Phylogeography of Coccoloba uvifera (Polygonaceae) Sampled across the Caribbean Basin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Lab Procedures
2.2. Sequence Alignment
2.3. Genetic Structure and Genetic Diversity
2.4. Evolutionary Relations and Time of Divergence between Haplotypes
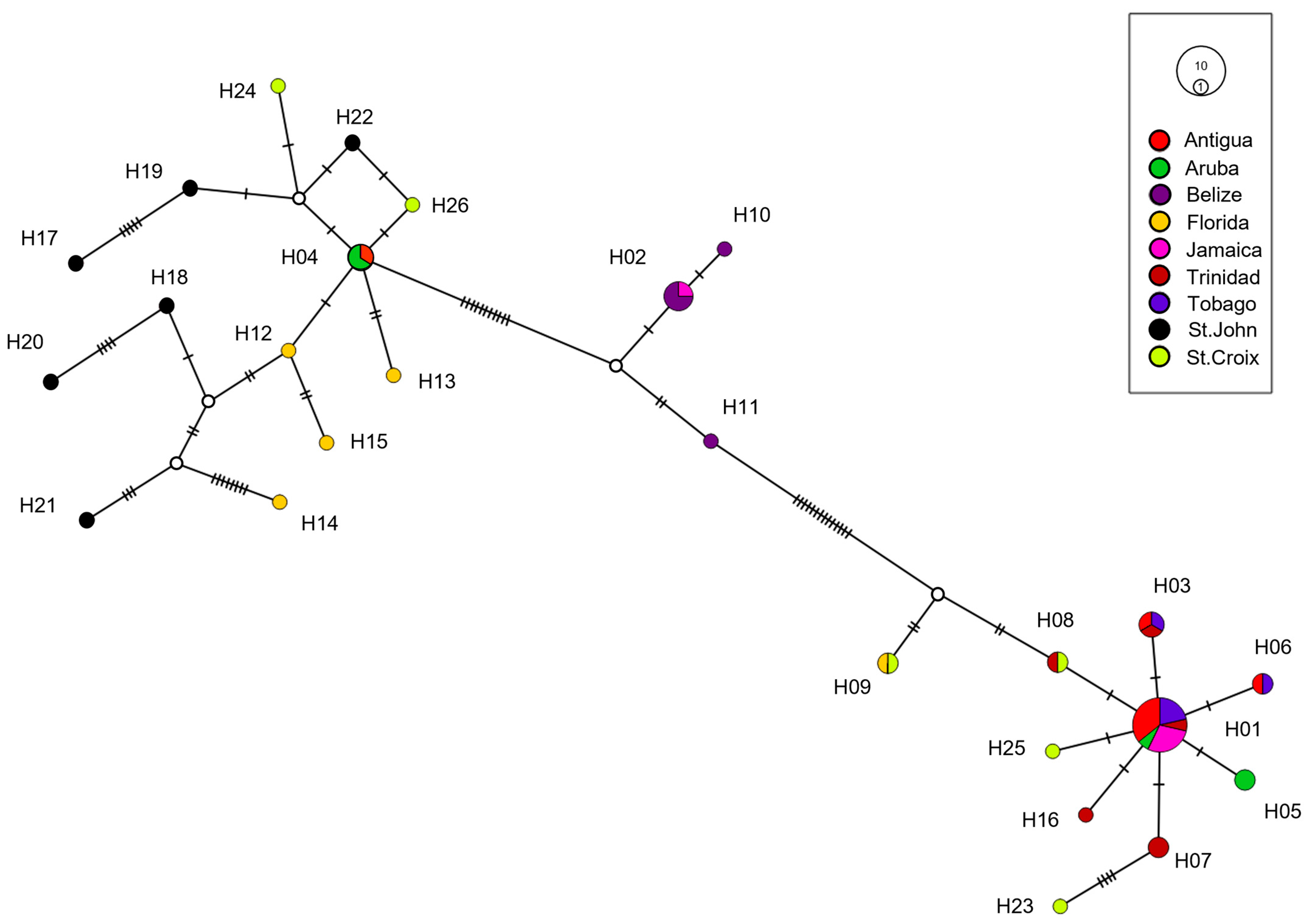
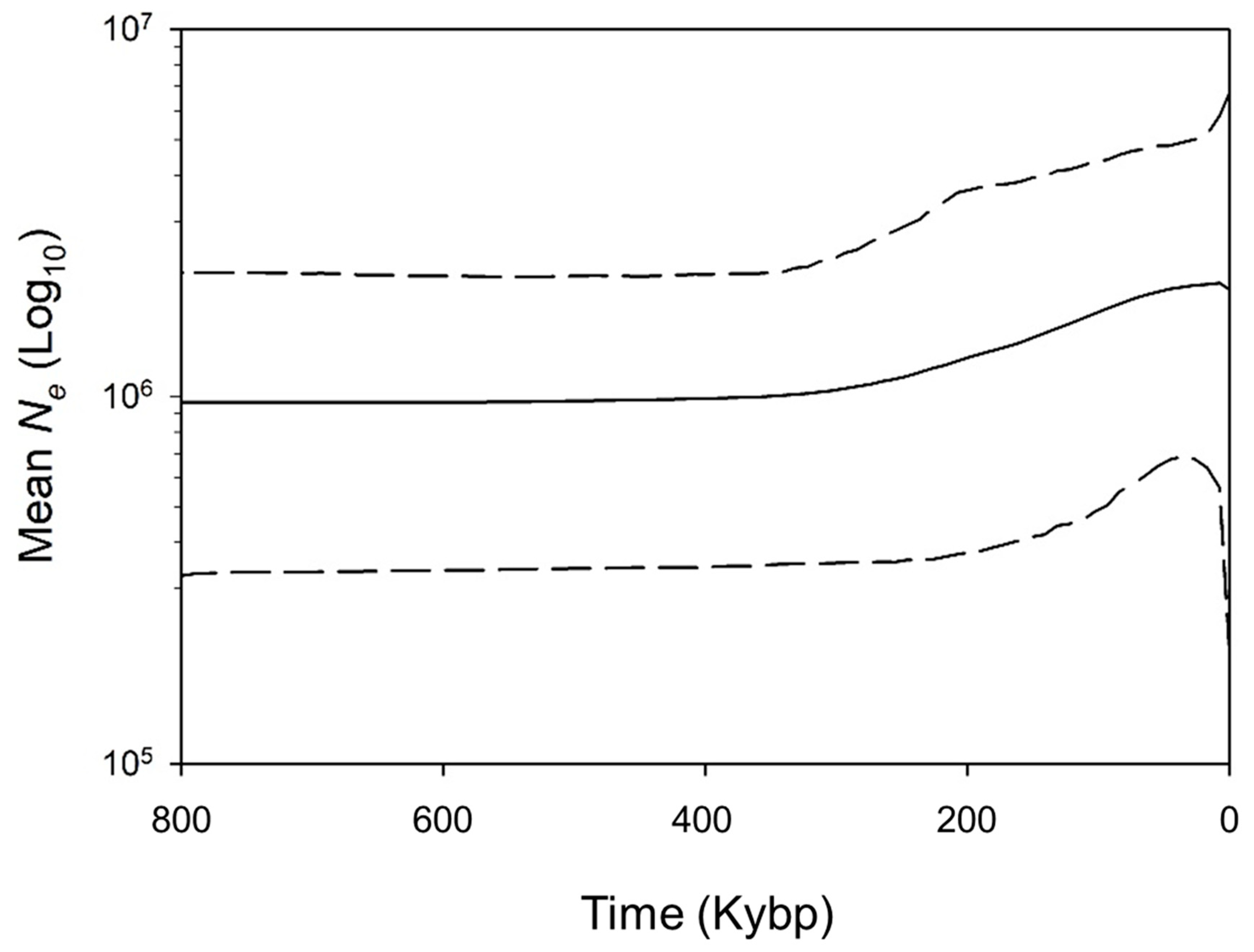
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flora of North America (FNA) Editorial Committee. Flora of North America North of Mexico; Flora of North America (FNA) Editorial Committee: New York, NY, USA; Oxford, UK, 1993; Volume 20. [Google Scholar]
- Parrota, J.A. Coccoloba uvifera (L.) Sea Grape, Uva de Playa; Research Notes SO-ITF-SM-74; U.S. Department of Agriculture, Forest Service, Southern Forest Experimental Station: New Orleans, LA, USA, 1994; 5p. [Google Scholar]
- Howard, R.A. The vegetation of the Antilles. In Vegetation and Vegetational History of Northern Latin America; Graham, A., Ed.; Elsevier Scientific Publishing: Amsterdam, The Netherlands, 1973; pp. 1–38. [Google Scholar]
- Little, E.L.; Wadsworth, F.H. Common Trees of Puerto Rico and the Virgin Islands; Agriculture Handbook #249; Department of Agriculture, Forest Service: Washington, DC, USA, 1964; 20250. [Google Scholar]
- Madriz, R.; Ramirez, N. Biologia reproductive de Coccoloba uvifera (Polygonaceae) una espicie poligamo-dioica. Rev. Biol. Trop. 1996–1997, 44/45, 105–115. [Google Scholar]
- Graham, S.A.; Wood, C.E. The genera of Polygonaceae in the southeastern U.S. J. Arnold Arbor. 1965, 46, 91–121. [Google Scholar] [CrossRef]
- Santiago-Valentin, E.; Olmstead, R.G. Historical biogeography of Caribbean plants: Introduction to current knowledge and possibilities form a phylogenetic perspective. Taxon 2004, 56, 299–319. [Google Scholar] [CrossRef]
- Berry, E.W. The Lower Eocene Floras of Southeastern North America; U. S. Geologic Survey Professional Paper 91; United States Government Publishing Office: Washington, DC, USA, 1916; pp. 1–481, (Coccoloba 212, 213, Pls. 8). [Google Scholar]
- Hollick, A. The Tertiary Flora of Alaska; U. S. Geologic Survey Professional Paper 182; United States Government Publishing Office: Washington, DC, USA, 1936; pp. 1–185, Pls. 1–122. (Coccoloba, 112, 113, Pls. 121, 122). [Google Scholar]
- Graham, A. Studies of Neotropical paleobotany. II. The Miocene communities of Veracruz, Mexico. Ann. Mo. Bot. Gard. 1976, 63, 787–842. [Google Scholar] [CrossRef]
- Koenemann, D.M.; Burke, J.M. A molecular phylogeny for the genus Coccoloba (Polygonaceae) with an assessment of biogeographic patterns. Syst. Bot. 2020, 45, 567–575. [Google Scholar] [CrossRef]
- Howard, R.A. Studies in the genus Coccoloba, Z. New species and a summary of the distribution in South America. J. Arnold Arbor. 1961, 42, 87–95. [Google Scholar] [CrossRef]
- Schuster, T.M.; Setaro, S.D.; Kron, K.A. Age estimates for the buckwheat family Polygonaceae based on sequence data calibrated by fossils and with a focus on the Amphi-Pacific Muehlenbeckia. PLoS ONE 2013, 8, e61261. [Google Scholar] [CrossRef]
- Lugo, A.E. Visible and invisible effects of hurricanes on forest ecosystems: An international review. Austral Ecol. 2008, 33, 368–398. [Google Scholar] [CrossRef]
- Eppinga, M.B.; Pucko, C.A. The impact of hurricanes Irma and Maria on the forest ecosystems of Saba and St. Eustatius, northern Caribbean. Biotropica 2018, 50, 723–728. [Google Scholar] [CrossRef]
- Uriarte, M.; Thompson, J.; Zimmerman, J.K. Hurricane Maria tripled stem breaks and doubled tree mortality relative to other major storms. Nat. Commun. 2019, 10, e1362. [Google Scholar] [CrossRef]
- Brokaw, N.V.; Walker, L.R. Summary of the effects of Caribbean hurricanes on vegetation. Biotropica 1991, 23, 442–447. [Google Scholar] [CrossRef]
- Milbrandt, E.C.; Greenawalt-Boswell, J.M.; Sokoloff, P.D.; Bortone, S.A. Impact and response of southwest Florida mangroves to the 2004 Hurricane season. Estuaries Coasts 2006, 29, 979–984. [Google Scholar] [CrossRef]
- Proffitt, C.E.; Milbrandt, E.C.; Travis, S.E. Red mangrove (Rhizophora mangle) reproduction and seedling colonization after Hurricane Charley: Comparisons of Charlotte Harbor and Tampa Bay. Estuaries Coasts 2006, 29, 972–978. [Google Scholar] [CrossRef]
- Comita, L.S.; Uriarte, M.; Thompson, J.; Jonckheere, I.; Canham, C.D.; Zimmerman, J.K. Abiotic and biotic drivers of seedling survival in a hurricane-impacted tropical forest. J. Ecol. 2009, 97, 1346–1359. [Google Scholar] [CrossRef]
- Ibanez, T.; Keppel, G.; Mankes, C.; Gillespie, T.W.; Lengaigne, M.; Mangeas, M.; Rivas-Torres, G.; Birnbaum, P. Globally consistent impact of tropical cyclones on the structure of tropical and subtropical forests. J. Ecol. 2019, 107, 279–292. [Google Scholar] [CrossRef]
- Paudel, S.; Battaglia, L.L. Linking responses of native and invasive plants to hurricane disturbances: Implications for coastal plant community structure. Plant Ecol. 2021, 222, 133–148. [Google Scholar] [CrossRef]
- Shiflett, S.A.; Backstrom, J.T. Impacts of Hurricane Isaias (2020) on geomorphology and vegetation communities of natural and planted dunes in North Carolina. J. Coast. Res. 2023, 39, 587–609. [Google Scholar] [CrossRef]
- Banks, S.C.; Cary, G.J.; Smith, A.L.; Davies, I.D.; Driscoll, D.A.; Gill, A.M.; Lindenmayer, D.B.; Peakall, R. How does ecological disturbance influence genetic diversity? Trends Ecol. Evol. 2013, 28, 670–679. [Google Scholar] [CrossRef]
- McMahon, K.M.; Evans, R.D.; van Dijk, K.; Hernawan, U.; Kendrick, G.A.; Lavery, P.S.; Lowe, R.; Puotinen, M.; Waycott, M. Disturbance is an important driver of clonal richness in tropical seagrasses. Front. Plant Sci. 2017, 8, e2026. [Google Scholar] [CrossRef]
- Connolly, R.M.; Smith, T.M.; Maxwell, P.S.; Olds, A.D.; Macreadie, P.I.; Sherman, C.D.H. Highly disturbed populations of seagrass show increased resilience by lower genotypic diversity. Front. Plant Sci. 2018, 9, 894. [Google Scholar] [CrossRef]
- Kennedy, J.P.; Dangremond, E.M.; Hayes, M.A.; Preziosi, R.F.; Rowntree, J.K.; Feller, I.C. Hurricanes overcome migration lag and shape intraspecific genetic variation beyond a poleward mangrove range limit. Mol. Ecol. 2020, 29, 2583–2597. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography—The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000; p. 447. [Google Scholar]
- Avise, J.C. Molecular Markers, Natural History, and Evolution, 2nd ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2004; p. 684. [Google Scholar]
- Lopez, L.; Barreiro, R. Patterns of chloroplast DNA polymorphism in the endangered polyploid Centaurea borjae (Asteraceae): Implications for preserving genetic diversity. J. Syst. Evol. 2013, 51, 451–460. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hair IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef]
- Gustafson, D.J.; Major, C.; Jones, D.; Synovec, J.; Baer, S.G.; Gibson, D.J. Genetic Sorting of Subordinate Species in Grassland Modulated by Intraspecific Variation in Dominant Species. PLoS ONE 2014, 9, e91511. [Google Scholar] [CrossRef]
- Burke, J.M.; Koenemann, D.M. The complete annotated plastome sequences of six genera in the tropical woody Polygonaceae. BMC Plant Biol. 2024, 24, 417. [Google Scholar] [CrossRef] [PubMed]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analysis. Syst. Biol. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Muller, K. SeqState—Primer design and sequence statistics for phylogenetic DNA datasets. Appl. Bioinform. 2005, 4, 65–69. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Bouckaert, R. Phylogeography by diffusion on a sphere: Whole world phylogeography. PeerJ 2016, 4, e2406. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.R.; Drummond, A.J. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017, 17, 42. [Google Scholar] [CrossRef]
- Yamane, K.; Yano, K.; Kawahara, T. Pattern and rate of indel evolution inferred from whole chloroplast intergenic regions in sugarcane, maize and rice. DNA Res. 2006, 13, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian Phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Draper, G.; Jackson, T.A.; Donovan, S.K. Geologic provinces of the Caribbean region. In Caribbean Geology; Donovan, S.K., Jackson, T.A., Eds.; The University of West Indies Publishers’ Association (UWIPA): Kingston, Jamaica, 1994; pp. 3–12. [Google Scholar]
- Speed, R.C.; Smith-Horowitz, P.L. The Tobago Terrane. Int. Geol. Rev. 1998, 40, 805–830. [Google Scholar] [CrossRef]
- White, R.V.; Tarney, J.; Kerr, A.C.; Saunders, A.D.; Kempton, P.D.; Pringle, M.S.; Klaver, G.T. Modification of an oceanic plateau, Aruba, Dutch Caribbean: Implications for the generation of continental crust. Lithos 1999, 46, 43–68. [Google Scholar] [CrossRef]
- Hughes, P.D.; Gibbard, P.L.; Ehlers, J. Timing of glaciation during the last glacial cycle: Evaluating the concept of a global ‘Last Glacial Maximum’ (LGM). Earth Sci. Rev. 2013, 125, 171–198. [Google Scholar] [CrossRef]
- Howard, R.A. Studies in the genus Coccoloba, VI. The species from the Lesser Antilles, Trinidad and Tobago. J. Arnold Arbor. 1959, 40, 68–93. [Google Scholar] [CrossRef]
- Negeve, M.N.; Engelhardt, K.A.M.; Gray, M.; Neel, M.C. Calm after the storm? Similar patterns of genetic variation in a riverine foundation species before and after severe disturbance. Ecol. Evol. 2023, 13, e10670. [Google Scholar] [CrossRef] [PubMed]
- Klingbeil, W.H.; Montecinos, G.J.; Alberto, F. Giant kelp genetic monitoring before and after disturbance reveals stable genetic diversity in Southern California. Front. Mar. Sci. 2022, 9, e947393. [Google Scholar] [CrossRef]


| Region | Primer Sequence |
|---|---|
| rpl32-trnL | 5’-TCT CTT TCT ACC GGC AAT TCA-3’ |
| 5’-TCA TAA TTT CAA CAA ACC GAT TAA A-3’ | |
| ndhA intron | 5’-TCG TTG AGG CAT AAA TTT TCC AA-3’ |
| 5’-ACC TCA TAC GGC TCC TCG AG-3’ | |
| rps16-trnK | 5’-TGT ATC ACA GCA AAT TCA ACG AA-3’ |
| 5’-TTC TTG AAA GGG GCG CTC AA-3’ | |
| psbD-trnT | 5’-TCC GAT AAG GGG CTT TTT ACT-3’ |
| 5’-GCA CCT GAC CCA TGA ATT GT-3’ | |
| psbE-petL | 5’-TCA GAC ATG CTC AGC TCC AC-3’ |
| 5’-TTT TGT GAA AGA TAG GAG CGA AA-3’ |
| Statistics. | Antigua | Aruba | Belize | Florida | Jamaica | Trinidad | Tobago | St.John | St.Croix |
|---|---|---|---|---|---|---|---|---|---|
| Sample size | 7 | 5 | 5 | 5 | 5 | 6 | 5 | 7 | 6 |
| No. haplotypes | 3 | 3 | 3 | 5 | 2 | 5 | 3 | 7 | 6 |
| No. of transitions | 1 | 6 | 0 | 13 | 2 | 3 | 1 | 5 | 10 |
| No. of transversions | 0 | 13 | 3 | 19 | 12 | 0 | 0 | 8 | 17 |
| No. of substitutions | 1 | 19 | 3 | 32 | 14 | 3 | 1 | 13 | 27 |
| No. of indels | 1 | 6 | 2 | 7 | 8 | 2 | 1 | 5 | 8 |
| No. of polymorphic sites | 2 | 25 | 5 | 39 | 22 | 5 | 2 | 18 | 35 |
| No. private subst. sites | 0 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 3 |
| Mean No. Pairwise Differences | 0.57 | 15 | 2 | 16.6 | 8.8 | 1.9 | 0.8 | 6.7 | 17.1 |
| pairwise differences (SD) | 0.52 | 8.1 | 1.3 | 8.9 | 4.9 | 1.2 | 0.7 | 3.6 | 8.9 |
| Nucleotide Diversity (p) | 0.000122 | 0.003207 | 0.004280 | 0.003549 | 0.001882 | 0.000399 | 0.000171 | 0.001425 | 0.003663 |
| Nucleotide Diversity (SD) | 0.000128 | 0.002028 | 0.003360 | 0.002235 | 0.001225 | 0.000305 | 0.000170 | 0.000877 | 0.002200 |
| Source of | Sum of | Variance | Percentage | |
|---|---|---|---|---|
| Variation | d.f. | Squares | Components | of Variation |
| Among | ||||
| Populations | 8 | 225.76 | 4.34 Va | 53.93 |
| Within | ||||
| Populations | 42 | 155.61 | 3.71 Vb | 46.07 |
| Total | 50 | 381.37 | 8.04 |
| Latitude | Pairwise | Nucleotide | Haplotypes | Transitions | Transversions | Substitutions | TS-Cat. 5 | Cat. 3-5 | |
|---|---|---|---|---|---|---|---|---|---|
| Differences | Diversity | (TS) | (TV) | (Sub) | |||||
| Latitude | - | 0.520 | 0.520 | 0.251 | 0.648 | 0.660 | 0.679 | 0.862 | 0.400 |
| 0.152 | 0.152 | 0.515 | 0.059 | 0.053 | 0.044 | 0.003 | 0.287 | ||
| 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | ||
| Pairwise Differences | - | 1.000 | 0.304 | 0.881 | 0.972 | 0.972 | 0.242 | 0.095 | |
| <0.001 | 0.426 | 0.002 | <0.001 | <0.001 | 0.53 | 0.809 | |||
| 9 | 9 | 9 | 9 | 9 | 9 | 9 | |||
| Nucleotide Diversity | - | 0.308 | 0.882 | 0.972 | 0.972 | 0.242 | 0.096 | ||
| 0.420 | 0.001 | <0.001 | <0.001 | 0.531 | 0.807 | ||||
| 9 | 9 | 9 | 9 | 9 | 9 | ||||
| Haplotypes | - | 0.572 | 0.283 | 0.403 | 0.351 | 0.430 | |||
| 0.107 | 0.461 | 0.283 | 0.354 | 0.248 | |||||
| 9 | 9 | 9 | 9 | 9 | |||||
| Transitions (TS) | - | 0.856 | 0.941 | 0.416 | 0.127 | ||||
| 0.003 | <0.001 | 0.266 | 0.744 | ||||||
| 9 | 9 | 9 | 9 | ||||||
| Transversions (TV) | - | 0.980 | 0.381 | 0.187 | |||||
| <0.001 | 0.313 | 0.629 | |||||||
| 9 | 9 | 0.9 | |||||||
| Substitution (Sub) | - | 0.407 | 0.171 | ||||||
| 0.277 | 0.66 | ||||||||
| 9 | 9 | ||||||||
| TS-Cat. 5 | - | 0.745 | |||||||
| 0.021 | |||||||||
| 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gustafson, D.J.; Dix, L.A.; Webster, D.P.; Scott, B.K.; Gustafson, I.E.; Farrell, A.D.; Koenemann, D.M. Phylogeography of Coccoloba uvifera (Polygonaceae) Sampled across the Caribbean Basin. Diversity 2024, 16, 562. https://doi.org/10.3390/d16090562
Gustafson DJ, Dix LA, Webster DP, Scott BK, Gustafson IE, Farrell AD, Koenemann DM. Phylogeography of Coccoloba uvifera (Polygonaceae) Sampled across the Caribbean Basin. Diversity. 2024; 16(9):562. https://doi.org/10.3390/d16090562
Chicago/Turabian StyleGustafson, Danny J., Logan A. Dix, Derek P. Webster, Benjamin K. Scott, Isabella E. Gustafson, Aidan D. Farrell, and Daniel M. Koenemann. 2024. "Phylogeography of Coccoloba uvifera (Polygonaceae) Sampled across the Caribbean Basin" Diversity 16, no. 9: 562. https://doi.org/10.3390/d16090562






