Complete Mitochondrial Genomes of the Leptocircini Species Iphiclides podalirius and I. podalirinus (Lepidoptera: Papilionidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Molecular Work
2.3. Genomic Analysis
2.4. Phylogeny, Genetic Distances, and BOLD BINs
3. Results
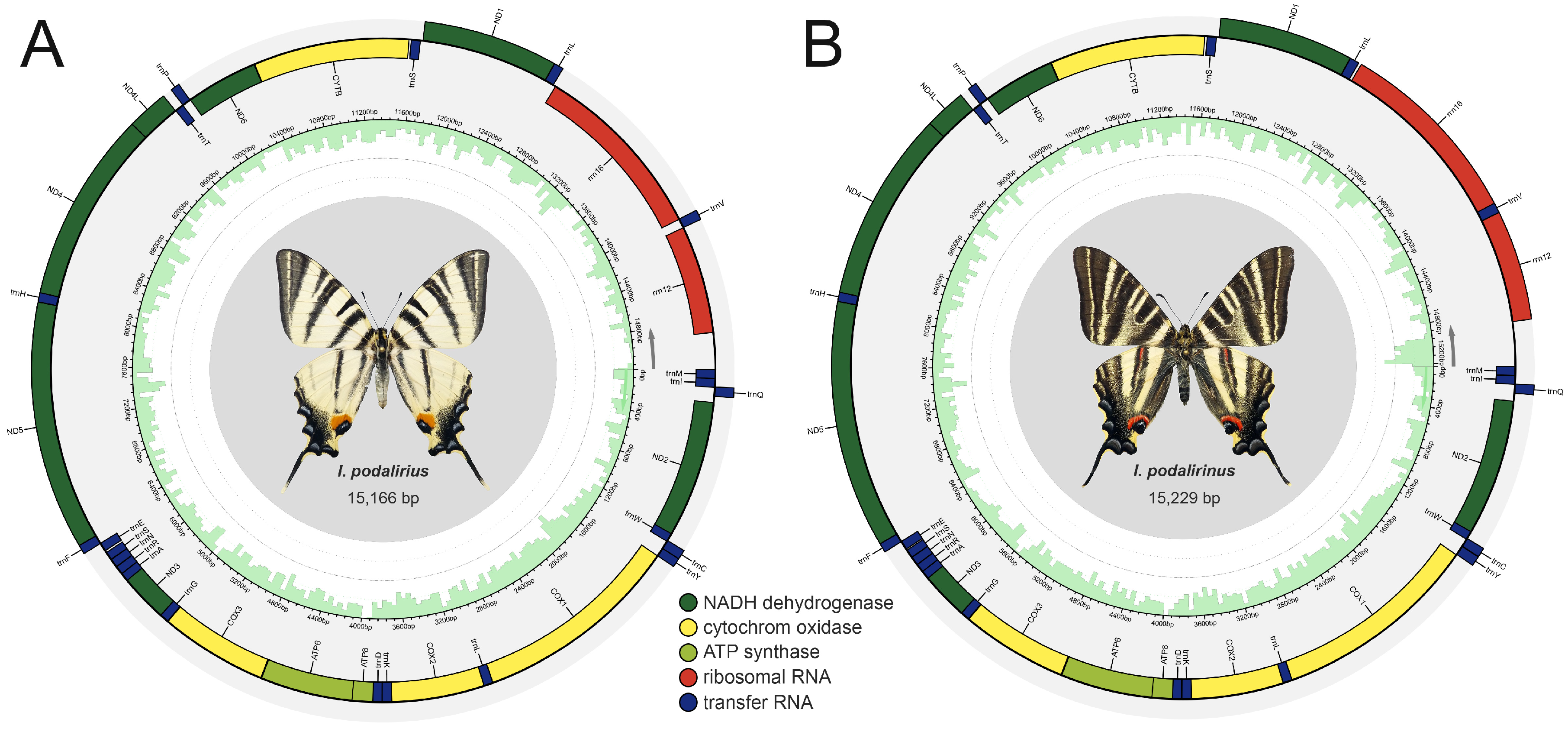
3.1. Sequencing Quality and Genomic Components
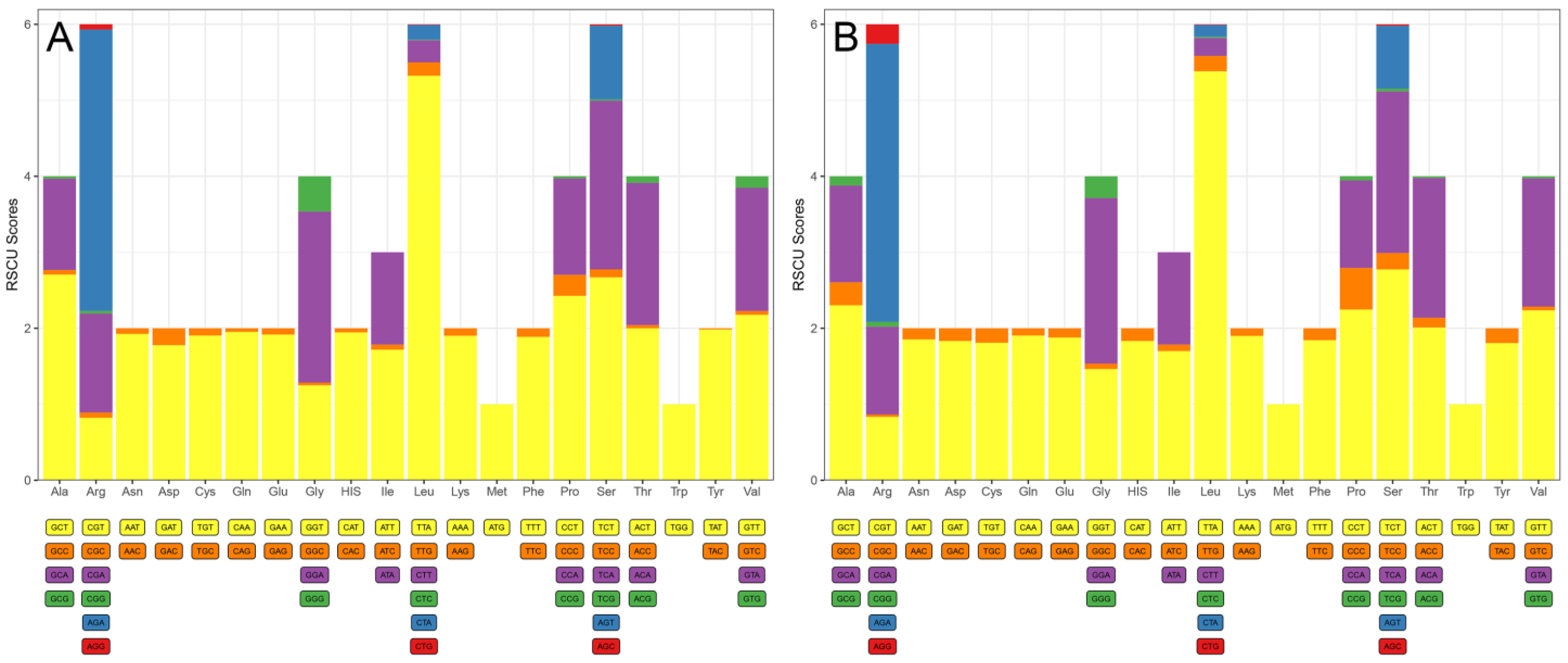
3.2. Codon Usage, Preference, and Substitution
3.3. Phylogenetic Validation, Genetic Distance, and BINs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Racheli, T.; Cotton, A.M. Guide to the Butterflies of the Palearctic Region. Papilionidae Part I. Subfamily Papilioninae. Tribes Leptocircini Teinopalpini; Omnes Artes: Milano, Italy, 2009; p. 69. [Google Scholar]
- Condamine, F.L.; Nabholz, B.; Clamens, A.-L.; Dupuis, J.R.; Sperling, F.A.H. Mitochondrial phylogenomics, the origin of swallowtail butterflies, and the impact of the number of clocks in Bayesian molecular dating. Syst. Entomol. 2018, 43, 460–480. [Google Scholar] [CrossRef]
- Tsukada, E.; Nishiyama, Y. Butterflies of the South East Asian Islands. Part I. Papilionidae; Plapac Co.: Tokyo, Japan, 1980; p. 457. [Google Scholar]
- Hu, S.J.; Cotton, A.M.; Lamas, G.; Duan, K.; Zhang, X. Checklist of Yunnan Papilionidae (Lepidoptera: Papilionoidea) with nomenclatural notes and descriptions of new subspecies. Zootaxa 2023, 5362, 1–69. [Google Scholar] [CrossRef] [PubMed]
- Wiemers, M.; Gottsberger, B. Discordant patterns of mitochondrial and nuclear differentiation in the Scarce Swallowtail Iphiclides podalirius feisthamelii (Duponchel, 1832) (Lepidoptera: Papilionidae). Entomol. Z. Stuttg. 2010, 120, 111–115. [Google Scholar]
- Coutsis, J.G.; Oorschot, H.v. Differences in the male and female genitalia between Iphiclides podalirius and Iphiclides feisthamelii, further supporting species status for the latter (Lepidoptera: Papilionidae). Phegea 2011, 39, 12–22. [Google Scholar]
- Nakae, M. Papilionidae of the World; Roppon-Ashi: Tokyo, Japan, 2021; p. 336. [Google Scholar]
- Racheli, T.; Bozano, G.C. Guide to the Butterflies of the Palearctic Region. Papilionidae Part I. Subfamily Papilioninae. Tribes Leptocircini Teinopalpini, 2nd ed.; Omnes Artes: Milano, Italy, 2024. [Google Scholar]
- Mackintosh, A.; Laetsch, D.R.; Baril, T.; Ebdon, S.; Jay, P.; Vila, R.; Hayward, A.; Lohse, K. The genome sequence of the scarce swallowtail, Iphiclides podalirius. G3 2022, 12, jkac193. [Google Scholar] [CrossRef]
- IUCN. The IUCN Red List of Threatened SpeciesTM. Available online: https://www.iucnredlist.org/search?query=Iphiclides&searchType=species (accessed on 5 April 2024).
- CITES. The CITES Appendices I, II, and III. Available online: https://cites.org/eng/app/appendices.php (accessed on 5 April 2024).
- Collins, N.M.; Morris, M.G. Threatened Swallowtail Butterflies of the World: The IUCN Red Data Book; IUCN: Gland, Switzerland; Cambridge, UK, 1985; p. 331. [Google Scholar]
- NFGA. List of Terrestrial Wildlife under State Protection for Ecological, Economic and Scientific Values (Bulletin of the State Forestry Administration of P. R. China 2000-07); National Forestry and Grassland Administration: Beijing, China, 2000.
- Wang, W.L.; Suman, D.O.; Zhang, H.H.; Xu, Z.B.; Ma, F.Z.; Hu, S.J. Butterfly conservation in China: From science to action. Insects 2020, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.R.; Yu, H.; Xu, J. The complete mitochondrial genome of the Papilio memno [sic!] (Lepidoptera: Papilionidae). Mitochondrial DNA Part B 2020, 5, 2159–2160. [Google Scholar] [CrossRef]
- Condamine, F.L.; Allio, R.; Reboud, E.L.; Dupuis, J.R.; Toussaint, E.F.A.; Mazet, N.; Hu, S.J.; Lewis, D.S.; Kunte, K.; Cotton, A.M.; et al. A comprehensive phylogeny and revised taxonomy illuminate the origin and diversification of the global radiation of Papilio (Lepidoptera: Papilionidae). Mol. Phylogenetic Evol. 2023, 183, 107758. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.; Nikolenko, S.; Pham, S.; Prjibelski, A.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Massouras, A.; Hens, K.; Gubelmann, C.; Uplekar, S.; Decouttere, F.; Rougemont, J.; Cole, S.T.; Deplancke, B. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods 2010, 7, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.M.; Guan, Q.X.; Li, H.M.; Zhang, Y.; Liu, Y.J.; Guo, D.N. The complete mitogenome of Lamproptera curia (Lepidoptera: Papilionidae) and phylogenetic analyses of Lepidoptera. Front. Biol. 2015, 10, 458–472. [Google Scholar] [CrossRef]
- Chen, X.; Hao, J.S. The complete mitochondrial genome of Graphium chironides (Lepidoptera: Papilionidae: Papilioninae). Mitochondrial DNA 2015, 27, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.J.; Hou, Y.; Zhang, G.T.; Yang, T.; Xiao, R.H. Complete mitochondrial genome of Graphium doson (Papilioninae: Leptocircini). Mitochondrial DNA Part B 2019, 4, 698–699. [Google Scholar] [CrossRef]
- Duan, K.; Zhang, X.; Zhang, H.H.; Hu, S.J. Complete mitochondrial genome of the subalpine swordtail butterfly Graphium (Pazala) parus (Nicéville, 1900) (Lepidoptera: Papilionidae). Mitochondrial DNA Part B 2020, 5, 1903–1904. [Google Scholar] [CrossRef]
- Hu, S.J.; Zhang, X.; Duan, K. Complete mitochondrial genomes of two insular races of Pazala swordtails from Taiwan, China (Lepidoptera: Papilionidae: Graphium). Mitochondrial DNA Part B 2021, 6, 1557–1559. [Google Scholar] [CrossRef] [PubMed]
- He, F.R.; Zhang, X.; Hu, S.J. Complete mitochondrial genome of the recently discovered multivoltine Graphium (Pazala) confucius Hu, Duan & Cotton, 2018 (Lepidoptera: Papilionidae). Mitochondrial DNA Part B 2022, 7, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef]
- Cui, X.F.; Liu, Z.W.; Wang, S.; Wang, J.J.-Y.; Gao, X. CMsearch: Simultaneous exploration of protein sequence space and structure space improves not only protein homology detection but also protein structure prediction. Bioinformatics 2016, 32, i332–i340. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Argasinska, J.; Quinones-Olvera, N.; Finn, R.D.; Bateman, A.; Petrov, A.I. Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinform. 2018, 62, e51. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.L.; Liu, J.X.; Li, H.Z.; Liu, B.Y.; Zhao, B.Q.; Ning, Z.Y. Comprehensive analysis of synonymous codon usage patterns and influencing factors of porcine epidemic diarrhea virus. Arch. Virol. 2021, 166, 157–165. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Gelman, A.; Rubin, D.B. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992, 7, 457–472. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree ver 1.4.4. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kim, M.I.; Baek, J.Y.; Kim, M.J.; Jeong, H.C.; Kim, K.G.; Bae, C.H.; Han, Y.S.; Jin, B.R.; Kim, I. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol. Cells 2009, 28, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, J.C.; Tóth, J.P.; Monasterio, Y.; Mesa, L.S.; Sariot, M.G.M.; Escobés, R.; Vila, R. Integrative taxonomy reveals a new Melitaea (Lepidoptera: Nymphalidae) species widely distributed in the Iberian Peninsula. Insect Syst. Divers. 2022, 6, ixac004. [Google Scholar] [CrossRef]
- Duponchel, P.A.J. Supplement. Diurnes. In Histoire naturelle des Lépidoptèrs ou Papillons de France; Godart, J.B., Ed.; Méquignon-Marvis, Libraire-Éditeur: Paris, France, 1832; pp. 1–466. [Google Scholar]
- Gaunet, A.; Dincă, V.; Dapporto, L.; Montagud, S.; Vodă, R.; Schär, S.; Badiane, A.; Font, E.; Vila, R. Two consecutive Wolbachia-mediated mitochondrial introgressions obscure taxonomy in Palearctic swallowtail butterflies (Lepidoptera, Papilionidae). Zool. Scr. 2019, 48, 507–519. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Hurst, G.D.D.; Jiggins, F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effects of inherited symbionts. Proc. R. Soc. B Biol. Sci. 2005, 272, 1525–1534. [Google Scholar] [CrossRef]
- Baldo, L.; Werren, J.H. Revisiting Wolbachia supergroup typing based on WSP: Spurious lineages and discordance with MLST. Curr. Microbiol. 2007, 55, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Hu, S.J.; Ma, X.Y.; Chen, J.M.; Li, Q.Q.; Ye, H. Origin and expansion of the Yunnan Shoot Borer, Tomicus yunnanensis (Coleoptera: Scolytinae): A mixture of historical natural expansion and contemporary human-mediated relocation. PLoS ONE 2014, 9, e111940. [Google Scholar] [CrossRef]

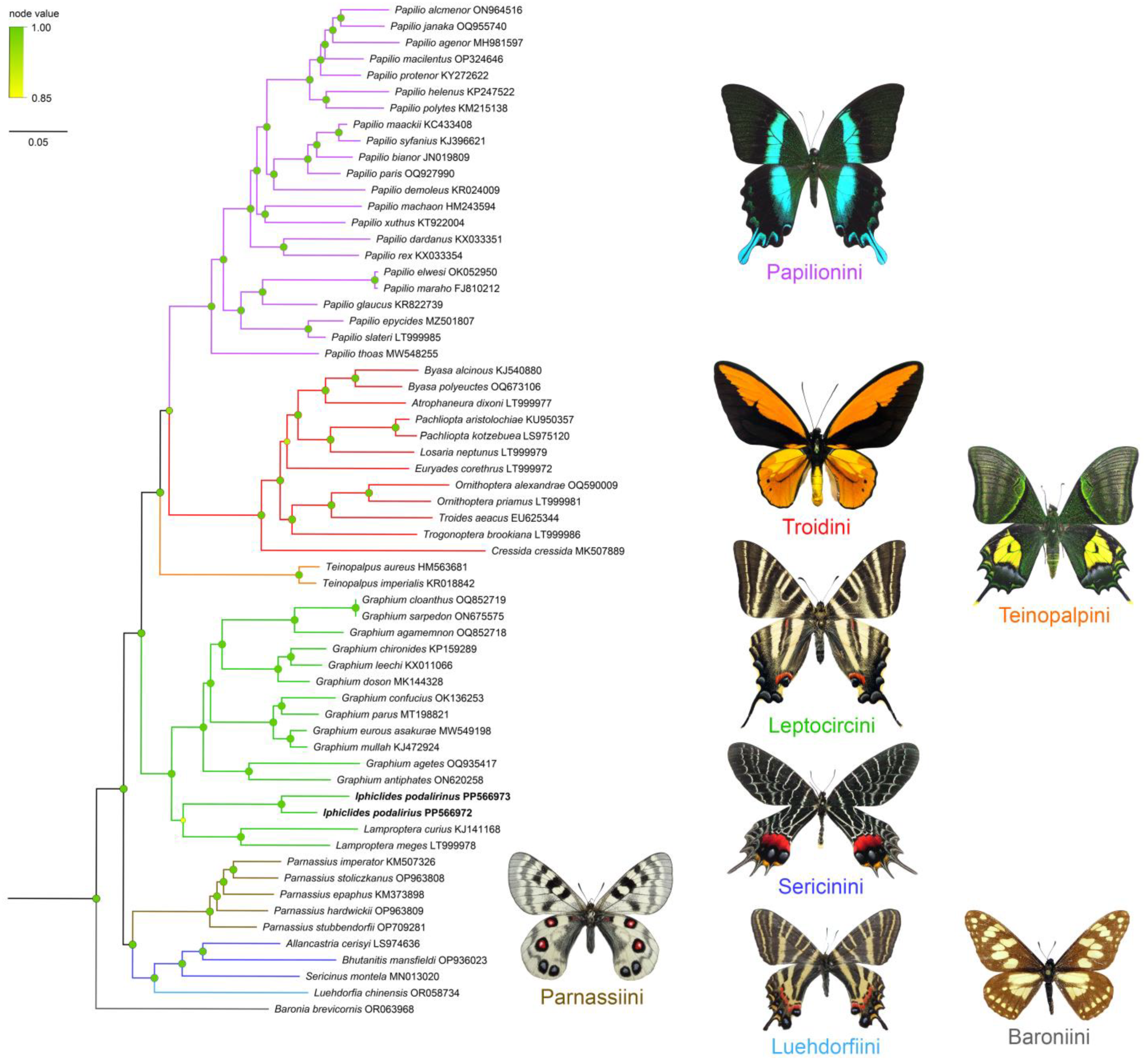
| Subfamily | Tribe | Species | GenBank Accession |
|---|---|---|---|
| Baroniinae | Baroniini | Baronia brevicornis | OR063968 |
| Parnassiinae | Sericinini | Allancastria cerisy | LS974636 |
| Bhutanitis mansfieldi | OP936023 | ||
| Sericinus montela | MN013020 | ||
| Luehdorfiini | Luehdorfia chinensis | OR058734 | |
| Parnassiini | Parnassius epaphus | KM373898 | |
| Parnassius stubbendorfii | OP709281 | ||
| Parnassius hardwickii | OP963809 | ||
| Parnassius stoliczkanus | OP963808 | ||
| Parnassius imperator | KM507326 | ||
| Papilioninae | Leptocircini | Lamproptera meges | LT999978 |
| Lamproptera curius | KJ141168 | ||
| Graphium parus | MT198821 | ||
| Graphium mullah | KJ472924 | ||
| Graphium eurous asakurae | MW549198 | ||
| Graphium confucius | OK136253 | ||
| Graphium antiphates | ON620258 | ||
| Graphium agetes | OQ935417 | ||
| Graphium leechi | KX011066 | ||
| Graphium chironides | KP159289 | ||
| Graphium doson | MK144328 | ||
| Graphium agamemnon | OQ852718 | ||
| Graphium sarpedon | ON675575 | ||
| Graphium cloanthus | OQ852719 | ||
| Teinopalpini | Teinopalpus imperialis | KR018842 | |
| Teinopalpus aureus | HM563681 | ||
| Troidini | Cressida cressida | MK507889 | |
| Euryades corethrus | LT999972 | ||
| Losaria neptunus | LT999979 | ||
| Pachliopta kotzebuea | LS975120 | ||
| Pachliopta aristolochiae | KU950357 | ||
| Atrophaneura dixoni | LT999977 | ||
| Byasa polyeuctes | OQ673106 | ||
| Byasa alcinous | KJ540880 | ||
| Trogonoptera brookiana | LT999986 | ||
| Troides aeacus | EU625344 | ||
| Ornithoptera priamus | LT999981 | ||
| Ornithoptera alexandrae | OQ590009 | ||
| Papilionini | Papilio machaon | HM243594 | |
| Papilio xuthus | KT922004 | ||
| Papilio rex | KX033354 | ||
| Papilio dardanus | KX033351 | ||
| Papilio demoleus | KR024009 | ||
| Papilio paris | OQ927990 | ||
| Papilio bianor | JN019809 | ||
| Papilio syfanius | KJ396621 | ||
| Papilio maackii | KC433408 | ||
| Papilio polytes | KM215138 | ||
| Papilio helenus | KP247522 | ||
| Papilio protenor | KY272622 | ||
| Papilio macilentus | OP324646 | ||
| Papilio agenor 1 | MH981597 | ||
| Papilio janaka 2 | OQ955740 | ||
| Papilio alcmenor | ON964516 |
| Taxon | Country | BOLD Accession |
|---|---|---|
| I. podalirius | Portugal | BDE369-19, EULEP5983-18, EULEP5984-18 |
| Spain | EULEP5951-18, EULEP5952-18, EULEP5980-18, EULEP5994-18, WMB3432-14 | |
| France | BIBSA922-15, BIBSA2258-20, BIBSA2271-20, EULEP4099-16, EULEP5979-18, EULEP5986-18, EULEP5987-18, EULEP5988-18, EULEP5989-18, EULEP5992-18, EZSPM448-09, OXB1065-15, OXB1259-15, OXB1366-15, OXB1388-15, WMB354-11, WMB1173-13, WMB1730-13, WMB1800-13, WMB3465-14, WMB3893-14, WMB3912-14, WMB3916-14, WMB3945-14, WMB3971-14, WMB4637-14, WMB5327-14 | |
| Switzerland | EULEP2164-15, EULEP4098-16 | |
| Germany | GWORA2430-09 | |
| Austria | ABOLC133-16, ABOLD084-16, LEASS703-17, LEASS856-17 | |
| Italy | BIBSA349-15, BIBSA580-15, BIBSA701-15, BIBSA970-15, BIBSA1031-15, BIBSA1140-15, BIBSA1171-15, BIBSA1329-15, BIBSA1423-15, BIBSA1600-16, BIBSA1613-16, BIBSA1618-16, BIBSA1667-16, BIBSA1766-16, EULEP5662-17, EULEP5953-18, EULEP5961-18, EULEP5965-18, EULEP5982-18, EULEP5991-18, EULEP5999-18, EULEP6000-18, LEATD016-13, LEATG462-14, LEASZ969-22, OXB587-15, OXB888-15, OXB910-15, OXB974-15, OXB1028-15, WMB127-11, WMB259-11, WMB641-11, WMB1895-13, WMB2120-13, WMB2216-13, WMB2360-13, WMB2361-13, WMB2442-13, WMB3834-14, WMB3853-14, WMB4131-14, WMB4271-14, WMB4288-14, WMB4342-14, WMB4765-14, WMB4804-14, WMB4855-14, WMB4968-14, WMB5040-14, WMB5084-14WMB5497-14, WMB6563-18 | |
| Slovakia | EULEP5022-16 | |
| Ukraine | EULEP4835-16 | |
| Romania | EULEP001-14, EULEP5973-18, EZRMN332-08, EZROM219-08, EZROM220-08, EZROM351-08, EZROM1028-08, EZROM1029-08, EZROM1030-08 | |
| Bulgaria | EULEP897-15, EULEP1197-15 | |
| Greece | EULEP701-15, EULEP1306-15, EULEP1367-15, EULEP1442-15, EULEP1600-15, EULEP1767-15, EULEP5962-18, EULEP5971-18, EULEP5972-18, EULEP5974-18, EULEP5975-18, EULEP5978-18 | |
| Turkey | EULEP5950-18 | |
| Iran | IRANB315-08 | |
| Kazakhstan | EULEP5956-18, EULEP5957-18, EULEP5958-18, EULEP5959-18, EULEP5960-18 | |
| Russia | LEFIL147-10 | |
| I. feisthamelii | Morocco | EULEP5954-18, EULEP5955-18, EULEP5990-18, EULEP5996-18, EULEP5997-18, WMB245-11, WMB373-11, WMB399-11, WMB421-11, WMB3604-14, BDE293-19, BDE293-19, BDE294-19 |
| Algeria | EULEP5964-18, EULEP5981-18, EULEP6001-18, WMB1472-13, WMB1473-13, WMB1476-13 | |
| Tunisia | EULEP5970-18, WMB1405-13, WMB1406-13, WMB6562-18 | |
| Portugal | WMB4400-14, BDE553-19 | |
| Spain | EULEP5949-18, EULEP5966-18, EULEP5967-18, EULEP5968-18, EULEP5969-18, EULEP5993-18, EULEP5995-18, EULEP5998-18, EULEP6002-18, EZSPC449-09, EZSPC450-09, EZSPC451-09, EZSPM099-09, EZSPM275-09, EZSPM362-09, EZSPM419-09, EZSPM450-09, EZSPM611-12, EZSPM629-12, EZSPN330-09, EZSPN403-09, EZSPN477-09, EZROM725-08, WMB3126-14, WMB3315-14, WMB3481-14, WMB3777-14, WMB3995-14, WMB4046-14 | |
| France | EULEP002-14, EULEP003-14, EULEP5985-18 |
| Gene | Strand | Span | Anti-Codon | Start Codon | Stop Codon | |||
|---|---|---|---|---|---|---|---|---|
| A | B | A | B | A | B | |||
| tRNA-Met | + | 1–66 | 1–67 | CAU | — | — | — | — |
| tRNA-Ile | + | 67–130 | 68–131 | GAU | — | — | — | — |
| tRNA-Gln | − | 128–196 | 129–197 | UUG | — | — | — | — |
| ND2 | + | 243–1256 | 256–1296 | — | ATT | ATT | TAA | TAA |
| tRNA-Trp | + | 1255–1319 | 1268–1332 | UCA | — | — | — | — |
| tRNA-Cys | − | 1312–1379 | 1325–1391 | GCA | — | — | — | — |
| tRNA-Tyr | − | 1380–1444 | 1394–1464 | GUA | — | — | — | — |
| COX1 | + | 1445–2978 | 1468–2998 | — | ATG | CGA 1 | T-tRNA | T-tRNA |
| tRNA-Leu | + | 2979–3045 | 2999–3065 | UAA | — | — | — | — |
| COX2 | + | 3046–3727 | 3066–3747 | — | ATG | ATG | T-tRNA | T-tRNA |
| tRNA-Lys | + | 3728–3797 | 3748–3817 | CUU | — | — | — | — |
| tRNA-Asp | + | 3798–3865 | 3818–3884 | GUC | — | — | — | — |
| ATP8 | + | 3866–4024 | 3885–4043 | — | ATC | ATA | TAA | TAA |
| ATP6 | + | 4018–4695 | 4037–4714 | — | ATG | ATG | TAA | TAA |
| COX3 | + | 4701–5492 | 4720–5508 | — | ATA | ATA | TAA | TAA |
| tRNA-Gly | + | 5496–5561 | 5512–5577 | UCC | — | — | — | — |
| ND3 | + | 5562–5915 | 5578–5931 | — | ATT | ATA | TAG | TAA |
| tRNA-Ala | + | 5914–5976 | 5931–5993 | UGC | — | — | — | — |
| tRNA-Arg | + | 5977–6039 | 5994–6057 | UCG | — | — | — | — |
| tRNA-Asn | + | 6041–6105 | 6059–6123 | GUU | — | — | — | — |
| tRNA-Ser | + | 6107–6167 | 6125–6187 | UCU | — | — | — | — |
| tRNA-Glu | + | 6183–6247 | 6197–6263 | UUC | — | — | — | — |
| tRNA-Phe | − | 6246–6310 | 6262–6326 | GAA | — | — | — | — |
| ND5 | − | 6311–8050 | 6327–8066 | — | TTG | TTG | TAA | TAA |
| tRNA-His | − | 8051–8116 | 8067–8132 | GUG | — | — | — | — |
| ND4 | − | 8116–9455 | 8132–9471 | — | ATG | ATG | T-tRNA | T-tRNA |
| ND4L | − | 9456–9746 | 9472–9762 | — | ATG | ATG | TAA | TAA |
| tRNA-Thr | + | 9749–9813 | 9765–9828 | UGU | — | — | — | — |
| tRNA-Pro | − | 9814–9878 | 9829–9893 | UGG | — | — | — | — |
| ND6 | + | 9881–10,411 | 9896–10,426 | — | ATT | ATT | TAA | TAA |
| CYTB | + | 10,416–11564 | 10,431–11,579 | — | ATG | ATG | TAA | TAA |
| tRNA-Ser | + | 11,580–11,647 | 11,597–11,665 | UGA | — | — | — | — |
| ND1 | − | 11,665–12,603 | 11,683–12,621 | — | ATG | ATG | TAG | TAG |
| tRNA-Leu | − | 12,605–12,672 | 12,623–12,689 | UAG | — | — | — | — |
| 16S-rRNA | + | 12,691–14,029 | 12,709–14,056 | — | — | — | — | — |
| tRNA-Val | − | 14,028–14,097 | 14,055–14,126 | UAC | — | — | — | — |
| 12S-rRNA | + | 14,098–14,895 | 14,127–14,900 | — | — | — | — | — |
| 1 | 2a | 2b | 3 | |
|---|---|---|---|---|
| 1. I. podalirius | ||||
| 2a. I. feisthamelii North Africa | 2.234 | |||
| 2b. I. feisthamelii Iberia | 0.134 | 2.184 | ||
| 3. I. podalirinus | 5.309 | 5.530 | 5.280 |
| BIN Number | No. of Sequences | Species Included | Country/Region |
|---|---|---|---|
| BOLD:AAB2272 | 198 | I. podalirius I. feisthamelii | Italy (58); Spain (40); France (34); Greece (16); Romania (10); Portugal (7); Austria (6); Switzerland (5); Kazakhstan (5); Czech Republic (3); Iran (3); Russia (2); Slovakia (2); Bulgaria (2); Turkey (2); Germany (1); Ukraine (1); Unknown (1) |
| BOLD:AEA3098 | 25 | I. feisthamelii | Morocco (13); Algeria (7); Tunisia (4); Unknown (1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Y.; Zhang, X.; Cotton, A.M.; Hu, S.-J. Complete Mitochondrial Genomes of the Leptocircini Species Iphiclides podalirius and I. podalirinus (Lepidoptera: Papilionidae). Diversity 2024, 16, 392. https://doi.org/10.3390/d16070392
Pan Y, Zhang X, Cotton AM, Hu S-J. Complete Mitochondrial Genomes of the Leptocircini Species Iphiclides podalirius and I. podalirinus (Lepidoptera: Papilionidae). Diversity. 2024; 16(7):392. https://doi.org/10.3390/d16070392
Chicago/Turabian StylePan, Yue, Xin Zhang, Adam M. Cotton, and Shao-Ji Hu. 2024. "Complete Mitochondrial Genomes of the Leptocircini Species Iphiclides podalirius and I. podalirinus (Lepidoptera: Papilionidae)" Diversity 16, no. 7: 392. https://doi.org/10.3390/d16070392
APA StylePan, Y., Zhang, X., Cotton, A. M., & Hu, S. -J. (2024). Complete Mitochondrial Genomes of the Leptocircini Species Iphiclides podalirius and I. podalirinus (Lepidoptera: Papilionidae). Diversity, 16(7), 392. https://doi.org/10.3390/d16070392








