The Complete Mitogenomes of Two Species of Snakehead Fish (Perciformes: Channidae): Genome Characterization and Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Identification
2.2. Sequence Analysis and Assembly and Mitochondrial Genome Annotation
2.3. Phylogenetic Analyses
3. Results and Discussion
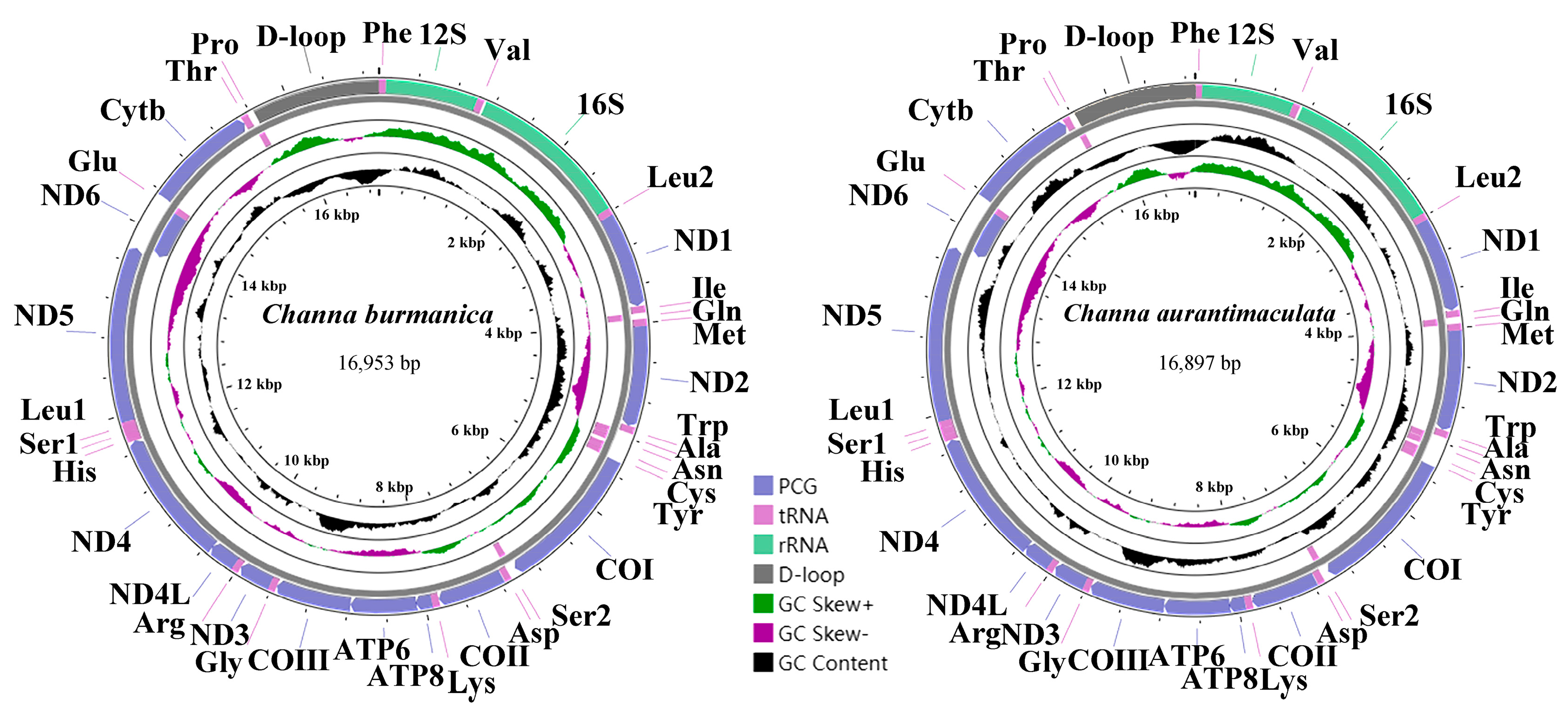
3.1. Mitogenome Organization and Structure
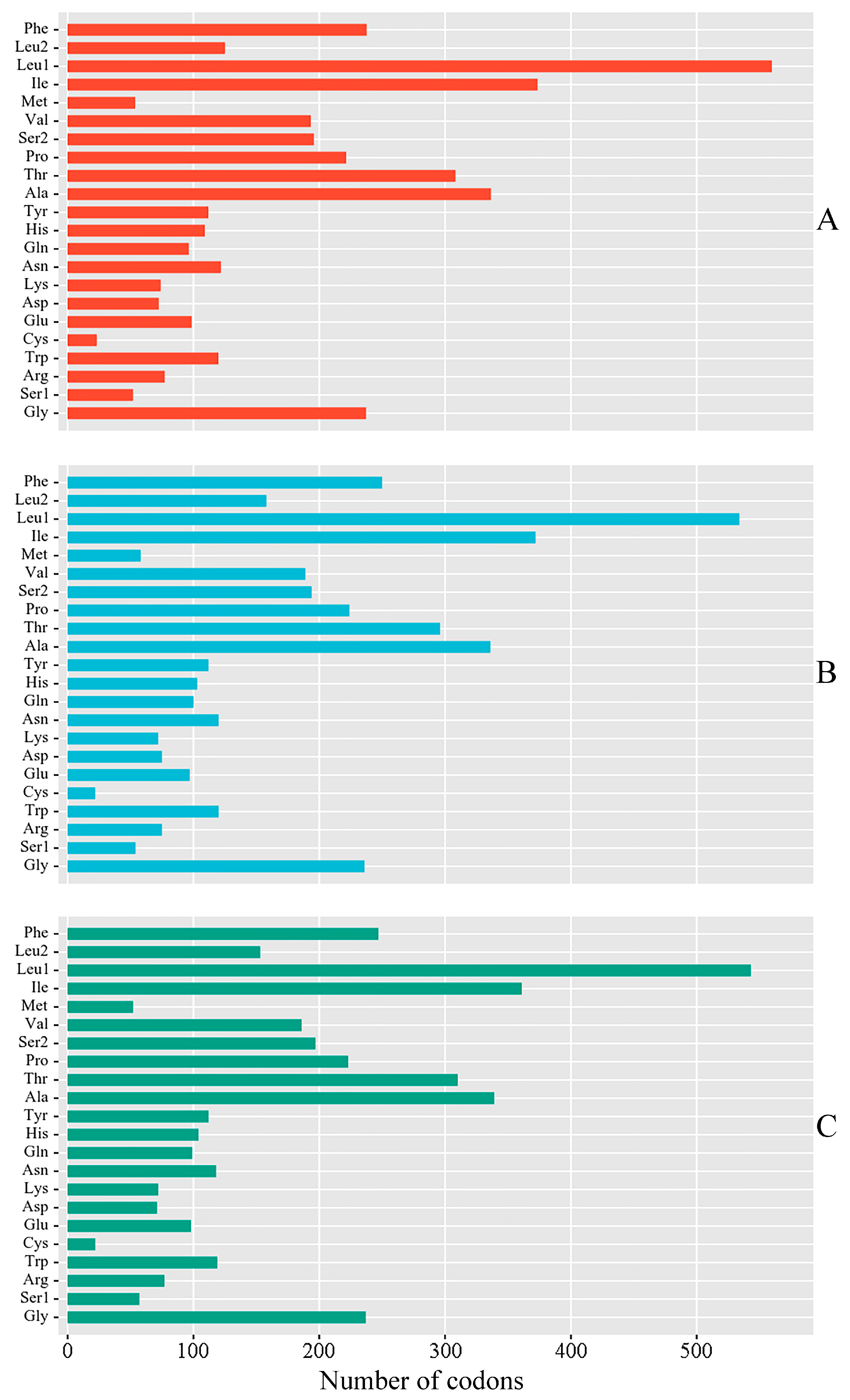
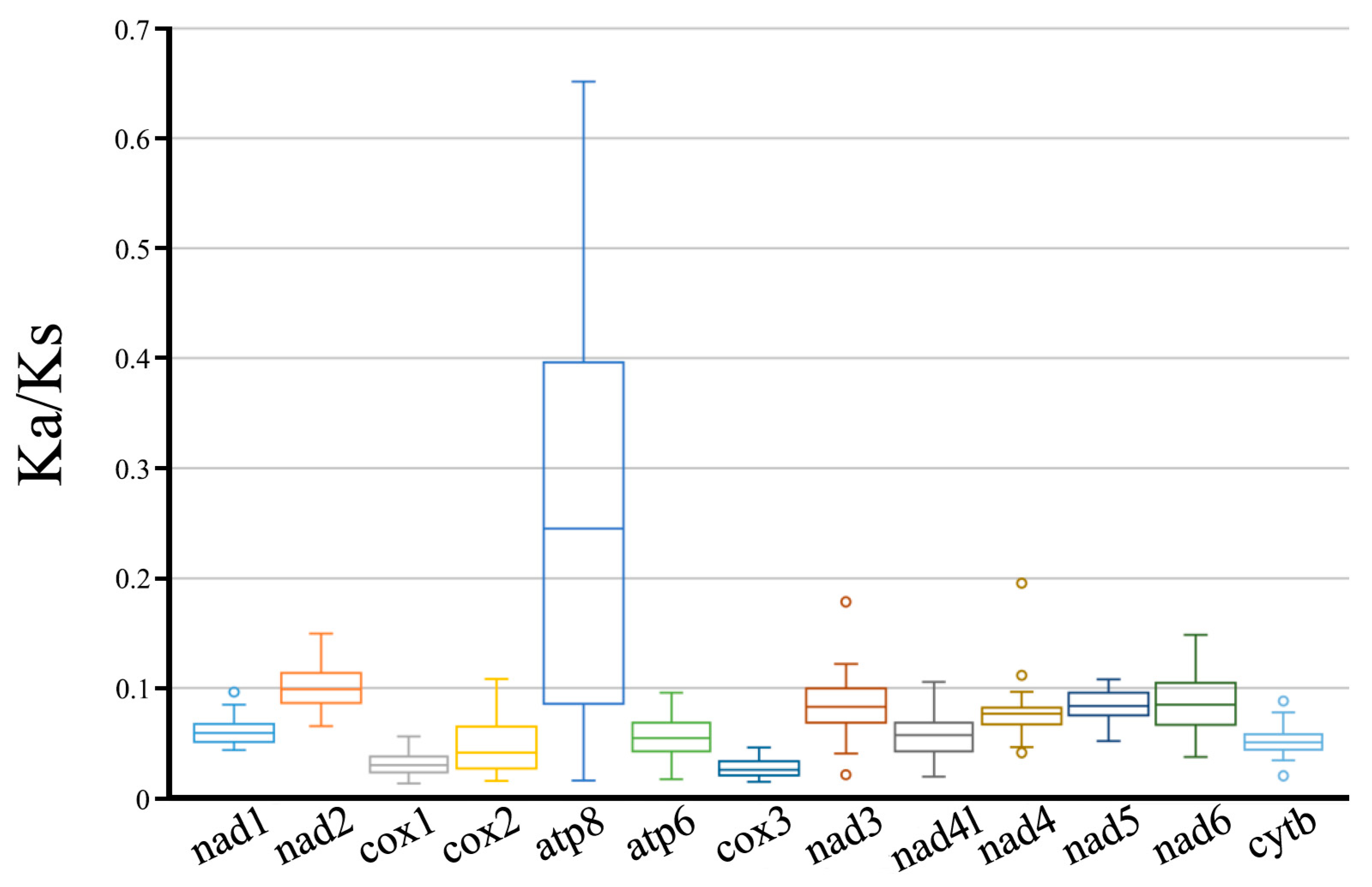
3.2. PCGs and Codon Usage
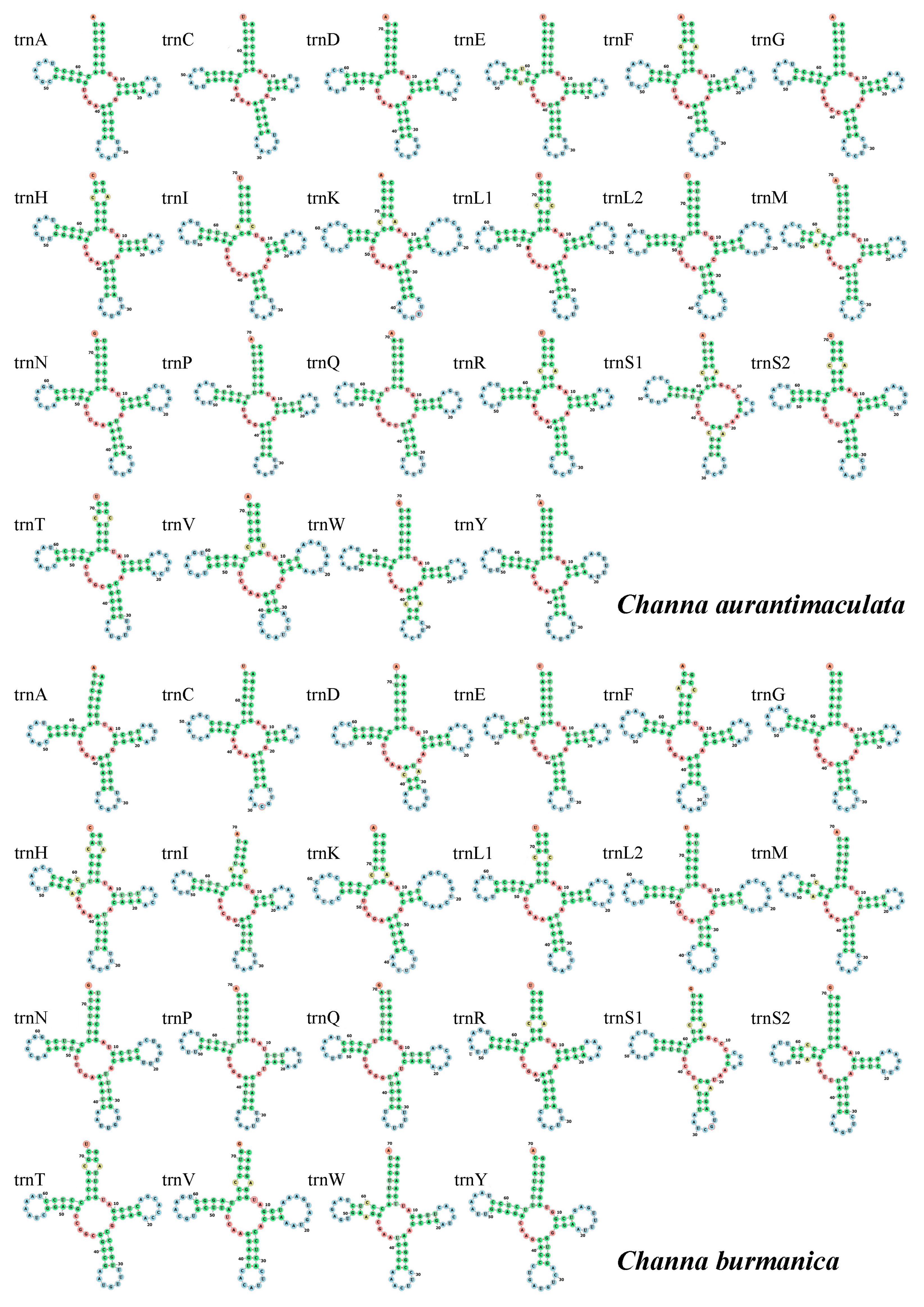
3.3. Transfer RNAs, Ribosomal RNAs, and Displacement Loop Region
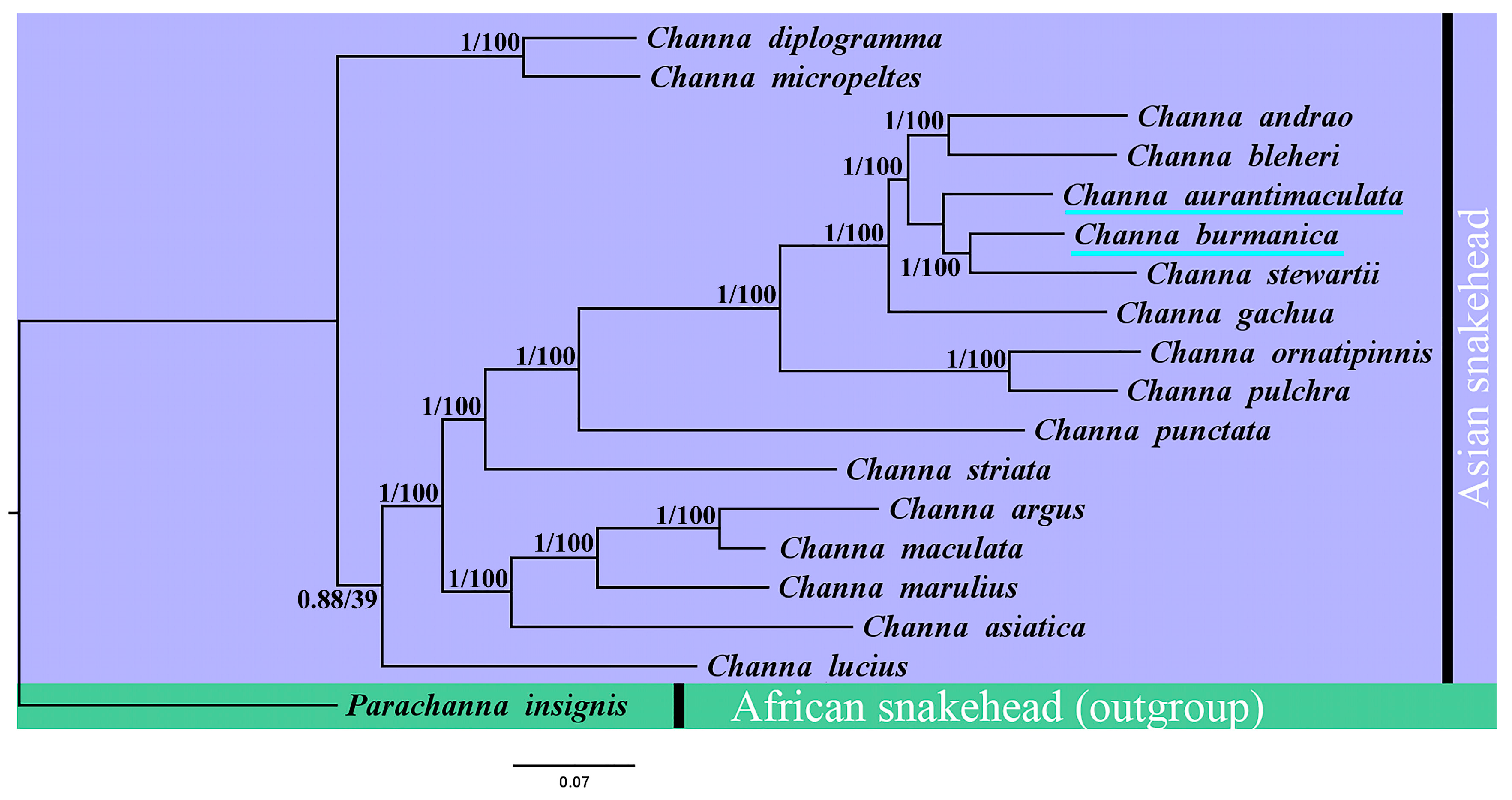
3.4. Phylogenetic Relationships within the Genus Channa
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Betancur, R.R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Ortí, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef]
- Bhat, A.A.; Haniffa, M.A.; Divya, P.R.; Gopalakrishnan, A.; Milton, M.J.; Kumar, R.; Paray, B.A. Molecular characterization of eight Indian Snakehead species (Pisces: Perciformes channidae) using RAPD markers. Mol. Biol. Rep. 2012, 39, 4267–4273. [Google Scholar] [CrossRef]
- Duan, T.; Shi, C.; Zhou, J.; Lv, X.; Li, Y.; Luo, Y. How does the snakehead Channa argus survive in air? The combined roles of the suprabranchial chamber and physiological regulations during aerial respiration. Biol. Open 2018, 7, bio029223. [Google Scholar] [CrossRef]
- Adamson, E.A.S.; Britz, R. The snakehead fish Channa aurolineata is a valid species (Teleostei: Channidae) distinct from Channa marulius. Zootaxa 2018, 4514, 542–552. [Google Scholar] [CrossRef]
- Praveenraj, J.; Knight, J.D.M.; Kiruba-Sankar, R.; Halalludin, B.; Raymond, J.J.A.; Thakur, V.R. Channa royi (Teleostei: Channidae): A new species of snakehead from Andaman Islands, India. Indian J. Fish. 2018, 65, 1–14. [Google Scholar] [CrossRef]
- Britz, R.; Anoop, V.; Dahanukar, N.; Raghavan, R. The subterranean Aenigmachanna gollum, a new genus and species of snakehead (Teleostei: Channidae) from Kerala, South India. Zootaxa 2019, 4603, 377–388. [Google Scholar] [CrossRef]
- Britz, R.; Dahanukar, N.; Anoop, V.; Ali, A. Channa rara, a new species of snakehead fish from the Western Ghats region of Maharashtra, India (Teleostei: Labyrinthici: Channidae). Zootaxa 2019, 4683, 589–600. [Google Scholar] [CrossRef]
- Praveenraj, J.; Uma, A.; Moulitharan, N.; Kannan, R. Channa brunnea, a new species of snakehead (Teleostei: Channidae) from West Bengal, India. Zootaxa 2019, 4624, 59–70. [Google Scholar] [CrossRef]
- Praveenraj, J.; Uma, A.; Moulitharan, N.; Singh, S.G. A new species of dwarf channa (Teleostei: Channidae) from Meghalaya, Northeast India. Copeia 2019, 107, 61–71. [Google Scholar] [CrossRef]
- Praveenraj, J.; Thackeray, T.; Singh, S.G.; Uma, A.; Moulitharan, N.; Mukhim, B.K. A New Species of Snakehead (Teleostei: Channidae) from East Khasi Hills, Meghalaya, Northeastern India. Copeia 2020, 108, 938–947. [Google Scholar] [CrossRef]
- Conte-Grand, C.; Britz, R.; Dahanukar, N.; Raghavan, R.; Pethiyagoda, R.; Tan, H.H.; Hadiaty, R.K.; Yaakob, N.S.; Rüber, L. Barcoding snakeheads (Teleostei, Channidae) revisited: Discovering greater species diversity and resolving perpetuated taxonomic confusions. PLoS ONE 2017, 12, e0184017. [Google Scholar] [CrossRef]
- Rüber, L.; Tan, H.H.; Britz, R. Snakehead (Teleostei: Channidae) diversity and the Eastern Himalaya biodiversity hotspot. J. Zoöl. Syst. Evol. Res. 2020, 58, 356–386. [Google Scholar] [CrossRef]
- Sudasinghe, H.; Ranasinghe, R.H.T.; Pethiyagoda, R.; Meegaskumbura, M.; Britz, R. Genetic diversity and morphological stasis in the Ceylon Snakehead, Channa orientalis(Teleostei: Channidae). Ichthyol. Res. 2020, 68, 67–80. [Google Scholar] [CrossRef]
- Landis, A.M.G.; Lapointe, N.W. First record of a northern snakehead (Channa argus cantor) nest in North America. Northeast. Nat. 2010, 17, 325–332. [Google Scholar] [CrossRef]
- Liu, M.; Yin, J.; Han, J.; Ren, J.; Yang, S. Channa argus BMH from Baima Hu Lake: Sequencing and phylogenetic analysis of the mitochondrial genome. Mitochondrial DNA Part B 2020, 5, 2413–2415. [Google Scholar] [CrossRef]
- Serrao, N.R.; Steinke, D.; Hanner, R.H. Calibrating snakehead diversity with DNA barcodes: Expanding taxonomic coverage to enable identification of potential and established invasive species. PLoS ONE 2014, 9, e99546. [Google Scholar] [CrossRef]
- Yu, R.; Wu, Q.; Li, F.; Zhan, A.; Zhou, J.; Li, S. Risk Screening of invasive aquatic species and a survey of fish diversity using environmental dna metabarcoding analysis in shanghai. Diversity 2024, 16, 29. [Google Scholar] [CrossRef]
- Herborg, L.-M.; Mandrak, N.E.; Cudmore, B.C.; MacIsaac, H.J. Comparative distribution and invasion risk of snakehead (Channidae) and Asian carp (Cyprinidae) species in North America. Can. J. Fish. Aquat. Sci. 2007, 64, 1723–1735. [Google Scholar] [CrossRef]
- Prosdocimi, F.; de Carvalho, D.C.; de Almeida, R.N.; Beheregaray, L.B. The complete mitochondrial genome of two recently derived species of the fish genus Nannoperca (Perciformes, Percichthyidae). Mol. Biol. Rep. 2012, 39, 2767–2772. [Google Scholar] [CrossRef] [PubMed]
- Paz, F.P.C.; Batista, J.d.S.; Porto, J.I.R. DNA Barcodes of Rosy Tetras and Allied Species (Characiformes: Characidae: Hyphessobrycon) from the Brazilian Amazon Basin. PLoS ONE 2014, 9, e98603. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A Mitochondrial Genome Database of Fish with an Accurate and Automatic Annotation Pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Wang, J.; Xu, W.; Liu, Y.; Bai, Y.; Liu, H. Comparative mitochondrial genomics and phylogenetics for species of the snakehead genus Channa Scopoli, 1777 (Perciformes: Channidae). Gene 2023, 857, 147186. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Pett, W. Animal mitochondrial DNA as we do not know it: Mt-Genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.; Li, M.; Li, Z.; Huang, J.; Chen, W.; Sun, J.; Liu, L.; Zou, K. Comparative analysis of complete mitochondrial genomes of three Gerres fishes (Perciformes: Gerreidae) and primary exploration of their evolution history. Int. J. Mol. Sci. 2020, 21, 1874. [Google Scholar] [CrossRef]
- Cooper, A.; Lalueza-Fox, C.; Anderson, S.G.; Rambaut, A.; Austin, J.J.; Ward, R. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 2001, 409, 704–707. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, T.; Lu, W. Complete mitochondrial DNA sequence of the endangered fish (Bahaba taipingensis): Mitogenome characterization and phylogenetic implications. ZooKeys 2015, 546, 181–195. [Google Scholar] [CrossRef]
- Li, R.; Wang, G.; Wen, Z.-Y.; Zou, Y.-C.; Qin, C.-J.; Luo, Y.; Wang, J.; Chen, G.-H. Complete mitochondrial genome of a kind of snakehead fish Channa siamensis and its phylogenetic consideration. Genes Genom. 2019, 41, 147–157. [Google Scholar] [CrossRef]
- Hughes, A.L.; Nei, M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 1988, 335, 167–170. [Google Scholar] [CrossRef]
- Hassanin, A.; Léger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Boil. Sci. 2003, 270 (Suppl. S1), S96–S99. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Yu, J.; Liu, J.; Li, C.; Wu, W.; Feng, F.; Wang, Q.; Ying, X.; Qi, D.; Qi, G. Characterization of the complete mitochondrial genome of Otus lettia: Exploring the mitochondrial evolution and phylogeny of owls (Strigiformes). Mitochondrial DNA Part B 2021, 6, 3443–3451. [Google Scholar] [CrossRef]
- Sun, C.-H.; Liu, H.-Y.; Xu, N.; Zhang, X.-L.; Zhang, Q.; Han, B.-P. Mitochondrial genome structures and phylogenetic analyses of two tropical characidae fishes. Front. Genet. 2021, 12, 627402. [Google Scholar] [CrossRef]
- Britz, R. Channa ornatipinnis and C. pulchra, two new species of dwarf snakeheads from Myanmar (Teleostei: Channidae). Ichthyol. Explor. Freshw. 2008, 18, 335–344. [Google Scholar]
- Li, X.; Musikasinthorn, P.; Kumazawa, Y. Molecular phylogenetic analyses of snakeheads (Perciformes: Channidae) using mitochondrial DNA sequences. Ichthyol. Res. 2006, 53, 148–159. [Google Scholar] [CrossRef]


| Family | Species | Accession No. | Length (bp) |
|---|---|---|---|
| Channa | Channa diplogramma | MG986721.1 | 16,571 |
| Channa lucius | MF804538.1 | 16,570 | |
| Channa marulius | KF420268.1 | 16,569 | |
| Channa micropeltes | KX129904.1 | 16,567 | |
| Channa gachua | MK371068.1 | 16,561 | |
| Channa argus | MG751766.1 | 16,558 | |
| Channa maculata | KC823606.1 | 16,559 | |
| Channa asiatica | KJ930190.1 | 16,550 | |
| Channa striata | KX177965.1 | 16,509 | |
| Channa punctata | MK007075.1 | 16,409 | |
| Channa ornatipinnis | OP271694.1 | 16,866 | |
| Channa pulchra | OP271693.1 | 16,895 | |
| Channa stewartii | OP402840.1 | 16,765 | |
| Channa andrao | OP402839.1 | 16,729 | |
| Channa bleheri | OP186040.1 | 16,714 | |
| Channa burmanica | OP954106.1 | 16,953 | |
| Channa aurantimaculata | OQ134162.1 | 16,897 | |
| Parachanna | Parachanna insignis | AP006042.1 | 16,607 |
| Gene | Location | Intergenic Nucleotides | Size | Codon | Stand | ||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| tRNA-Phe | 1/1 | 69/69 | 0/0 | 69/69 | H | ||
| 12S rRNA | 70/70 | 1017/1017 | 0/0 | 948/948 | H | ||
| tRNA-Val | 1018/1018 | 1089/1089 | 0/0 | 72/72 | H | ||
| 16S rRNA | 1113/1113 | 2768/2765 | 23/23 | 1656/1653 | H | ||
| tRNA-Leu2 | 2769/2766 | 2842/2839 | 0/0 | 74/74 | H | ||
| ND1 | 2843/2840 | 3817/3814 | 0/0 | 975/975 | ATG/ATG | TAA/TAA | H |
| tRNA-Ile | 3822/3819 | 3891/3888 | 4/4 | 70/70 | H | ||
| tRNA-Gln | 3889/3888 | 3959/3958 | −3/−1 | 71/71 | L | ||
| tRNA-Met | 3959/3958 | 4028/4027 | −1/−1 | 70/70 | H | ||
| ND2 | 4029/4028 | 5075/5074 | 0/0 | 1047/1047 | ATG/ATG | TAA/TAA | H |
| tRNA-Trp | 5075/5074 | 5144/5143 | −1/−1 | 70/70 | H | ||
| tRNA-Ala | 5146/5145 | 5214/5213 | 1/1 | 69/69 | L | ||
| tRNA-Asn | 5216/5215 | 5288/5287 | 1/1 | 73/73 | L | ||
| tRNA-Cys | 5327/5326 | 5391/5390 | 38/38 | 65/65 | L | ||
| tRNA-Tyr | 5392/5391 | 5461/5460 | 0/0 | 70/70 | L | ||
| COI | 5463/5462 | 7004/7003 | 1/1 | 1542/1542 | GTG/GTG | TAA/TAA | H |
| tRNA-Ser2 | 7013/7012 | 7083/7082 | 8/8 | 71/71 | L | ||
| tRNA-Asp | 7086/7086 | 7157/7157 | 2/2 | 72/72 | H | ||
| COII | 7165/7165 | 7855/7855 | 7/7 | 691/691 | ATG/ATG | T/T | H |
| tRNA-Lys | 7856/7856 | 7930/7930 | 0/0 | 75/75 | H | ||
| ATP8 | 7932/7932 | 8099/8099 | 1/1 | 168/168 | ATG/ATG | TAA/TAA | H |
| ATP6 | 8090/8090 | 8773/8773 | −10/−10 | 684/684 | ATG/ATG | TAA/TAA | H |
| COIII | 8773/8773 | 9558/9558 | −1/−1 | 786/786 | ATG/ATG | TAA/TAA | H |
| tRNA-Gly | 9558/9558 | 9626/9626 | −1/−1 | 69/69 | H | ||
| ND3 | 9627/9627 | 9975/9975 | 0/0 | 349/349 | ATA/ATA | T/T | H |
| tRNA-Arg | 9976/9976 | 10,044/10,044 | 0/0 | 69/69 | H | ||
| ND4L | 10,045/10,045 | 10,341/10,341 | 0/0 | 297/297 | ATG/ATG | TAA/TAA | H |
| ND4 | 10,335/10,335 | 11,715/11,715 | −7/−7 | 1381/1381 | ATG/ATG | T/T | H |
| tRNA-His | 11,716/11,716 | 11,784/11,784 | 0/0 | 69/69 | H | ||
| tRNA-Ser1 | 11,785/11,785 | 11,852/11,852 | 0/0 | 68/68 | H | ||
| tRNA-Leu1 | 11,855/11,855 | 11,927/11,927 | 2/2 | 73/73 | H | ||
| ND5 | 11,928/11,928 | 13,766/13,766 | 0/0 | 1839/1839 | ATG/ATG | TAA/TAA | H |
| ND6 | 13,763/13,763 | 14,284/14,284 | −4/−4 | 522/522 | ATG/ATG | TAG/TAG | L |
| tRNA-Glu | 14,285/14,285 | 14,353/14,353 | 0/0 | 69/69 | L | ||
| Cyt b | 14,358/14,358 | 15,498/15,498 | 4/4 | 1141/1141 | ATG/ATG | T/T | H |
| tRNA-Thr | 15,499/15,499 | 15,570/15,570 | 0/0 | 72/72 | H | ||
| tRNA-Pro | 15,570/15,570 | 15,639/15,639 | −1/−1 | 70/70 | L | ||
| D-loop | 15,640/15,640 | 16,953/16,897 | 0/0 | 1314/1258 | |||
| Species | Whole Mitogenome | AT Skew | GC Skew | PCGs | tRNAs | rRNAs | D-Loop | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | |||
| C. burmanica | 16,953 | 53.67 | 0.030 | −0.312 | 11,422 | 52.50 | 1550 | 55.16 | 2604 | 52.69 | 1314 | 65.14 |
| C. aurantimaculata | 16,897 | 55.68 | 0.024 | −0.315 | 11,420 | 54.40 | 1550 | 56.97 | 2601 | 53.94 | 1258 | 70.03 |
| C. diplogramma | 16,571 | 53.53 | 0.102 | −0.337 | 11,430 | 52.35 | 1557 | 54.66 | 2627 | 54.05 | 918 | 65.14 |
| C. lucius | 16,570 | 54.00 | 0.084 | −0.335 | 11,429 | 52.70 | 1560 | 55.13 | 2633 | 54.58 | 909 | 67.55 |
| C. marulius | 16,569 | 52.75 | 0.077 | −0.325 | 11,430 | 51.56 | 1556 | 55.21 | 2631 | 54.24 | 915 | 60.00 |
| C. micropeltes | 16,567 | 53.34 | 0.102 | −0.333 | 11,430 | 52.32 | 1558 | 55.13 | 2602 | 53.65 | 921 | 63.52 |
| C. gachua | 16,561 | 55.16 | 0.028 | −0.306 | 11,429 | 54.72 | 1548 | 55.36 | 2629 | 54.43 | 908 | 63.66 |
| C. argus | 16,558 | 51.43 | 0.059 | −0.301 | 11,426 | 49.91 | 1556 | 54.37 | 2632 | 53.12 | 907 | 61.30 |
| C. maculata | 16,559 | 51.86 | 0.041 | −0.360 | 11,426 | 51.30 | 1558 | 55.39 | 2631 | 54.12 | 908 | 63.66 |
| C. asiatica | 16,550 | 55.75 | 0.055 | −0.310 | 11,426 | 55.30 | 1555 | 56.01 | 2636 | 54.74 | 896 | 63.73 |
| C. striata | 16,509 | 54.98 | 0.055 | −0.296 | 11,422 | 54.31 | 1554 | 55.47 | 2625 | 55.01 | 870 | 63.56 |
| C. punctata | 16,409 | 53.67 | 0.026 | −0.308 | 11,428 | 52.63 | 1549 | 54.74 | 2623 | 54.02 | 774 | 66.41 |
| C. ornatipinnis | 16,866 | 53.71 | 0.035 | −0.336 | 11,412 | 52.83 | 1547 | 55.07 | 2624 | 53.24 | 1244 | 62.14 |
| C. pulchra | 16,895 | 53.90 | 0.045 | −0.350 | 11,411 | 52.97 | 1550 | 54.39 | 2630 | 53.54 | 1263 | 63.66 |
| C. stewartii | 16,765 | 53.30 | 0.017 | −0.297 | 11,420 | 51.78 | 1550 | 55.81 | 2625 | 53.14 | 1126 | 66.43 |
| C. andrao | 16,729 | 56.55 | 0.022 | −0.307 | 11,420 | 56.26 | 1550 | 56.52 | 2628 | 55.06 | 1086 | 63.90 |
| C. bleheri | 16,714 | 53.08 | 0.012 | −0.302 | 11,420 | 51.74 | 1549 | 55.78 | 2620 | 53.59 | 1081 | 63.09 |
| P. insignis | 16,607 | 52.88 | 0.058 | −0.325 | 11,426 | 51.98 | 1552 | 54.96 | 2660 | 53.08 | 923 | 61.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.; Zhang, W.; Li, Y.; Wang, J.; Bai, Y.; Liu, H. The Complete Mitogenomes of Two Species of Snakehead Fish (Perciformes: Channidae): Genome Characterization and Phylogenetic Analysis. Diversity 2024, 16, 346. https://doi.org/10.3390/d16060346
Xu T, Zhang W, Li Y, Wang J, Bai Y, Liu H. The Complete Mitogenomes of Two Species of Snakehead Fish (Perciformes: Channidae): Genome Characterization and Phylogenetic Analysis. Diversity. 2024; 16(6):346. https://doi.org/10.3390/d16060346
Chicago/Turabian StyleXu, Tangjun, Wenwen Zhang, Yao Li, Jiachen Wang, Yawen Bai, and Hongyi Liu. 2024. "The Complete Mitogenomes of Two Species of Snakehead Fish (Perciformes: Channidae): Genome Characterization and Phylogenetic Analysis" Diversity 16, no. 6: 346. https://doi.org/10.3390/d16060346
APA StyleXu, T., Zhang, W., Li, Y., Wang, J., Bai, Y., & Liu, H. (2024). The Complete Mitogenomes of Two Species of Snakehead Fish (Perciformes: Channidae): Genome Characterization and Phylogenetic Analysis. Diversity, 16(6), 346. https://doi.org/10.3390/d16060346







