K.; Kinoshita, A.; Tokuhara, K.; Yukawa, T.; Ogura-Tsujita, Y. Symbiotic culture of three closely related Dendrobium species reveals a growth bottleneck and differences in mycorrhizal specificity at early developmental stages. Diversity 2022, 14, 1119. [Google Scholar] [CrossRef]
- Gehlert, R.; Kindl, H. Induced formation of dihydrophenanthrenes and bibenzyl synthase upon destruction of orchid mycorrhiza. Phytochemistry 1991, 30, 457–460. [Google Scholar] [CrossRef]
- Reinecke, T.; Kindl, H. Inducible enzymes of the 9,10-dihydro-phenanthrene pathway. Sterile orchid plants responding to fungal infection. Mol. Plant-Microbe Interact. 1994, 7, 449–454. [Google Scholar] [CrossRef]
- Chen, J.; Tang, Y.; Kohler, A.; Lebreton, A.; Xing, Y.; Zhou, D.; Li, Y.; Martin, F.M.; Guo, S. Comparative transcriptomics analysis of the symbiotic germination of D. officinale (Orchidaceae) with emphasis on plant cell wall modification and cell wall-degrading enzymes. Front. Plant Sci. 2022, 13, 880600. [Google Scholar] [CrossRef]
- Volpin, H.; Elkind, Y.; Okon, Y.; Kapulnik, Y. A vesicular arbuscular mycorrhizal fungus (Glomus intraradix) induces a defense response in Alfalfa roots. Plant Physiol. 1994, 104, 683–689. [Google Scholar] [CrossRef]
- Volpin, H.; Phillips, D.A.; Okon, Y.; Kapulnik, Y. Suppression of an isoflavonoid phytoalexin defense response in mycorrhizal Alfalfa roots. Plant Physiol. 1995, 108, 1449–1454. [Google Scholar] [CrossRef]
- da Trindade, R.; Almeida, L.; Xavier, L.; Lins, A.L.; Andrade, E.H.; Maia, J.G.; Mello, A.; Setzer, W.N.; Ramos, A.; da Silva, J.K. Arbuscular mycorrhizal fungi colonization promotes changes in the volatile compounds and enzymatic activity of lipoxygenase and phenylalanine ammonia lyase in Piper nigrum L. ‘Bragantina’. Plants 2019, 8, 442. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, G.; Li, B.; Guo, S.X. Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile Lindl. infected with mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 316. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).