Exploring the Genetic Structure and Phylogeographic Patterns of the Copepod Genus Eurytemora in Europe
Abstract
1. Introduction
2. Materials and Methods
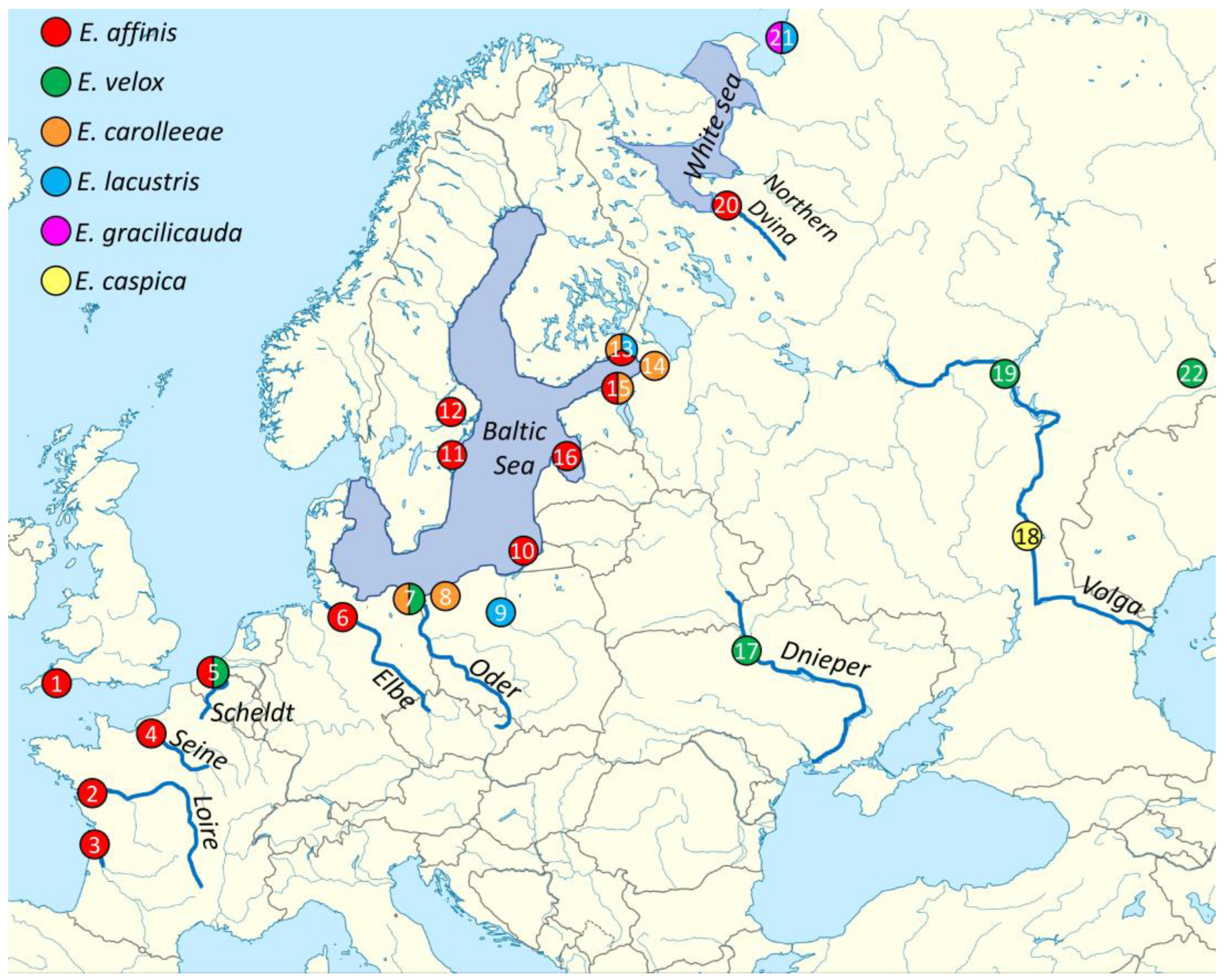
2.1. Sampling Strategy
2.2. Molecular Analysis
2.3. DNA Sequencing
2.4. Phylogenetic Reconstruction, Haplowebs, and Genetic Diversity
3. Results
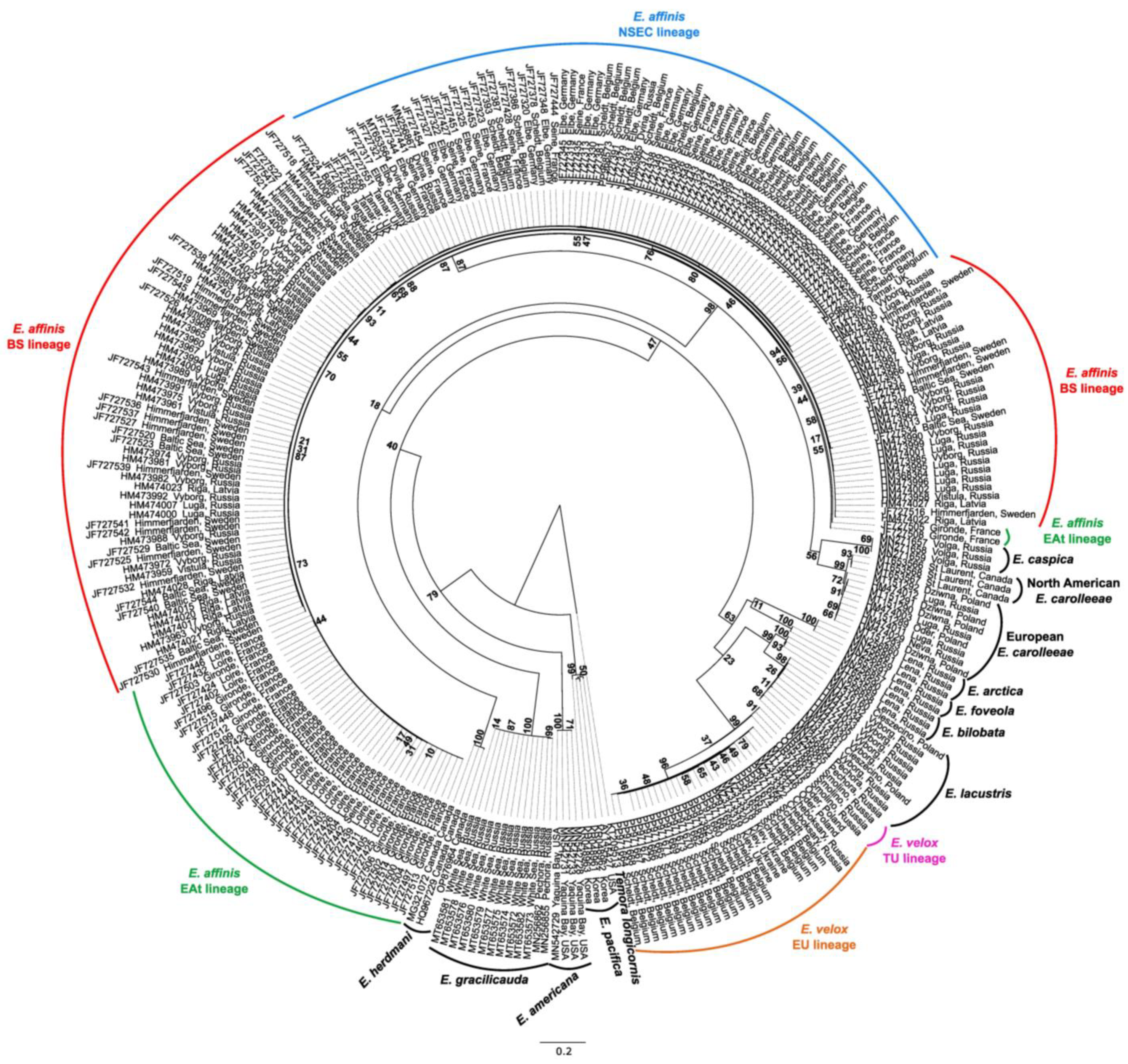
3.1. Phylogenetic Reconstruction and Genetic Diversity of the Eurytemora Genus in Geographic Europe
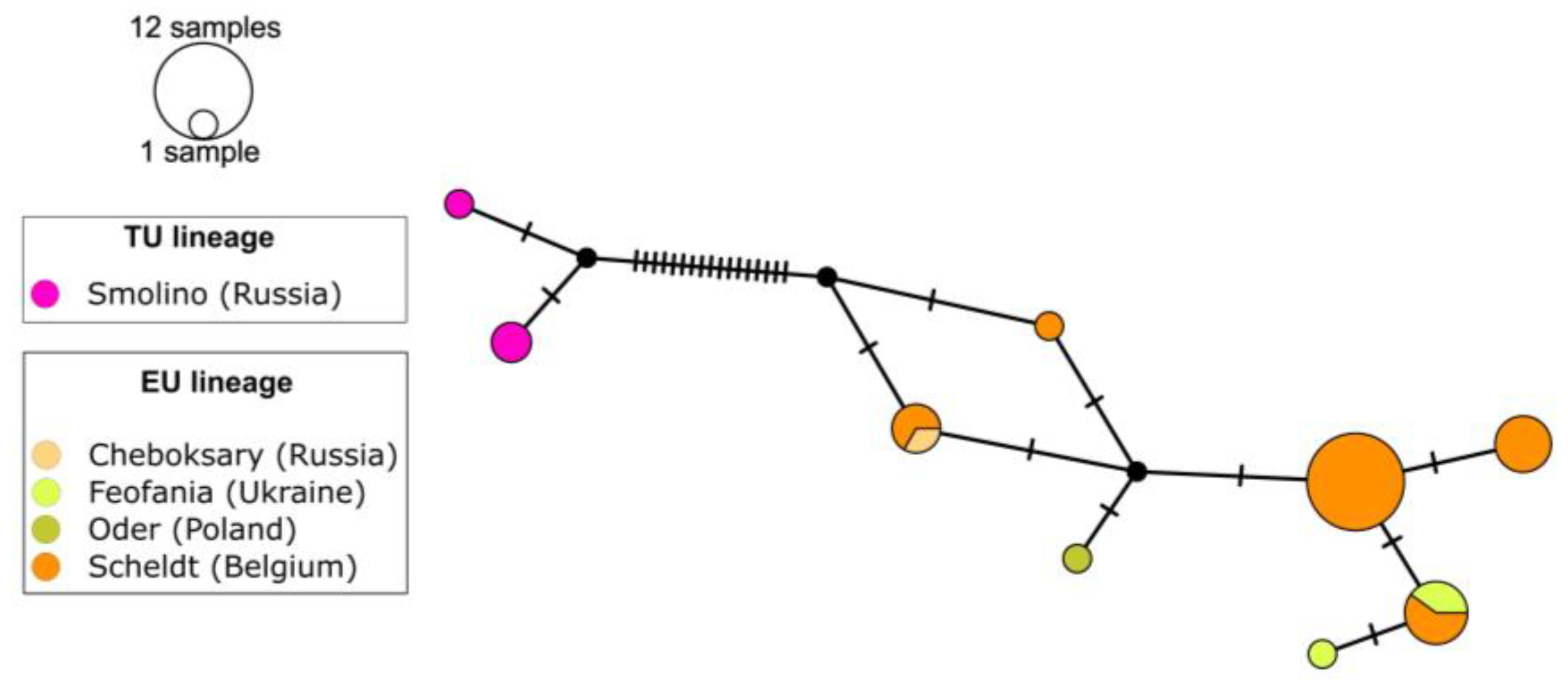
3.2. Haplotype Genetic Diversity and Network of E. affinis and E. velox

4. Discussion
4.1. Global Phylogeny of the Eurytemora Genus in Geographic Europe
4.2. Eurytemora affinis Population Structure
4.3. Eurytemora velox Population Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Sukhikh, N.; Abramova, E.; Holl, A.C.; Souissi, S.; Alekseev, V. A Comparative Analysis of Genetic Differentiation of the E. affinis Species Complex and Some Other Eurytemora Species, Using the CO1, NITS and 18SrRNA Genes (Copepoda, Calanoida). Crustaceana 2020, 93, 931–955. [Google Scholar] [CrossRef]
- Heinle, D.R.; Flemer, D.A. Carbon Requirements of a Population of the Estuarine Copepod Eurytemora affinis. Mar. Biol. 1975, 31, 235–247. [Google Scholar] [CrossRef]
- Devreker, D.; Souissi, S.; Molinero, J.C.; Beyrend-Dur, D.; Gomez, F.; Forget-Leray, J. Tidal and Annual Variability of the Population Structure of Eurytemora affinis in the Middle Part of the Seine Estuary during 2005. Estuar. Coast. Shelf Sci. 2010, 89, 245–255. [Google Scholar] [CrossRef]
- Mialet, B.; Gouzou, J.; Azémar, F.; Maris, T.; Sossou, C.; Toumi, N.; Van Damme, S.; Meire, P.; Tackx, M. Response of Zooplankton to Improving Water Quality in the Scheldt Estuary (Belgium). Estuar. Coast. Shelf Sci. 2011, 93, 47–57. [Google Scholar] [CrossRef]
- Dur, G.; Souissi, S. Ontogenetic Optimal Temperature and Salinity Envelops of the Copepod Eurytemora affinis in the Seine Estuary (France). Estuar. Coast. Shelf Sci. 2018, 200, 311–323. [Google Scholar] [CrossRef]
- Winkler, G.; Dodson, J.J.; Lee, C.E. Heterogeneity within the Native Range: Population Genetic Analyses of Sympatric Invasive and Noninvasive Clades of the Freshwater Invading Copepod Eurytemora. affinis. Mol. Ecol. 2008, 17, 415–430. [Google Scholar] [CrossRef]
- Sukhikh, N.; Souissi, A.; Souissi, S.; Winkler, G.; Castric, V.; Holl, A.C.; Alekseev, V. Genetic and Morphological Heterogeneity among Populations of Eurytemora affinis (Crustacea: Copepoda: Temoridae) in European Waters. Comptes Rendus Biol. 2016, 339, 197–206. [Google Scholar] [CrossRef]
- Lee, C.E. Global Phylogeography of a Cryptic Copepod Species Complex and Reproductive Isolation between Genetically Proximate “Populations”. Evolution 2000, 54, 2014–2027. [Google Scholar] [CrossRef]
- Lee, C.E.; Frost, B.W. Morphological Stasis in the Eurytemora affinis Species Complex (Copepoda: Temoridae). Hydrobiologia 2002, 480, 111–128. [Google Scholar] [CrossRef]
- Beyrend-Dur, D.; Souissi, S.; Devreker, D.; Winkler, G.; Hwang, J.S. Life Cycle Traits of Two Transatlantic Populations of Eurytemora affinis (Copepoda: Calanoida): Salinity Effects. J. Plankton Res. 2009, 31, 713–728. [Google Scholar] [CrossRef]
- Sukhikh, N.; Souissi, A.; Souissi, S.; Holl, A.C.; Schizas, N.V.; Alekseev, V. Life in Sympatry: Coexistence of Native Eurytemora affinis and Invasive Eurytemora carolleeae in the Gulf of Finland (Baltic Sea). Oceanologia 2019, 61, 227–238. [Google Scholar] [CrossRef]
- Winkler, G.; Souissi, S.; Poux, C.; Castric, V. Genetic Heterogeneity among Eurytemora affinis Populations in Western Europe. Mar. Biol. 2011, 158, 1841–1856. [Google Scholar] [CrossRef]
- Heron, G.A.; Damkaer, D.M. Eurytemora richingsi New Species of Deep Water Calanoid Copepod from the Arctic Ocean. Proc. Biol. Soc. Wash. 1976, 89, 127–136. [Google Scholar]
- Akatova, N.A. Zooplankton of the Kolyma River and Its Basins // Scientific Notes of Leningrad State University. Biol. Sci. Ser. 1949, 21, 341–367. [Google Scholar]
- Fefilova, E.B.; Sukhikh, N.M.; Rasova, E.E.; Velegzhaninov, I.O.; Abramova, E.N. New Data on the Expansion of Eurytemora giesbrecht (Copepoda: Calanoida) in the Russian Arctic Region. Dokl. Biol. Sci. 2020, 492, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Sukhikh, N.; Fefilova, E. Eurytemora gracilicauda (Copepoda: Calanoida) in the Russian Arctic. Proc. Zool. Inst. RAS 2023, 327, 25–40. [Google Scholar] [CrossRef]
- Maier, G.; Speth, B.; Arp, W.; Bahnwart, M.; Kasprzak, P. New Records of the Rare Glacial Relict Eurytemora lacustris (Poppe 1887) (Copepoda; Calanoida) in Atypical Lake Habitats of Northern Germany. J. Limnol. 2011, 70, 145–148. [Google Scholar] [CrossRef][Green Version]
- Sługocki, Ł.; Rymaszewska, A.; Kirczuk, L. Insights into the Morphology and Molecular Characterisation of Glacial Relict Eurytemora lacustris (Poppe, 1887) (Crustacea, Copepoda, Calanoida, Temoridae). Zookeys 2019, 2019, 15–33. [Google Scholar] [CrossRef]
- Minguez, J.; Maris, T.; Tackx, M.; Gers, C.; Meire, P.; Legal, L. Genetics of the Estuarine Copepod Eurytemora affinis with Regard to Improving Water Quality. Estuar. Coast. Shelf Sci. 2020, 246, 107037. [Google Scholar] [CrossRef]
- Gasmi, S.; Ferval, M.; Pelissier, C.; D’Amico, F.; Maris, T.; Tackx, M.; Legal, L. Genetic Diversity among the Eurytemora affinis Species Complex in the Scheldt Estuary and Its Tributaries Using ISSR-PCR Marker Assay. Estuar. Coast. Shelf Sci. 2014, 145, 22–30. [Google Scholar] [CrossRef]
- Forró, L.; Gulyás, P. Eurytemora velox (Lilljeborg, 1853) (Copepoda, Calanoida) in the Szigetköz Region of the Danube. Misc. Zool. Hung. 1992, 7, 53–58. [Google Scholar]
- Samchyshyna, L. Assumed Recent Invasion of Eurytemora velox (Lill.) (Copepoda, Calanoida) in the Dnieper River and Its Tributaries. Ecol. Moray 2000, 52, 52–55. [Google Scholar]
- Sukhikh, N.; Garibian, P.; Chertoprud, E. Resettlement of Eurytemora velox (Crustacea: Copepoda) in Europe, the Urals and Western Siberia. Diversity 2024, 16, 47. [Google Scholar] [CrossRef]
- Sukhikh, N.; Fefilova, E. The First Record of Eurytemora velox (Lilljeborg, 1853) (Crustacea, Calanoida) Outside of Europe, Genetic Identification with Surprise. Russ. J. Biol. Invasions 2024, 15, 134–140. [Google Scholar] [CrossRef]
- Sukhikh, N.M.; Lazareva, V.I. First Results of a Molecular Genetic Analysis of the European Invader Eurytemora velox (Crustacea, Calanoida). Inland Water Biol. 2022, 15, 201–203. [Google Scholar] [CrossRef]
- Mouth, C.; Tackx, M.; Azémar, F.; Bou, E.; Meire, P.; Maris, T.; Legal, L.; Bernard, A. Environmental Factors as Drivers of the Spatial Distribution of the Copepods Eurytemora affinis affinis and Eurytemora velox in the Scheldt Tributaries. Estuar. Coast. Shelf Sci. 2024, 303, 108802. [Google Scholar] [CrossRef]
- Lee, C. Rapid and Repeated Invasions of Fresh Water by the Copepod Eurytemora affinis. Evolution 1999, 53, 1423–1434. [Google Scholar] [CrossRef]
- Sukhikh, N.; Souissi, A.; Souissi, S.; Alekseev, V. Invasion of Eurytemora Sibling Species (Copepoda: Temoridae) from North America into the Baltic Sea and European Atlantic Coast Estuaries. J. Nat. Hist. 2013, 47, 753–767. [Google Scholar] [CrossRef]
- Vasquez, A.A.; Hudson, P.L.; Fujimoto, M.; Keeler, K.; Armenio, P.M.; Ram, J.L. Eurytemora carolleeae in the Laurentian Great Lakes Revealed by Phylogenetic and Morphological Analysis. J. Great Lakes Res. 2016, 42, 802–811. [Google Scholar] [CrossRef]
- Alekseev, V.; Sukhikh, N. Copepod Cryptic Species as Aquatic Invaders. Limnol. Freshw. Biol. 2022, 5, 1645–1655. [Google Scholar] [CrossRef]
- Bledzki, L.A.; Rybak, J.I. Freshwater Crustacean Zooplankton of Europe; Springer: Cham, Switzerland; Edinburgh, UK, 2016. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Sługocki, Ł.; Rymaszewska, A.; Kirczuk, L. To Fit or to Belong: Characterization of the Non-Native Invader Eurytemora carolleeae (Copepoda: Calanoida) in the Oder River System (Central Europe). Aquat. Invasions 2021, 16, 443–460. [Google Scholar] [CrossRef]
- Sukhikh, N.M.; Castric, V.; Polyakova, N.V.; Souissi, S.; Alekseev, V.R. Isolated Populations of Eurytemora americana Williams (Crustacea, Copepoda) in the White Sea Rock Pools—Postglacial Relicts or Anthropogenic Invasions? Russ. J. Biol. Invasions 2016, 7, 396–404. [Google Scholar] [CrossRef]
- Avila Romero, L. Contribution Des Espèces Cryptiques Eurytemora affinis et E. aarolleeae Au Régime Alimentaire Des Larves d’éperlan Arc-En-Ciel, Osmerus Mordax, Dans La Zone de Turbidité Maximale de l’estuaire de Saint-Laurent; Université du Québec à Rimouski, Institut des sciences de la mer de Rimouski: Rimouski, QC, Canada, 2023. [Google Scholar]
- Raupach, M.J.; Barco, A.; Steinke, D.; Beermann, J.; Laakmann, S.; Mohrbeck, I.; Neumann, H.; Kihara, T.C.; Pointner, K.; Radulovici, A.; et al. The Application of DNA Barcodes for the Identification of Marine Crustaceans from the North Sea and Adjacent Regions. PLoS ONE 2015, 10, e0139421. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.; Lindley, A.J.; Wootton, M.; Lee, C.M.; Kirby, R.R. Morphological and Genetic Variation in the North Atlantic Copepod, Centropages typicus. J. Mar. Biol. Assoc. UK 2012, 92, 99–106. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations: The Theory of Gene Frequencies; University of Chicago Press: London, UK, 1969. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA; Chichester, UK, 1987. [Google Scholar]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Perumal, P.; Rajthilak, C.; Perumal, S.; Raja, M.; Yoganandham Suman, T.; Balasubramani, G. Cladistic Phylogeny of Calanoid Copepods (Crustacea: Arthropoda) Using 18S RRNA Gene. Res. J. Biotechnol. 2019, 14, 79–87. [Google Scholar]
- Sabia, L.; Di Capua, I.; Percopo, I.; Uttieri, M.; Amato, A. ITS2 in Calanoid Copepods: Reconstructing Phylogenetic Relationships and Identifying a Newly Introduced Species in the Mediterranean. Eur. Zool. J. 2017, 84, 104–115. [Google Scholar] [CrossRef]
- Hill, R.S.; Allen, L.D.; Bucklin, A. Multiplexed Species-Specific PCR Protocol to Discriminate Four N. Atlantic Calanus Species, with an MtCOI Gene Tree for Ten Calanus Species. Mar. Biol. 2001, 139, 279–287. [Google Scholar] [CrossRef]
- Øines, Ø.; Heuch, P.A. Identification of Sea Louse Species of the Genus Caligus Using MtDNA. J. Mar. Biol. Assoc. UK 2005, 85, 73–79. [Google Scholar] [CrossRef]
- Bucklin, A.; Guarnieri, M.; Hill, R.S.; Bentley, A.M.; Kaartvedt, S. Taxonomic and Systematic Assessment of Planktonic Copepods Using Mitochondrial COI Sequence Variation and Competitive, Species-Specific PCR. Hydrobiologia 1999, 401, 239–254. [Google Scholar] [CrossRef]
- Sepahvand, V.; Shahabi, S. First Molecular Evidence for Two New Associate Copepods of Genus Clausidium Kossmann, 1874 (Copepoda: Cyclopoida: Clausidiidae) from the Persian Gulf and Gulf of Oman. Nauplius 2021, 29, 1–8. [Google Scholar] [CrossRef]
- Hamrová, E.; Krajicek, M.; Karanovic, T.; Černý, M.; Petrusek, A. Congruent Patterns of Lineage Diversity in Two Species Complexes of Planktonic Crustaceans, Daphnia longispina (Cladocera) and Eucyclops serrulatus (Copepoda), in East European Mountain Lakes. Zool. J. Linn. Soc. 2012, 166, 754–767. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P. DNA Barcoding Australia’s Fish Species. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Costa, F.O.; DeWaard, J.R.; Boutillier, J.; Ratnasingham, S.; Dooh, R.T.; Hajibabaei, M.; Hebert, P.D.N. Biological Identifications through DNA Barcodes: The Case of the Crustacea. Can. J. Fish. Aquat. Sci. 2007, 64, 272–295. [Google Scholar] [CrossRef]
- da Silva, J.M.; Creer, S.; dos Santos, A.; Costa, A.C.; Cunha, M.R.; Costa, F.O.; Carvalho, G.R. Systematic and Evolutionary Insights Derived from MtDNA COI Barcode Diversity in the Decapoda (Crustacea: Malacostraca). PLoS ONE 2011, 6, e19449. [Google Scholar] [CrossRef]
- Bucklin, A.; Frost, B.W.; Bradford-Grieve, J.; Allen, L.D.; Copley, N.J. Molecular Systematic and Phylogenetic Assessment of 34 Calanoid Copepod Species of the Calanidae and Clausocalanidae. Mar. Biol. 2003, 142, 333–343. [Google Scholar] [CrossRef]
- Alekseev, V.R.; Abramson, N.I.; Sukhikh, N.M. Introduction of Sibling Species to the Ecosystem of the Baltic Sea. Dokl. Biol. Sci. 2009, 429, 694–697. [Google Scholar] [CrossRef]
- Burton, R.S. Hybrid Breakdown in Developmental Time in the Copepod Tigriopus californicus. Evolution 1990, 44, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Parent, G.J.; Plourde, S.; Turgeon, J. Natural Hybridization between Calanus finmarchicus and C. Glacialis (Copepoda) Arct. Northwest Atlantic. Limnol. Ocean. 2012, 57, 1057–1066. [Google Scholar] [CrossRef]
- Chen, C.Y.; Folt, C.L.; Cook, S. The Potential for Hybridization in Freshwater Copepods. Oecologia 1997, 111, 557–564. [Google Scholar] [CrossRef]
- Baek, S.Y.; Jang, K.H.; Choi, E.H.; Ryu, S.H.; Kim, S.K.; Lee, J.H.; Lim, Y.J.; Lee, J.; Jun, J.; Kwak, M.; et al. DNA Barcoding of Metazoan Zooplankton Copepods from South Korea. PLoS ONE 2016, 11, e0157307. [Google Scholar] [CrossRef]
- Bucklin, A.; LaJeunesse, T.C. Molecular Genetic Variation of Calanus pacificus (Copepoda: Calanoida): Preliminary Evaluation of Genetic Structure and Subspecific Differentiation Based on MtDNA Sequences. Calif. Coop. Ocean. Fish. Investig. Rep. 1994, 35, 45–51. [Google Scholar]
- Avise, J.C.; Neigel, J.E.; Arnold, J. Demographic Influences on Mitochondrial DNA Lineage Survivorship in Animal Populations. J. Mol. Evol. 1984, 20, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Grant, W. Shallow Population Histories in Deep Evolutionary Lineages of Marine Fishes: Insights from Sardines and Anchovies and Lessons for Conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Dodson, J.J.; Tremblay, S.; Colombani, F.; Carscadden, J.E.; Lecomte, F. Trans-Arctic Dispersals and the Evolution of a Circumpolar Marine Fish Species Complex, the Capelin (Mallotus villosus). Mol. Ecol. 2007, 16, 5030–5043. [Google Scholar] [CrossRef]
- Rifkin, J.L.; Castillo, A.S.; Liao, I.T.; Rausher, M.D. Gene Flow, Divergent Selection and Resistance to Introgression in Two Species of Morning Glories (Ipomoea). Mol. Ecol. 2019, 28, 1709–1729. [Google Scholar] [CrossRef]
- Payseur, B.A.; Krenz, J.G.; Nachman, M.W. Differential Patterns of Introgression across the X Chromosome in a Hybrid Zone between Two Species of House Mice. Evolution 2004, 58, 2064–2078. [Google Scholar] [CrossRef]
- Teeter, K.C.; Payseur, B.A.; Harris, L.W.; Bakewell, M.A.; Thibodeau, L.M.; O’Brien, J.E.; Krenz, J.G.; Sans-Fuentes, M.A.; Nachman, M.W.; Tucker, P.K. Genome-Wide Patterns of Gene Flow across a House Mouse Hybrid Zone. Genome Res. 2008, 18, 67–76. [Google Scholar] [CrossRef]
- Maroja, L.S.; Andrés, J.A.; Harrison, R.G. Genealogical Discordance and Patterns of Introgression and Selection across a Cricket Hybrid Zone. Evolution 2009, 63, 2999–3015. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.L.; White, T.A.; Ross, C.L.; Harisson, R.G. Gene Flow and the Maintenance of Species Boundaries. Mol. Ecol. 2013, 23, 1668–1678. [Google Scholar] [CrossRef]
- Via, S.; Conte, G.; Mason-Foley, C.; Mills, K. Localizing FST Outliers on a QTL Map Reveals Evidence for Large Genomic Regions of Reduced Gene Exchange during Speciation-with-Gene-Flow. Mol. Ecol. 2012, 21, 5546–5560. [Google Scholar] [CrossRef]
- Rich, S.S.; Bell, A.E.; and Wilson, S.P. Genetic Drift in Small Populations of Tribolium. Evolutuion 1979, 33, 579–584. [Google Scholar] [CrossRef]
- Sautour, B.; Castel, J. Spring Zooplankton Distribution and Production of the Copepod Euterpina acutifrons in Marennes-Oléron Bay (France). Hydrobiologia 1995, 310, 163–175. [Google Scholar] [CrossRef]
- Karabanov, E.B.; Prokopenko, A.A.; Williams, D.F.; Kuzmin, M.I.; Gvozdkov, A.N.; Khursevich, G.K.; Bezrukova, E.V. High-Resolution MIS 11 Record from the Continental Sedimentary Archive of Lake Baikal, Siberia. Geophysical. Monogr. Ser. 2003, 137, 223–230. [Google Scholar]
- Porter, D.; Sriram, K.; Wolfarth, M.; Jefferson, A.; Schwegler-Berry, D.; Andrew, M.E.; Castranova, V. A Biocompatible Medium for Nanoparticle Dispersion. Nanotoxicology 2008, 2, 144–154. [Google Scholar] [CrossRef]
- Eronen, M. A Scrutiny of the Late Quaternary History of the Baltic Sea. In The Baltic Sea: Papers Prepared for a Colloquium on Baltic Sea Marine Geology in Parainen, Finland, 27–29 May 1987; Winterhalter, B., Ed.; Geological Survey of Finland: Espoo, Finland, 1988; Volume 6 of Special paper, pp. 11–18. [Google Scholar]
- Russell, G. Recent Evolutionary Changes in the Algae of the Baltic Sea. Br. Phycol. J. 1985, 20, 87–104. [Google Scholar] [CrossRef]
- Kochanova, E.; Nair, A.; Sukhikh, N.; Väinölä, R.; Husby, A. Patterns of Cryptic Diversity and Phylogeography in Four Freshwater Copepod Crustaceans in European Lakes. Diversity 2021, 13, 448. [Google Scholar] [CrossRef]
- Borutsky, E.S.; Stepanova, L.A.; Kos, M.S. Key to Calanoida Fresh Waters of the USSR; Nauka: Leningrad, Russia, 1991. [Google Scholar]
- Forró, L.; Gaviria Melo, S. Morphological Characterization of New Populations of the Copepod Eurytemora velox (Lilljeborg, 1853) (Calanoida, Temoridae) found in Austria and Hungary. Hydrobiologia 2000, 438, 205–216. [Google Scholar] [CrossRef]
- Bernatchez, L.; Wilson, C.W. Comparative Phylogeography of Neartic and Paleartic Fishes. Mol. Ecol. 1998, 7, 431–452. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, UK, 2000. [Google Scholar]
- Mangerud, J.; Jakobsson, M.; Alexanderson, H.; Astakhov, V.; Clarke, G.K.C.; Henriksen, M.; Hjort, C.; Krinner, G.; Lunkka, J.P.; Möller, P.; et al. Ice-Dammed Lakes and Rerouting of the Drainage of Northern Eurasia during the Last Glaciation. Quat. Sci. Rev. 2004, 23, 1313–1332. [Google Scholar] [CrossRef]
- Millette, K.L.; Xu, S.; Witt, J.D.S.; Cristescu, M.E. Pleistocene-Driven Diversification in Freshwater Zooplankton: Genetic Patterns of Refugial Isolation and Postglacial Recolonization in Leptodora kindtii (Crustacea, Cladocera). Limnol Ocean. 2011, 56, 1725–1736. [Google Scholar] [CrossRef]
- Ermolaeva, N.I. Zooplankton of Different Types of Water Bodies of the Yamal Peninsula in 2015. Sci. Bull. Yamalo-Nenets Auton. Okrug 2016, 2, 56–62. [Google Scholar]
- Abdullina, G.K.; Bondar, M.S. Zooplankton of Water Bodies of the Arctic Tundra of the Yamal Peninsula. In Proceedings of the XXI International Scientific and Practical Conference I, Moscow, Russia, 20 May 2019; pp. 297–304. [Google Scholar]
- Semenova, L.A.; Aleksyuk, V.A. Zooplankton of the Lower Ob. Bull. Ecol. For. Landsc. Sci. 2010, 156–169. (In Russian) [Google Scholar]
| Species Name | CO1 Accession Numbers | nITS Accession Numbers | Sources |
|---|---|---|---|
| E. affinis | MT653564, MT653565, MT653567 | KX401005-KX401023 | [1] |
| HM473958-HM473961, HM473963-HM474010 HM474013, HM474015-HM474018 HM474020-HM474023, HM474025-HM474028 | MT667429-MT667431 | [1] | |
| JF72731-JF727331, JF72734-JF727349 JF727376-JF727395, JF727402-JF727405 JF727410, JF727411, JF727417 JF727423-JF727430, JF727432-JF727446 JF727449-JF727454, JF727496-JF727525 JF727527, JF727529-JF727532, JF727534-JF727548 JF727550-JF727552, JF727555, JF727556 | [12] | ||
| PP968572, PP968573 | Present paper | ||
| E. velox | OR578619-OR578621 MZ373318-MZ373322 | MZ400499-MZ400504 OR583025-OR583031 | [24,25] |
| MT146445, MT146446 | MT787212-MT787214 | [36] | |
| PP971656-PP971674, PP973821 | Present paper | ||
| E. caspica | MN271657-MN271659 | MT667435-MT667438 | [1] |
| E. carolleeae | HM474003, HM474011, HM474012, HM474029 | MT752961-MT752963 | [30], unpublished |
| MT151289-MT151293 | KX401024-KX401038 | [30] | |
| MT653566-MT653568 | MN541395-MN541397 | [1] | |
| E. lacustris | MH316160, MH316161 | MT787215, MT787216 | [25,36] |
| HM474030-HM474035 | [37] | ||
| MN256864, MN256865 | [15] | ||
| E. gracilicauda | MN256855, MN256862 | [15] | |
| MT653572-MT653582 | [1] | ||
| E. americana | MN542728, MN542729, MN542731, MN542733 | [15] | |
| E. arctica | MN256859-MN256861 | MT667432-MT667434 | [1] |
| E. foveola | MN256857, MT653591 | [1] | |
| E. bilobata | MN256858, MT653589, MT653590 | [1] | |
| E. pacifica | AY145427, KR048961- KR048963 | unpublished | |
| E. herdmani | OP876964, MG321072, HQ967229 | [38], unpublished | |
| Temora longicornis | KT209513 | [39] | |
| Centropages typicus | GU132316 | GU125729 | [40] |
| E. affinis | E. velox | E. lacustris | E. gracilicauda | E. caspica | E. carolleeae | |
|---|---|---|---|---|---|---|
| E. affinis | 0.037 | 0.037 | 0.038 | 0.052 | 0.057 | |
| E. velox | 0.931 *** | 0.026 | 0.018 | 0.028 | 0.024 | |
| E. lacustris | 0.930 *** | 0.950 *** | 0.006 | 0.009 | 0.012 | |
| E. gracilicauda | 0.928 *** | 0.964 *** | 0.988 *** | 0.002 | 0.007 | |
| E. caspica | 0.904 *** | 0.947 *** | 0.981 * | 0.994 *** | 0.014 | |
| E. carolleeae | 0.897 *** | 0.953 *** | 0.975 *** | 0.985 *** | 0.971 * |
| Source of Variation | Sum of Squares | Variance Components | % Variation | p-Value | |
|---|---|---|---|---|---|
| A | Among lineages | 588.361 | 4.299 | 80.575 | <0.0001 |
| Among populations within lineages | 22.984 | 0.089 | 1.666 | <0.0001 | |
| Within populations | 191.042 | 0.948 | 17.759 | <0.0001 | |
| Total | 802.387 | 5.336 | 100 | ||
| B | Among lineages | 54.177 | 9.285 | 87.363 | <0.0001 |
| Among populations within lineages | 11.169 | 0.839 | 7.901 | <0.0001 | |
| Within populations | 12.428 | 0.503 | 4.736 | <0.0001 | |
| Total | 77.774 | 10.627 | 100 |
| Populations | Vistula | Vyborg | Riga | Luga | Baltic Sea | Himmerfjarden | Gironde | Loire | Elbe | N. Dvina | Tamar | Scheldt | Seine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vistula | 0.089 | 0.079 | 0 | 0 | 0 | 7.059 | 5.434 | 11.096 | 6.607 | 6.336 | 15.520 | 5.156 | |
| Vyborg | 0.082 | 0.100 | 0.041 | 0.079 | 0.027 | 8.359 | 7.272 | 9.402 | 7.754 | 7.039 | 10.485 | 6.322 | |
| Riga | 0.074 | 0.091 * | 0.012 | 0 | 0.078 | 6.880 | 5.758 | 8.945 | 5.414 | 5.186 | 10.234 | 5.191 | |
| Luga | −0.111 | 0.039 ** | 0.012 | 0 | 0.012 | 1.499 | 1.437 | 1.899 | 0.763 | 0.802 | 1.713 | 1.359 | |
| Baltic Sea | −0.026 | 0.073 * | −0.016 | −0.014 | 0.043 | 5.038 | 4.376 | 6.549 | 2.809 | 2.886 | 6.919 | 3.804 | |
| Himmerfjarden | −0.001 | 0.026 | 0.072 * | 0.012 | 0.041 | 6.045 | 5.313 | 7.195 | 4.850 | 4.548 | 7.692 | 4.727 | |
| Gironde | 0.876 *** | 0.893 *** | 0.873 *** | 0.599 *** | 0.834 *** | 0.858 *** | 0.283 | 15.908 | 11.989 | 9.918 | 17.832 | 9.325 | |
| Loire | 0.845 *** | 0.879 *** | 0.852 *** | 0.590 *** | 0.814 *** | 0.841 *** | 0.221 *** | 12.455 | 8.355 | 6.974 | 13.248 | 7.486 | |
| Elbe | 0.917 *** | 0.904 *** | 0.899 *** | 0.655 | 0.867 *** | 0.878 *** | 0.941 *** | 0.926 *** | 0 | 2.555 | 0.001 | 0.014 | |
| N. Dvina | 0.868 * | 0.886 *** | 0.844 *** | 0.433 | 0.737 * | 0.829 *** | 0.923 ** | 0.893 ** | −0.016 | 2.104 | 0.117 | 0 | |
| Tamar | 0.864 ** | 0.876 *** | 0.838 *** | 0.445 *** | 0.743 * | 0.819 *** | 0.908 *** | 0.874 *** | 0.719 *** | 0.678 * | 3.772 | 1.056 | |
| Scheldt | 0.939 *** | 0.913 *** | 0.911 *** | 0.631 *** | 0.874 *** | 0.885 *** | 0.947 *** | 0.929 *** | 0.001 | 0.104 | 0.790 *** | 0.004 | |
| Seine | 0.837 *** | 0.863 *** | 0.838 *** | 0.576 *** | 0.792 *** | 0.825 *** | 0.903 *** | 0.882 *** | 0.014 | −0.114 | 0.514 *** | 0.004 |
| Populations | Cheboksary | Feofania | Oder | Scheldt | Smolino |
|---|---|---|---|---|---|
| Cheboksary | 0.076 | 0 | 0.217 | 0.024 | |
| Feofania | 0.868 | 0.259 | 0.545 | 0.026 | |
| Oder | 0 | 0.659 | 0.352 | 0.042 | |
| Scheldt | 0.697 ** | 0.479 ** | 0.587 * | 0.021 | |
| Smolino | 0.953 | 0.949 | 0.922 | 0.959 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mouth, C.; Ferreira, F.; Sukhikh, N.; Bou, E.; Bernard, A.; Tackx, M.; Azémar, F.; Meire, P.; Maris, T.; Legal, L. Exploring the Genetic Structure and Phylogeographic Patterns of the Copepod Genus Eurytemora in Europe. Diversity 2024, 16, 483. https://doi.org/10.3390/d16080483
Mouth C, Ferreira F, Sukhikh N, Bou E, Bernard A, Tackx M, Azémar F, Meire P, Maris T, Legal L. Exploring the Genetic Structure and Phylogeographic Patterns of the Copepod Genus Eurytemora in Europe. Diversity. 2024; 16(8):483. https://doi.org/10.3390/d16080483
Chicago/Turabian StyleMouth, Céleste, Flavien Ferreira, Natalia Sukhikh, Elisa Bou, Anaëlle Bernard, Michèle Tackx, Fréderic Azémar, Patrick Meire, Tom Maris, and Luc Legal. 2024. "Exploring the Genetic Structure and Phylogeographic Patterns of the Copepod Genus Eurytemora in Europe" Diversity 16, no. 8: 483. https://doi.org/10.3390/d16080483
APA StyleMouth, C., Ferreira, F., Sukhikh, N., Bou, E., Bernard, A., Tackx, M., Azémar, F., Meire, P., Maris, T., & Legal, L. (2024). Exploring the Genetic Structure and Phylogeographic Patterns of the Copepod Genus Eurytemora in Europe. Diversity, 16(8), 483. https://doi.org/10.3390/d16080483








