Abstract
During the present study, DNA sequence and morphological data were used to delineate species boundaries in the velvet worm, Peripatopsis sedgwicki species complex. The combined mitochondrial cytochrome c oxidase subunit one (COI) and the nuclear 18S rRNA loci were phylogenetically analyzed using Bayesian inference and maximum likelihood platforms that both demonstrated the presence of four, statistically well-supported clades (A–D). In addition, five species delimitation methods (ASAP, bPTP, bGMYC, STACEY and iBPP) were used on the combined DNA sequence data to identify possible novel lineages. All five species delimitation methods supported the distinction of the Fort Fordyce Nature Reserve specimens in the Eastern Cape province, however, in the main P. sedgwicki s.l. species complex, the species delimitation methods revealed a variable number of novel operational taxonomic units. Gross morphological characters were of limited utility, with only the leg pair number in the Fort Fordyce Nature Reserve specimens and the white head-collar of the Van Stadens Wildflower Nature Reserve specimens being diagnostic. The RADseq results from the earlier study of P. sedgwicki s.l. provided highly congruent results with the four clades observed in the present study. The distribution of P. sedgwicki s.s. (clade B) is restricted to the western portions of its distribution in the Afrotemperate forested regions of the Western Cape Province, South Africa. Three novel species, P. collarium sp. nov., (clade C) P. margaritarius sp. nov., (clade A) and P. orientalis sp. nov., (clade D) are described, of which the first two species are narrow range endemics. The present study, along with several recent systematic studies of velvet worms affirms the importance of fine-scale sampling to detect and document the alpha taxonomic diversity of Onychophora.
1. Introduction
Onychophora are ancient euarthropods and that lives in or under decaying logs of wood or stones where they are predators on other invertebrates. Two families occur globally, the southern hemisphere Peripatopsidae Bouvier, 1905, and the circum-tropical Peripatidae Audouin & Milne-Edwards, 1832. In the southern hemisphere velvet worms are present on Gondwanic continental landmasses including Australia, Chile, New-Zealand and South Africa [1]. In South Africa two velvet worm genera, Peripatopsis Pocock, 1904 and Opisthopatus Purcell, 1899 are present [1]. Historically, Peripatopsis, contained seven species [1]. However, most species were poorly defined based on morphological criteria and traditional alpha taxonomic characteristics such as leg pair numbers and colour have been demonstrated to be of limited diagnostic value [2,3,4,5,6,7,8]. Following the first DNA based phylogeny of Peripatopsis, five species complexes were identified; these include P. capensis Grube, 1866, P. clavigera Purcell, 1899, P. balfouri Sedgwick, 1885, P. moseleyi Wood-Mason, 1879 and P. sedgwicki Purcell, 1899 [2]. Fine-scale sampling of the first four species complexes, followed by DNA sequencing of a combination of mitochondrial and nuclear loci, gross morphological and scanning electron microscopy studies (SEM) resulted in the description of 17 new species [3,4,5,6,7,8]. Collectively, these studies resulted in a marked increase in the species diversity of Peripatopsis, and several of the novel species identified are now characterised by narrower geographic distribution ranges.
Of the five aforementioned species complexes, P. sedgwicki has remained devoid of fine-scale systematic scrutiny. Peripatopsis sedgwicki is distributed along the Afrotemperate forest belt of the southern Cape coast of South Africa, from Diepwalle Forest Reserve outside of Knysna in the Western Cape province, to Gqeberha (formerly Port Elizabeth) where the species range extends into the adjacent interior towards Makhanda (formerly Grahamstown) [1] and more recently Fort Fordyce Nature Reserve in the Eastern Cape province, South Africa. Within its distribution range P. sedgwicki primarily occurs in closed canopy Afrotemperate forests, along streams and rivers where it is found under or inside of decaying logs of wood [1; Daniels pers. com.]. Recently [9] revealed the presence of three allopatric clades in the P. sedgwicki species complex and identified specimens from Fort Fordyce as a close relative. A 2024 study [10] using Sanger sequence data for the cytochrome oxidase one subunit (COI) in combination with RADseq data corroborated the presence of four discrete clades (A–D), of which three correspond to novel species. In the main P. sedgwicki species complex, two additional novel lineages could be identified. The latter study [10] did not apply scanning electron microscopy or gross morphological data to delineate the new species. Consequently, the three new species have remained undescribed. During the present study we describe the novel species within the P. sedgwicki species complex with the use of DNA sequence, morphological and SEM.
2. Materials and Methods
2.1. Sample Collection
During the winter of June 2019 a total of 123 Peripatopsis sedgwicki specimens were collected from the southern Cape Afrotemperate and riverine forests in the Western and Eastern Cape Provinces of South Africa [2,9,10] (Figure 1). Specimens of P. sedgwicki, collected and sequenced in three earlier studies [2,9,10] were combined, and formed the basis for the present study. All velvet worm specimens were collected by active searching within saproxylic environments, generally consisting of decaying logs and leaf litter in forested areas. A handheld GPS was used to record the latitude and longitude of sample localities and live specimens were photographed in the field and preserved in absolute ethanol. Vouchers were deposited in the Iziko Museums Natural History Collection Cape Town, South Africa (SAM-ENW-C).
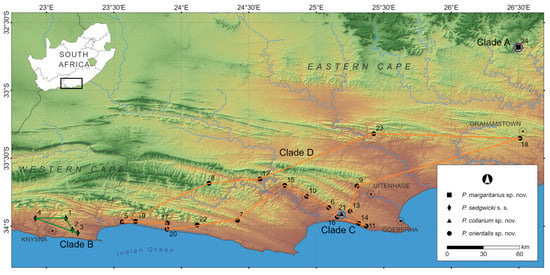
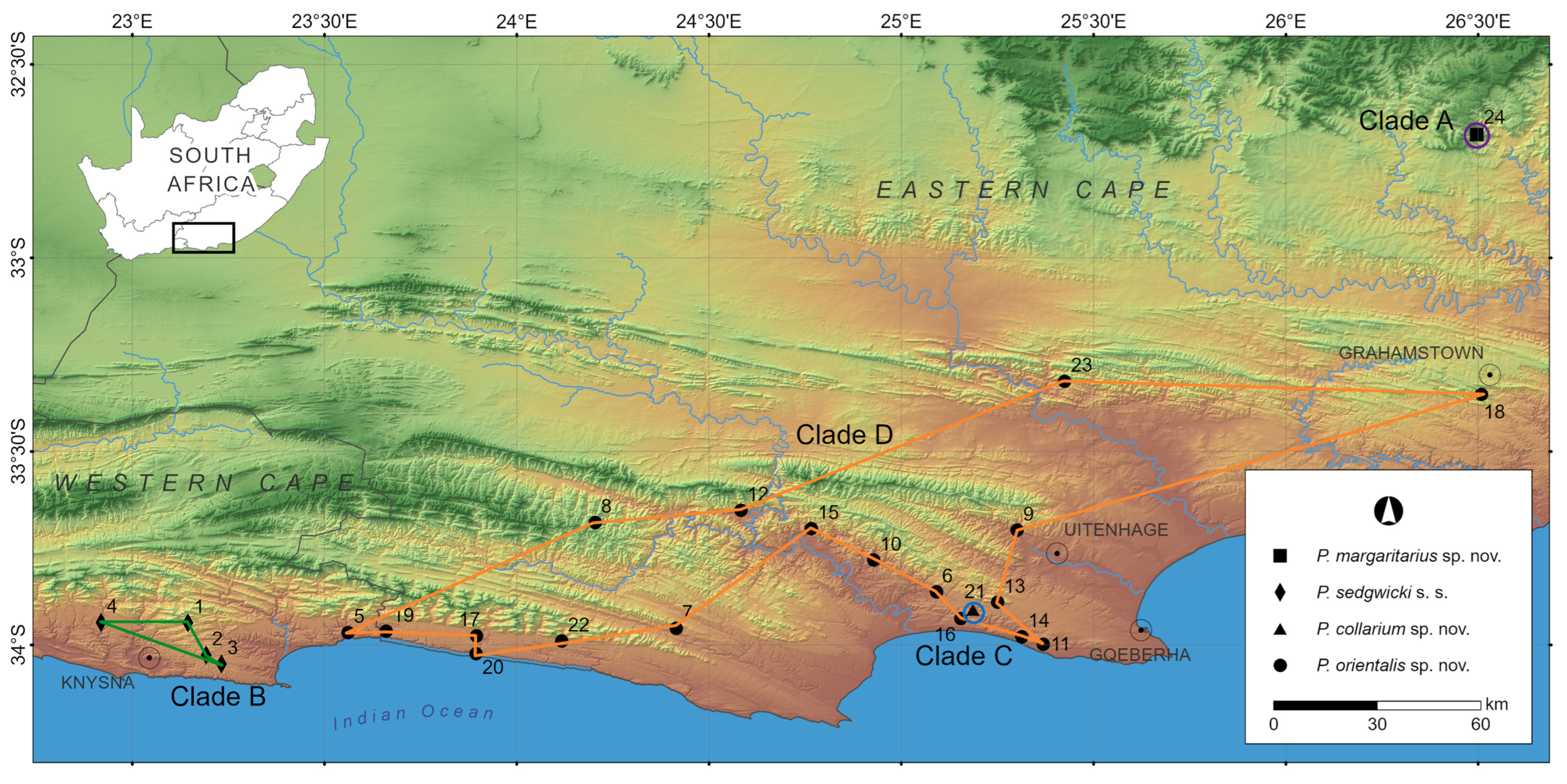
Figure 1.
Map showing localities in the southern Western Cape and Eastern Cape provinces of South Africa indicating the four genetic clades retrieved in the Peripatopsis sedgwicki species complex revealed with the use of Sanger sequences (COI) and RADseq [10]. Localities 1–4 represents the distribution of P. sedgwicki s.s. (clade B); localities 5–20, 22–23 represents the distribution of P. orientalis sp. nov., (clade D) while localities 21 and 24 represents the distribution of the two narrow point endemics, P. collarium sp. nov., (clade C) and P. margaritarius sp. nov., (clade A) respectively. Clades correspond to Figure 2.
2.2. DNA Extraction and Sequencing
Standard DNA extraction was performed on the velvet worm species. The 18S rRNA locus was amplified using the primer pairs from [11]. For details on the PCR conditions and sequencing see [7]. The nuclear DNA marker was combined with the corresponding COI locus from [10]. All phylogenetic analyses were performed on the combined DNA sequence data.
2.3. Phylogenetic Analyses
Forward and reverse strands were used to compute a consensus sequence and to check for base ambiguities using SEQUENCE NAVIGATOR (Applied Biosystems, Foster City, CA, USA). Sequence alignment was computed using CLUSTAL X (ver. 2.1, University College Dublin, Dublin, Ireland, http://www.clustal.org (accessed on 10 January 2024), [12]). The GenBank accession numbers from [2,9,10] were incorporated into the present study. Maximum likelihood (ML) and Bayesian inference (BI) were used to infer phylogenetic relationships. For details on phylogenetic reconstruction using the DNA sequence data see [10]. Uncorrected, ‘p’ distances were calculated for both the COI and 18S rRNA loci using PAUP (vers. 4.0b10) [13].
2.4. Species Delimitation Using ASAP, bPTP, bGMYC and iBPP
The first method employed for the species delimitation comprised the newly developed assemble species by automatic partitioning (ASAP) [14]. ASAP uses genetic distances to hierarchically cluster species partitions (https://bioinfo.mnhn.fr/abi/public/asap (accessed on 10 January 2024)). ASAP first assigns a probability that each new clustering is a new species and then computes the relative width of the barcode gap of a partition in relation to the previous partitions. These metrics are combined into an ASAP score to rank all partitions detected in the analyses. Since ASAP is an exploratory method that does not consider the evolutionary history among sequences, we report the first two partitions ranked by ASAP score, using p-distances and the default setting splitting groups below a probability of <0.01. Additionally, a Bayesian implementation of the poison tree processes (PTP) analysis was run on the online bPTP web server (https://species.h-its.org/ptp/ (accessed on 10 January 2024)) for its ability to delimit species without a priori knowledge of population parameters [15]. The PTP method is based on the differences between sequences, but contrary to the generalised mixed Yule-coalescent (GMYC) models, it does not use a calibrated tree [15]. As input, a maximum-likelihood phylogeny of the complete COI dataset excluding outgroups was used, running the analysis for 500,000 MCMC generations with a thinning value = 100 and burn-in = 0.20. The convergence of the MCMC chain was visually confirmed as recommended by [15].
The mitochondrial dataset was used to hypothesise species limits using the R package bGMYC (vers. 4.3.3) under a Bayesian implementation of the GMYC [16]. To account for error in phylogenetic estimation, 500 post-burn-in trees were randomly selected for analysis. A Markov chain was run for 50,000 generations, sampling the chain every 1000th generation and 4000 generations were discarded as burn-in. A uniform prior for the number of species was applied, with a lower bound of seven (six outgroup taxa and the ingroup) and an upper bound of 195 (the total number of terminals in the analysis). Convergence was assessed visually by examining the performance of the chain. The ‘check rates’ function was used to determine the rate of branching of the coalescent model to that of the Yule model.
Analyses were also conducted using a total-evidence DNA sequence dataset to assess the validity of candidate species identified in the phylogenetic analyses. Due to incongruencies and discordance between analyses using single locus datasets, we employed a multilocus, coalescent model using Species Tree and Classification Estimation, Yarely (STACEY) for its evidential success in species boundaries validation [17,18,19,20]. Said discordances arose from the faster mutation rate of the COI locus, whereby single locus delimitation analyses oversplit putative lineages. This was based on shallow clades corresponding to geographic proximity as opposed to genetic variation at a level significant enough to distinguish between operational taxonomic units (Figure 2). The incorporation of the 18S RNA locus with its slower mutation rate accommodated for this issue. Thus, species tree-estimation and species-delimitation analysis was performed in STACEY (ver. 1.2.1, http://www.beast2.org/ (accessed on 10 January 2024) [21]), in BEAST2, with both the COI and 18S rRNA loci, with one to two samples from each locality sequenced for both markers. The estimated number of putative species in STACEY ranges from one to the number of putative clusters specified. Each locality was defined as a taxon set or minimal cluster, without a priori species definition. The input files (.xml) were created using BEAUti, implementing a Yule Model before estimating the species tree [priors: Collapse Height = 0.0001, Collapse Weight = 0.5 using a beta prior (1.1) around [0.1], bdcGrowthRate = log normal (M = 4.6, S = 1.5); pop-PriorScale = log normal (M = −7, S = 2); relativeDeathRate = beta (alpha = 1.0, beta = 1.0) and an uncorrelated Lognormal Model to describe the relaxed molecular clock. For the species-delimitation analysis, we set the ploidy level for the COI locus to 0.5 and for the 18S rRNA locus to 2.0. Equal ploidy settings for all loci represents a more robust approach by avoiding the disproportionate influence of mitochondrial partial-sequence data and the overestimation of the number of putative species [22]. Equal ploidy levels were attempted but were deemed too conservative for the present study as a lack of delineation was experienced between putative lineages, in contrast to the discordance issue explained at the start of this paragraph. Thus, the mitochondrial partial-sequence influence was incorporated into this analysis by setting varied ploidy levels at a ratio of 1:4 to create an optimal balance between the varied mutation rates of the two loci incorporated. The MCMC analysis was run for 100 million generations, sampling every 1000 generations. The latter parameters were chosen to ensure convergence of the chain while storing as much data as possible within computational limits. The obtained log files were analysed with Tracer to verify convergence (ESS > 200) of the analysis and SpeciesDelimitationAnalyser (vers. 1.0) (http://indriid.com/software.html, (accessed on 10 January 2024) [23]) was used to process the log files and to examine the clusters of species assignments. Posterior probabilities of localities belonging in the same cluster were visualized in a similarity matrix constructed in R Studio (ver. 1.2.5001, RStudio: Integrated Development for R Studio, Inc., Boston, MA, USA. http://www.R-project.org/, R Studio Team 2015 (accessed on 10 January 2024)).
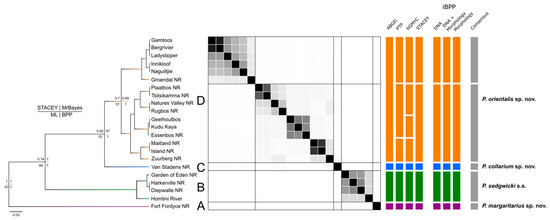
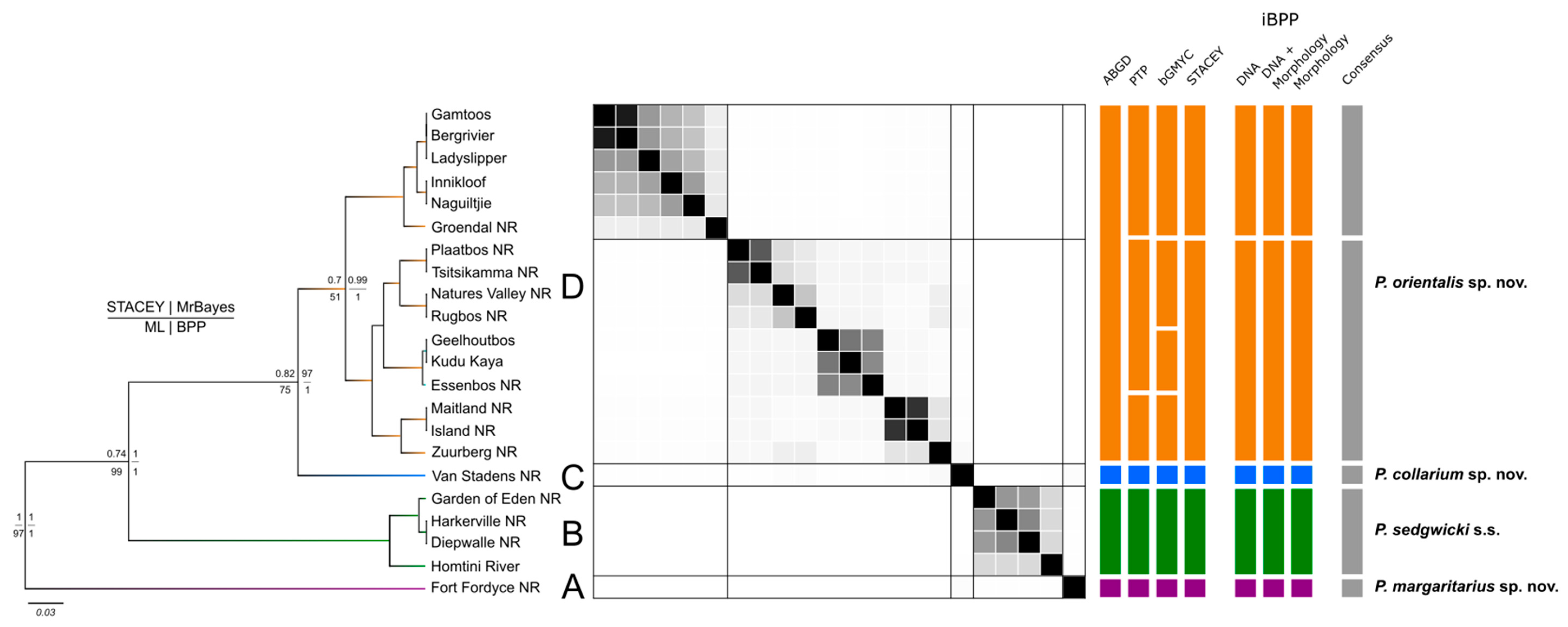
Figure 2.
Species delimitation tree based on COI and 18S. Total evidence species tree of concatenated COI + 18S rRNA sequences for Peripatopsis sedgwicki s.s. produced by the STACEY analysis. Nodal support presentation for each tree produced by each of these analyses follows the of the key on the left of the figure (top left = STACEY; top right = MrBayes [BEAST]; bottom left = Maximum likelihood; bottom right = Bayesian phylogenetics and phylogeography). The similarity matrix represents the results of the STACEY maximum clade credibility tree minimum clusters from the total evidence dataset (COI + 18S rRNA). Black squares represent posterior probabilities (white = 0, black = 1) for pairs of individuals (sample localities) belonging to the same cluster. The lines in the matrix separate putative species boundaries based on the observed clusters. The seven vertical multi-coloured bars represent alternative taxonomies with each segment of these bars representing distinct species according to the respective approach. The final bar (right) represents the species consensus between these methods.
As additional validation to the putative species recovered from previous analyses, both morphological and molecular data were analysed in a Bayesian framework using the program integrative Bayesian Phylogenetics and Phylogeography (iBPP) (ver. 2.1.3, [24]). The program iBPP is an extension of the program Bayesian Phylogenetics and Phylogeography (BPP) [25] and improves the accuracy of species delimitation by integrating quantitative data with molecular data [25]. As with the earlier versions of BPP, iBPP makes use of a user specified guide tree and a prior definition of species boundaries. Species were defined based on the largest number of species estimated from the delimitation results to test which cladogenic events are statistically supported. The guide tree was the topology retrieved from the minimum cluster species tree obtained from STACEY. The analysis was performed with molecular data only (COI + 18S rRNA), molecular and morphological data combined and morphological data only. For the morphological data, dorsal and ventral primary scale rank counts and leg pair numbers were included. The BPP coalescent model is sensitive to model violations and prone to detecting population structure and over splitting species [26]. Therefore, to obtain a more conservative species delimitation when implementing iBPP, the population size (θ) and the divergence time at the root of the tree (τ) were assigned a gamma prior of G (1, 10) and G (2, 2000) respectively. The latter values suggest large ancestral population sizes and shallow divergence times [25,27]. For analyses including morphological data, a default noninformative prior distribution was used for the trait variances and ancestral means V0 = 0 and K0 = 0. Each analysis was performed twice to confirm convergence, implementing Algorithm 0 with ε = 5 for 500,000 post burn-in rjMCMC generations after a burn-in period of 100,000 generations, sampling every 50 generations.
2.5. Gross Morphology and Scanning Electron Microscopy
A Canon EOS 650D DSLR camera (Canon, South Africa) with a Canon EF-S 15–85 mm lens was used to capture images of live specimens to demonstrate colour variation. Gross morphological characters were examined under a Leica MZ 7.5 stereomicroscope (Leica, Germany), specifically noting the following characters: number of leg pairs, dorsal and ventral integument colour, sex and the presence of any unique head structures or colour variations. In addition, using a digital calliper we measured (in millimetres) two standard dimensions of preserved specimens to avoid hydrostatic variability: total length from the anterior most point of the head to the posterior end of the body and the diameter of the body in line with oncopod ten. Specimens underwent analysis of dorsal and ventral integument structure by scanning electron microscopy (SEM) due to the success of these characters as diagnostic features [4,5,6]. Specifically, the scale ranks and shape of the integumentary primary and accessory papillae were examined. Scale ranks were counted from the sensory bristle down to the base of the papilla following the angle formed by the scale rank organisation rather than counting straight down in order to include rows that may not conform to a uniform arrangement. Specimens were dehydrated firstly in absolute ethanol, followed by partial dehydration in a 1:1 solution of hexamethyldisilazane (HMDS) and absolute ethanol for 30 min. Specimens were finally dehydrated in 100% HMDS in a glass petri dish for an additional 30 min, before being left to dry inside a drying oven at 55 °C overnight. Images were captured using a Zeiss MERLIN Field Emission SEM at the Electro Microbeam Unit of Stellenbosch University’s Central Analytical Facility in the Department of Geology. Prior to imaging, the samples were mounted on aluminium stubs with double-sided carbon tape. The samples were then gold coated to 10 nm, using an Edwards (Kolzer, Milan, Italy) S150A Gold Sputter Coater. Beam conditions during surface analysis were 5 kV and approximately 250 nA, with a working distance of 6 mm. In accordance with the ICZN act, the names of the three new species were deposited in ZooBank.
3. Results
3.1. Phylogenetic Analyses of the COI + 18S rRNA Data
A 650 base pair fragment of the 18S rRNA locus was sequenced and combined with a corresponding COI sequence from the same specimen in the Peripatopsis sedgwicki species complex [10]. The new 18S rRNA sequences were deposited in GenBank (Accession numbers MW557345-MW557355; MW557361-MW557369). As with the COI phylogeny [10], the combined COI + 18S rRNA topology yielded four clades (A–D) (Figure 2) congruent with the RADseq results [10]. Fixed base pair differences for the 18S rRNA locus are shown in Table 1. Clades A and C were narrow endemics, and restricted to Fort Fordyce and Van Stadens Wild Flower Nature reserves, and are herein described as P. margaritarius sp. nov. and P. collarium sp. nov., respectively, while clades B and D represent P. sedgwicki s.s. and P. orientalis sp. nov. respectively.

Table 1.
Diagnostic molecular characters in 18S rRNA for distinction of P. orientalis sp. nov., P. collarium sp. nov., P. sedgwicki s.s. and P. margaritarius sp. nov. Numbering refers to the starting base pair for each sequence.
3.2. Species Delimitation Using ASAP, bPTP, bGMYC, STACEY and iBPP
The first two partitions retrieved by ASAP differed minimally in the amount of putative species identified, with the first and second partitions retrieving 13 and 15 putative species, respectively, with partition into two being more sensitive to divergence among localities (Figure 2). The Bayesian implementation of the Poison Tree Processes (bPTP) analysis identified 18 putative species (Figure 2). Among both former analyses, the clear overestimation of species when compared to the phylogenetic results can be attributed to intraspecific diversity whereby population variation both between and within localities was recognised such as at Diepwalle and Groendal NR. However, both P. margaritarius sp. nov., (clade A) and P. collarium sp. nov., (clade C) were retrieved as distinct species (Figure 2).
To delimit species based on the generalised mixed Yule-coalescent (GMYC) output, a threshold needs to be established at which individuals could be considered conspecific. However, as the threshold increases so does the number of conspecifics retrieved which can be skewed by high intraspecific diversity. Thus, selecting a threshold at which to assign putative species status can be troublesome, especially among closely related lineages. The bGMYC results were interpreted at a threshold of p = 0.9–0.95 which retrieved 16 putative species (Figure 2). At this threshold, a similar recognition of putative species was made to that of the bPTP and ASAP analyses. Again, oversplitting was exhibited on the basis of intraspecific diversity, often recognising all specimens from a single locality as a putative species (Figure 2). However, at a threshold of p = 0.05–0.5, four putative species were identified, aligning with the results interpreted from the phylogenetic analyses (Figure 2).
When defining localities as minimum clusters, the multi-locus species-delimitation analysis with Species Tree and Classification Estimation, Yarely (STACEY) performed using the combined COI + 18S rRNA dataset, retrieved five putative species (Figure 2). Upon visual inspection of the similarity matrix (Figure 2), the result of the five clades identifying as independent clusters is well supported (pP > 0.95). The repeated identification of the samples from Van Stadens Wildflower NR as an independent cluster shows supports the novel species designation as P. collarium sp. nov. (Table 1). A second analysis was performed using the five initially identified clusters which confirmed the result of five putative species for this analysis (pP > 0.95).
Under a total evidence approach, the integrative Bayesian Phylogenetics and Phylogeography (iBPP) analysis supported an assignment of five putative species, retrieving the same tree topology as the COI analyses with significant support at each major node (pP > 0.95). This included support for the bifurcation of subclades from the eastern distribution of clade A, offering conflict to designations. The species delimitation based on molecular data only was consistent with the total-evidence approach (pP > 0.95). However, when using the morphological data alone, there was a lack of significant nodal support for the bifurcation of the clade 1 subclades, due to the lack of morphological variation between them, thus showing support for a single species in this regard.
3.3. Morphological Examination and Scanning Electron Microscopy
Gross morphological features including leg pair counts and colour were non-diagnostic for the P. sedgwicki species complex, except for clade A of Fort Fordyce. Leg pair counts varied between 18–20 amongst the three clades (B–D) (Figure 3A–H; Table 2). In P. margaritarius sp. nov, from Fort Fordyce leg pair numbers ranged between 21 and 23 with a mean of 22, making the character diagnostic for clade A (Figure 3A–H; Table 1). Colour among the entirety of the sample set varied stochastically and included shades of yellow to orange, brown, grey, and blue (Table 2). Papillae colour also varied between black, brown, grey, red and orange (Table 2).

Figure 3.
Live images of dorsal and ventral surfaces of the four velvet worm species. Live photographs of a single specimen representative from each of the four clades in the Peripatopsis sedgwicki species complex. Peripatopsis sedgwicki s.s. (A) dorsal and (B) ventral view of live specimen (clade B); P. margaritarius sp. nov., (C) dorsal and (D) ventral view of live specimen (clade A); P. collarium sp. nov., (E) dorsal and (F) ventral image of live specimen (clade C); finally, P. orientalis sp. nov., (clade D) is represented by (G) dorsal and (H) ventral images of live specimens. Scale bar = 10 mm. Photo credit: A. Barnes.

Table 2.
Summary of the morphological characters of specimens in the velvet worm, Peripatopsis sedgwicki s.s. species complex. All measurements are in millimeters (mm). Number of samples = n. The column labelled “D” refers to specimens that could not be sexed. Clade designations follow the phylogenetic designations in Figure 2 based on the combined DNA sequence data (COI + 18S rRNA).
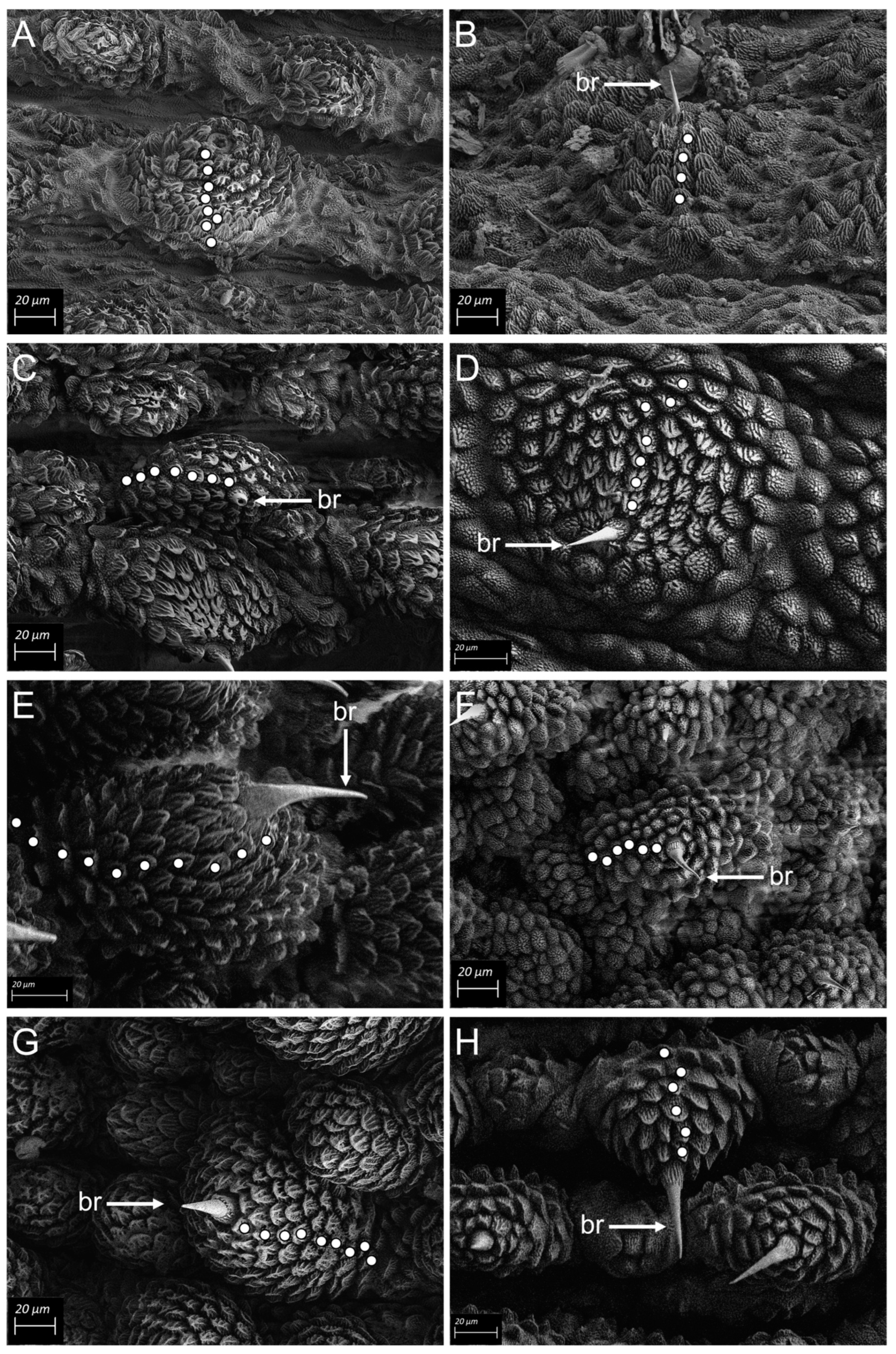
Scanning electron microscopy (SEM) revealed morphological diagnostic differences distinguishing between the three clades of the complex. Peripatopsis orientalis sp. nov., (clade D) exhibited nine scale ranks on the dorsal primary papillae and six scale ranks on the ventral primary papillae, P. collarium sp. nov., (clade C) exhibited ten scale ranks on the dorsal primary papillae and six scale ranks on the ventral primary papillae and specimens in P. sedgwicki s.s. (clade B) eight scale ranks were present on the dorsal primary papillae and four scale ranks on the ventral primary papillae. In P. margaritarius sp. nov., specimens exhibited seven scale ranks on both the dorsal and ventral primary papillae. Dorsal accessory papillae for P. orientalis sp. nov., and P. collarium sp. nov., (clades D and C respectively) were identical at five scale ranks, similarly, the dorsal accessory papillae were identical for P. sedgwicki s.s. (clade B) and the P. margaritarius sp. nov., specimens at four scale ranks. Ventral accessory papillae were identical across all four species (Figure 4A–H).
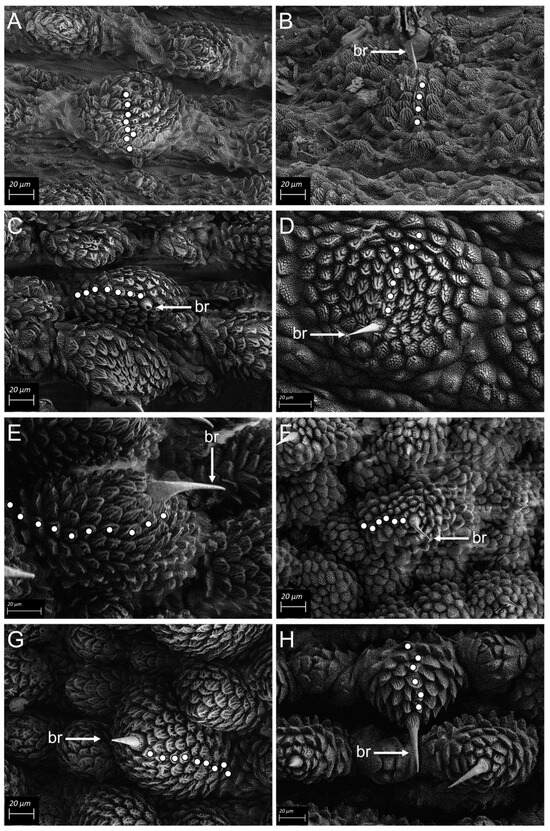
Figure 4.
SEM micrographs Scanning electron micrographs (SEM) of primary and accessory dermal papillae for the four clades in the Peripatopsis sedgwicki species complex. Each white dot represents a scale rank. (A) dorsal papilla and (B) ventral papilla of P. sedgwicki s.s. (clade B). (C) dorsal papilla and (D) ventral papilla of P. margaritarius sp. nov., (clade A). (E) dorsal papilla and (F) ventral papilla of P. collarium sp. nov., (clade C). (G) dorsal papilla and (H) ventral papilla of P. orientalis sp. nov., (clade D). Scale bar = 20 µm.
4. Discussion
Our phylogenetic results based on mitochondrial and nuclear DNA sequence dataset, gross morphology and SEM, together with the species delimitation results, clearly demonstrates the presence of four lineages nested within Peripatopsis sedgwicki species complex, three of which are novel lineages. The latter pattern supported the RADseq sequence study of the P. sedgwicki species complex [10]. However, the 18S rRNA data was less informative in comparison to the RADseq data set, providing clear nuclear DNA data for the corroboration of the three novel lineages within the P. sedgwicki species complex. Furthermore, SEM was used to verify the distinction of these four species and proved successful, by contrast, traditional gross morphological characters such as leg pair number could only be applied with limited success to differentiate P. margaritarius sp. nov., from the remaining three species. Among the three novel species, one is regionally widespread (P. orientalis sp. nov.), two are narrow endemics, P. collarium sp. nov. from Van Stadens Wildflower NR and P. margaritarius sp. nov. from Fort Fordyce Nature, while P. sedgwicki s.s. is now restricted to the eastern most corner of the complex’s distribution, from where the type material was presumably collected in the region of Knysna [1,28]. Our results corroborate the power of fine-scale sampling in deciphering alpha taxonomic diversity in saproxylic organisms such as velvet worms. For example, two recent fine-scale velvet worm studies corroborate the importance of sampling intensity. While sampling the P. birgeri species complex, [10] detected a novel sister species P. polychroma Grobler, Myburgh, Barnes & Daniels, 2023. Similarly, [8] while studying the phylogeography of P. lawrencei, collected a sympatric highly divergent lineage at Oubos in the Western Cape, and described the species as P. aureus Daniels, 2024. Furthermore, these results demonstrate the importance of sampling previously neglected areas in an attempt to document alpha taxonomic diversity among habitat specialists such as velvet worms.
All of the novel species detected in the P. sedgwicki species complex are distributed within conservation areas and managed by three distinct conservation authorities. Most of these areas are managed by the South African National Parks such as for example, Diepwalle, Garden of Eden and Plaatbos NR; Fort Fordyce Nature Reserve is managed by Eastern Cape Parks; and Van Stadens Wildflower Nature Reserve is managed by Nelson Mandela Metropol Municipality. While most of the conservation areas for the species are relatively well-managed and represent a large conservation area when combined, continuous pressure from human developments, both formal and informal, together with a large-scale timber industry in the southern Cape are impacting the fragment sizes of these Afrotemperate forest patches (Daniels pers. obs.). There is very little interconnectivity between several of the conservation areas in these nature reserves, limiting natural migration of these less vagile animals. However, the small size of the Van Stadens Wildflower Nature Reserve where P. collarium sp. nov., occurs as a narrow endemic is of particular concern. The Van Stadens Wildflower Nature Reserve is only 600 ha with alien vegetation encroaching on the small patch of forest contained in the gorge. To date, this is one of the most narrow endemic peripatopsid velvet worm species we have discovered, similar to for example, the P. alba IUCN Red Listed as vulnerable since it is restricted to the Wynberg Cave system on Table Mountains, and P. leonine IUCN Red Listed as extinct in the wild since the species was present on Signal Hill in Cape Town. In the latter two instances the species are listed as critically endangered and extinct, respectively [1]. Similarly, while revising the P. moseleyi species complex, two new species, P. hamerae Ruhberg & Daniels, 2013 and P. storchi Ruhberg & Daniels 2013, both of which are narrow endemics confined to high altitude mountainous regions [6]. While we did not extensively sample along the Afrotemperate forest belt in the Eastern Cape, for P. collarium sp. nov., we never encountered the species again, suggesting the species is highly endemic. Ideally, species should be described based on large sample sizes, however, velvet worm species are frequently exceptionally rare and very difficult to collect, consequently we describe the latter lineage based on limited sample sizes. Similar trends are evident in the literature. For example both, P. hamerae Ruhberg & Daniels, 2013 and P. aereus Daniels, 2024 were described based on two specimens [6,8]. The conservation status of such narrow endemic velvet worm species such as for example, P. collarium sp. nov., need further illumination in future studies, by potentially undertaking more extensive systematic surveys in previously neglected areas such as for example in the region of the Van Stadens Bridge area, a deep gorge may potentially yield additional specimens. We are planning a formal IUCN conservation assessment for all the South African Peripatopsis species, to potentially aid the conservation management of the fauna. We performed a taxonomic revision of the P. sedgwicki s.s. species complex in the present study and describe three novel species.
5. Taxonomy
Phylum Onychophora GRUBE 1853
Family Peripatopsidae BOUVIER 1905
Genus Peripatopsis POCOCK 1894
Peripatopsis sedgwicki PURCELL 1899
Peripatopsis sedgwicki RUHBERG 1985
Holotype: Not designated.
Following ICZN article 75, a specimen from Knysna is herewith designated to be the type (‘neotype’) of Peripatopsis sedgwicki Purcell 1899 since no original type material exists.
5.1. Peripatopsis sedgwicki s.s.
Material Examined. Neotype. SOUTH AFRICA; 1 F; Diepwalle NR, Knysna, Western Cape province; 33°56.576 S, 23°08.670 E; collected 11 December 2006 by S. R. Daniels and H. van den Worm; SAM-ENW-C011102.
Paraneotypes. SOUTH AFRICA; 3 F; Diepwalle NR, Knysna, Western Cape province; 33°56.576 S, 23°08.670 E; collected 11 December 2006 by S. R. Daniels and H. van den Worm; SAM-ENW-C011097.
Additional material examined. SOUTH AFRICA; 2 F and 2 D; Garden of Eden NR, Harkerville Forest, Plettenberg Bay, Western Cape province; 33°56.576 S, 23°08.670 E; collected 11 June 2006 by S. R. Daniels and H. van den Worm; SAM-ENW-C011095. SOUTH AFRICA; 4 F and 4 M; Garden of Eden NR, Harkerville Forest, Plettenberg Bay, Western Cape province; 33°56.576 S, 23°08.670 E; collected 20 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011100. SOUTH AFRICA; 2 F and 3 M; Harkerville NR, Harkerville Forest, Plettenberg Bay, Western Cape province; 33°56.576 S, 23°08.670 E; collected 19 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011096. SOUTH AFRICA; 1 F and 2 D; Homtini River, Rheenendal, Western Cape province; 33°56.510 S, 22°55.120 E; collected, 10 Dec. 2006 by S. R. Daniels and H. van den Worm; SAM-ENW-C011098. SOUTH AFRICA; 3 F, 1 M and 1 D; Homtini River, Rheenendal, Western Cape province; 33°56.510 S, 22°55.120 E; collected, 22 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011101. SOUTH AFRICA; 1 F; Harkerville NR, Harkerville Forest, Plettenberg Bay, Western Cape province; 33°56.576 S, 23°08.670 E; collected by A. Moussalli and D. Stuart Fox; SAM-ENW-C011099.
Description. Measurements. Neotype (F): length: 33.48 mm, other material (F, n = 3): 12–25 mm (Table 2).
Colour and patterning. Dorsal integument varies from orange to grey (fading to dark brown or blue when preserved) with black and orange dermal papillae. No head collar present (Figure 3A,B).
Legs. 20 leg pairs (Table 2).
Integument. Sparsely spaced primary dermal papillae with 1–2 accessory papillae, dome-shaped dermal papillae with eight scale ranks on dorsal papillae (Figure 4A) and moderately spaced conical ventral papillae with five scale ranks (Figure 4B).
Genital opening. Male genital pore cruciform, female genital pore a horizontal and small vertical slit.
GenBank. COI: Diepwalle C1–C4: EU855253-EU855250 [2]; Garden of Eden C1–C4: EU855373-EU855370 [2]; Garden of Eden 1–8: MW519067-MW519074 [10]; Harkerville 1–5: MW519075-MW519079 [10]; Homtini River C1–C3: EU855266-EU855264 [2]; Homtini River 1–5: MW519080-MW519084 [10].
18S rRNA: Diepwalle C1: EU855552 [2]; Garden of Eden C1: EU855553 [2]; Harkerville: EU855559 [2]; Harkerville 5: MW557369 [present study]; Homtini: EU855558 [2].
Distribution. Narrow range endemic occurring within the Afrotemperate forests of Diepwalle NR, Harkerville NR and at Homtini River, Western Cape province, South Africa (Figure 1, localities 1–4).
Habitat. Found under or inside decaying indigenous logs of wood or in leaf-litter.
5.2. Peripatopsis margaritarius sp. nov.
urn:lsid:zoobank.org:act:79F50F8F-3A74-415D-A8DE-854D356B45F4
Material examined. Holotype. SOUTH AFRICA; 1 F; Fort Fordyce NR, Eastern Cape province; 32°40.877 S, 26°29.787 E; collected between 31 March and 3 April 2016 by S. R. Daniels; SAM-ENW-C011105.
Paratypes. SOUTH AFRICA; 13 F, 8 M and 4 D; Fort Fordyce NR, Eastern Cape province; 32°40.877 S, 26°29.787 E; collected between 31 March and 3 April 2016 by S. R. Daniels; SAM-ENW-C011505.
Diagnosis. Seven scale ranks on the dorsal and ventral primary papillae (Figure 4C,D). Fixed base pair differences derived from the 18S rRNA marker were retrieved (Table 1).
Description. Measurements. Holotype (F): length: 31.61 mm, other material (F, n = 13; M, n = 8; D, n = 4): 17–47 mm.
Colour and patterning. Dorsal integument varies from dark brown to black (remaining these colours or fading to darker shades when preserved) with black dermal papillae. No head collar present (Figure 3C,D).
Legs. 22–23 leg pairs (Table 2).
Integument. Moderately spaced primary dermal papillae with 1–2 accessory papillae, pyramidal or dome-shaped dermal papillae with seven scale ranks on dorsal papillae (Figure 4C) and moderately spaced, low-rise pyramidal ventral papillae with seven scale ranks (Figure 4 D).
Genital opening. Male genital pore cruciform, female genital pore a horizontal and small vertical slit.
GenBank. COI: Fort Fordyce C1–27: MF327491—MF327465 [9].
18S rRNA: Fort Fordyce C1: MF327500 [9].
Etymology. Named in honour of the late Prof. Margaretha Hofmeyr who lectured the senior author and who was a passionate biologist.
Distribution. Point endemic found exclusively to Fort Fordyce Nature Reserve, a high altitude plateau in the Eastern Cape province, South Africa (Figure 1, endemic to locality 24).
Habitat. Found under or inside decaying indigenous logs of wood in Afrotemperate forest patches.
Remarks. P. margaritarius sp. nov. is genetically discrete from P. sedgwicki s.s. exhibiting an 8.08% sequence divergence using the COI locus [10].
5.3. Peripatopsis orientalis sp. nov.
urn:lsid:zoobank.org:act:55416D4B-CA3C-438A-BE0B-014964737AC5
Material examined. Holotype. SOUTH AFRICA; 1 F; Nature’s Valley NR, Nature’s Valley, Eastern Cape province; 33°58.070 S, 23°33.643 E; collected 19 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011107.
Paratypes. SOUTH AFRICA; 2 F, 2 M and 2 J; Nature’s Valley NR, Nature’s Valley, Eastern Cape province; 33°58.070 S, 23°33.643 E; collected, 19 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011492.
Additional material examined. SOUTH AFRICA; 5 F; Rugbos NR, Eastern Cape province; 33°57.894 S, 23°39.635 E; collected, 13 July 2006 by S. R. Daniels and H. van den Worm; SAM-ENW-C011494. SOUTH AFRICA; 4 F and 1 M; Nature’s Valley NR, Nature’s Valley, Eastern Cape province; 33°58.070 S, 23°33.643 E; collected 12 July 2006 by S. R. Daniels and H. van den Worm; SAM-ENW-C013542. SOUTH AFRICA; 3 F and 2 J; Essenbos Farm, Kareedouw, Eastern Cape province; 33°57.320 S, 25°36.590 E; collected 12 Dec. 2006 by S.R. Daniels and H. van den Worm; SAM-ENW-C011504. SOUTH AFRICA; SOUTH AFRICA; 3 F and 3 J; Rivendell Farm, Makhanda (formerly Grahamstown), Eastern Cape province; 33°21.200 S, 26°30.490 E; collected by S. R. Daniels, N. Solomons and H. Ruhberg; SAM-ENW-C011108. SOUTH AFRICA; 1 F and 2 M; Rivendell Farm, Makhanda (formerly Grahamstown), Eastern Cape province; 33°21.200 S, 26°30.490 E; collected 8 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011501. SOUTH AFRICA; 5 F, 1 M and 2 D; The Island NR, Gqeberha (formerly Port Elizabeth), Eastern Cape province; 33°59.900 S, 25°22.170 E; collected 11 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011500. SOUTH AFRICA; 6 F and 3 M; Maitland NR, Port Elizabeth, Eastern Cape province; 33°58.722 S, 25°18.801 E; collected 12 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011499. SOUTH AFRICA; 2 F and 4 M; Gamtoos River Mouth, Eastern Cape province; 33°55.877 S, 25°09.263 E; collected 12 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011495. SOUTH AFRICA; 6 F and 4 M; Innikloof, Hankey, Eastern Cape province; 33°46.856 S, 24°55.701 E; collected, 13 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011496. SOUTH AFRICA; 4 M; Groendal NR, Eastern Cape province; 33°42.207 S, 25°18.100 E; collected 14 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011498. SOUTH AFRICA; 1 F and 7 M; Bergrivier Eco Retreat, Thornhill, Eastern Cape province; 33°51.804 S, 25°05.468 E; collected 15 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011502. SOUTH AFRICA; 7 F, 2 M and 1 J; Naguiltjie, Patensie, Eastern Cape province; 33°41.975 S, 24°45.927 E; collected 16 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C013540. SOUTH AFRICA; 5 F and 3 M; Rugbos NR, Eastern Cape province; 33°57.894 S, 23°39.635 E; collected, 17 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C013544. SOUTH AFRICA; 7 F and 3 M; Essenbos Farm, Kareedouw, Eastern Cape province; 33°57.320 S, 25°36.590 E; collected 18 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C013543. SOUTH AFRICA; 3 F and 2 M; Plaatbos NR, Storms River, Eastern Cape province; 33°58.599 S, 23°53.705 E; collected, 18 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011503. SOUTH AFRICA; 1 F and 5 M; Tsitsikamma NR, Storms River Mouth, Storms River, Eastern Cape province; 34°01.380 S, 23°53.624 E; collected, 18 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011497. SOUTH AFRICA; 4 F and 2 M; Kudu Kaya, Baviaanskloof NR, Eastern Cape province; 33°39.098 S, 24°34.960 E; collected 4 August 2019 by A. Barnes and T. Busschau; SAM-ENW-C011493. SOUTH AFRICA; 1 F; 2 F and 3 M; Witelsbos State Forest, Witelsbos, Eastern Cape province; 33°59.240 S, 24°06.590 E; collected 17 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C013541.
Description. Measurements. Holotype (F): length: 28.24 mm, other material (F, n = 2; M, n = 2; J, n = 2): 9–33 mm (Table 2).
Colour and patterning. Dorsal integument varies from dark blue to grey to orange (fading to dark brown when preserved) with black and orange dermal papillae. No head collar present (Figure 3G,H).
Legs. 20 leg pairs (Table 2).
Integument. Highly dense primary dermal papillae with 1–2 accessory papillae, conical or dome-shaped dermal papillae with nine scale ranks on dorsal papillae (Figure 4G) and densely spaced conical or pyramidal ventral papillae with nine scale ranks (Figure 4H).
Genital opening. Male genital pore cruciform, female genital pore a horizontal and small vertical slit.
GenBank. COI: Bergrivier 1–8: MW518961-MW518968 [10]; Essenbos Farm C1–C5: EU855256-EU855260 [2]; Essenbos Farm: 1–9: MW519040-MW519048 [10]; Gamtoos 1–6: MW518998-MW519002 [10]; Geelhoutbos 1: MW519021 [10]; Groendal 1–4: MW518969-MW51972 [10]; The Island Nature Reserve, Gqeberha (formerly Port Elizabeth) C1–C3: EU855369-EU855367 [2]; The Island Nature Reserve 1–6: MW519003-MW519008; Innikloof MW518973-MW518982 [10]; Kudu Kaya 1–6: MW519023-MW519028 [10]; Ladyslipper C1: EU855366 [2]; Maitland 1–9: [10]; Naguiltjie 1–10: MW518983-MW518992 [10]; Nature’s Valley NR: 1–7: MW519049-MW519055 [10]; Nature’s Valley NR: C1–C5: EU855386-EU855382 [2]; Plaatbos 1–5: MW519029-MW519033 [10]; Rivendell Farm, Makhanda (formerly Grahamstown) C1–C6: MF327497-MF327492 [9]; Rivendell Farm 1–3: MW519018-MW519020 [10]; Rugbos C1–C5: EU855381-EU855377 [2]; Rugbos 1–9: MW519056-MW519063 [10]; Tsitsikamma 1–6: MW519034-MW519039 [10]; Witelsbos 1–5: MW518993-MW518997 [10]; Zuurberg C1: MF327498 [9]; Zuurberg 1: MW519022 [10].
18S rRNA: Bergrivier 1–2: MW557361-MW557362 [present study]; Essenbos C1: EU855549 [2]; Gamtoos 1–2: MW557366-MW557367 [present study]; Geelhoutbos 1: MW557363 [present study]; Groendal 1–2: MW557364-MW55735 [present study]; Innikloof 1–2: MW557346, MW557348 [present study]; Kudu Kaya 5: MW557348 [present study]; Ladyslipper: EU855551 [2]; Maitland 1–2: MW557349-MW557350 [present study]; Naguiltjie 1–2: MW557351-MW557352 [present study]; Nature’s Valley C1: EU855547 [2]; Plaatbos 1–2: MW557353-MW557354 [10]; Rugbos C1: EU855548 [2]; Tsitsikamma 3: MW557355; The Island Nature Reserve, Gqeberha (formerly Port Elizabeth) C1: EU855550; Zuurberg C1; MF327499 [9].
Etymology. Name prescribed on the basis that this species holds the most eastern distribution in the complex.
Distribution. Broad distribution inhabiting forest patches ranging from Nature’s Valley in the West to Grahamstown in the east (Figure 1).
Habitat. Found under or inside decaying indigenous logs of wood.
Remarks. P. orientalis sp. nov. is genetically discrete from P. sedgwicki s.s. exhibiting a 7.03% sequence divergence using the COI locus [10].
5.4. Peripatopsis collarium sp. nov.
urn:lsid:zoobank.org:act:832D5A0E-2F89-4E2A-8677-1A221B70F06F
Material examined. Holotype. SOUTH AFRICA; 1 F; Van Stadens Wildflower NR, Gqeberha, (formerly Port Elizabeth), Eastern Cape province; 33°54.430 S, 25°11.150 E; collected, 11 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011103.
Paratypes. SOUTH AFRICA; 1 F and 1 M; Van Stadens Wildflower NR, Gqeberha, (formerly Port Elizabeth), Eastern Cape province; 33°54.430 S, 25°11.150 E; collected, 11 June 2019 by A. Barnes and S. R. Daniels; SAM-ENW-C011104.
Diagnosis. Prominent, white head collar (Figure 3E,F), 10 scale ranks on the dorsal primary papillae (Figure 4E).
Description. Measurements. Holotype (F): length: 28.93 mm, other material (F, n = 1; M, n = 1): 26–29 mm.
Colour and patterning. Dorsal integument varies from slate grey and blue-grey to orange (fading to dark purple when preserved) with black dermal papillae. White head collar present in all individuals (Figure 3E,F).
Legs. 20 leg pairs (Table 2).
Integument. Primary dermal papillae with 1–2 accessory papillae, dome-shaped dermal papillae with ten scale ranks on dorsal papillae (Figure 4E) and low pyramidal shaped ventral papillae with concentrated scale ridges with six scale ranks (Figure 4F).
Genital opening. Male genital pore cruciform, female genital pore a horizontal and small vertical slit.
GenBank. COI: Van Stadens Wildflower NR 1–3: MW519064-MW519066 [10].
18S rRNA: Van Stadens Wildflower NR 1: MW557368 [present study].
Etymology. Name given due to the white collar found around the head in all individuals. A white collar around the head has also been found to be diagnostic for P. birgeri Ruhberg & Daniels 2013.
Distribution. Point endemic found exclusively at Van Stadens Wildflower NR, Eastern Cape province, South Africa (Figure 1, restricted to locality 21).
Habitat. Found under or inside decaying indigenous logs of wood.
Remarks. P. collarium sp. nov. is genetically distinct from P. sedgwicki s.s. exhibiting a 5.60% sequence divergence using the COI locus [10].
Author Contributions
A.B. and S.R.D. undertook the sampling for the project. S.R.D. conceived the idea for the project and provided the financial resources. A.B. produced the initial draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable for invertebrates.
Data Availability Statement
All genetic data were submitted to GenBank (accession numbers provided).
Acknowledgments
SANParks are thanked for permits that allowed the collection of the velvet worm specimens used in the present study. The University of Stellenbosch is thanked for logistic support. Aisha Mayekiso of the Iziko Museums of Cape Town is thanked for accessioning the specimens into the museum collection.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| SAM ENW | South African Museum |
| Cape Town | Entomological Collection (Iziko Museums of Cape Town) |
| SEM | scanning electron microscopy |
| COI | cytochrome c oxidase subunit I |
| M | male specimens |
| F | female specimens |
| J | juvenile specimens |
| D | Indiscernible |
References
- Hamer, M.L.; Samways, M.J.; Ruhberg, H. A review of the Onychophora of South Africa, with discussion of their conservation. Ann. Natl. Mus. 1997, 38, 283–312. [Google Scholar]
- Daniels, S.R.; Picker, M.D.; Cowlin, R.M.; Hamer, M.L. Unravelling evolutionary lineages among South African velvet worms (Onychophora: Peripatopsis) provides evidence for widespread cryptic speciation. Biol. J. Lin. Soc. 2009, 97, 200–216. [Google Scholar] [CrossRef]
- Barnes, A.; Reiss, T.; Daniels, S.R. Systematics of the Peripatopsis clavigera species complex (Onychophora: Peripatopsidae) reveals cryptic cladogenic patterning, with the description of five new species. Invert. Syst. 2020, 34, 569–590. [Google Scholar] [CrossRef]
- Daniels, S.R.; McDonald, D.E.; Picker, M.D. Evolutionary insight into the Peripatopsis balfouri sensu lato species complex (Onychophora: Peripatopsidae) reveals novel lineages and zoogeographic patterning. Zool. Scri. 2013, 42, 656–674. [Google Scholar] [CrossRef]
- McDonald, D.E.; Ruhberg, H.; Daniels, S.R. Two new Peripatopsis species (Onychophora: Peripatopsidae) from the Western Cape province, South Africa. Zootaxa 2012, 3380, 55–68. [Google Scholar] [CrossRef]
- Ruhberg, H.; Daniels, S.R. Morphological assessment supports the recognition of four novel species in the widely distributed velvet worm Peripatopsis moseleyi (Onychophora: Peripatopsidae). Invert. Syst. 2013, 27, 131–145. [Google Scholar] [CrossRef]
- Grobler, P.C.J.; Myburgh, A.M.; Barnes, A.; Daniels, S.R. Integrative taxonomy provides evidence for a cryptic lineage in the velvet worm Peripatopsis birgeri species complex (Onychophora: Peripatopsidae) in KwaZulu-Natal, South Africa. Syst. Biodiv. 2023, 21, 2207574. [Google Scholar] [CrossRef]
- Lawrence, J.A.N.; Daniels, S.R. Sample design in biodiversity studies matters: A fine-scale study of Lawrence’s velvet worm, Peripatopsis lawrencei (Onychophora: Peripatopsidae), reveals hidden diversity. Invert. Syst. 2024, 38, IS23051. [Google Scholar] [CrossRef]
- Daniels, S.R.; Dreyer, M.; Sharma, P.P. Contrasting the population genetic structure of two velvet worm taxa (Onychophora: Peripatopsidae: Peripatopsis) in forest fragments along the south-eastern Cape, South Africa. Invert. Syst. 2017, 31, 781–796. [Google Scholar] [CrossRef]
- Myburgh, A.; Barnes, A.; Henriques, R.; Daniels, S.R. Congruent patterns of cryptic cladogenesis revealed using RADseq and Sanger sequencing in a velvet worm species complex (Onychophora: Peripatopsidae: Peripatopsis sedgwicki). Mol. Phylogenet. Evol. 2024, 198, 108132. [Google Scholar] [CrossRef]
- Giribet, G.; Carranza, S.; Baguñà, J.; Riutort, M.; Ribera, C. First molecular evidence for the existence of a Tardigrada + Arthropoda clade. Mol. Biol. Evol. 1996, 13, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acid Res 1997, 24, 4876–4882. [Google Scholar] [CrossRef]
- Swofford, D. PAUP* Phylogenetic Analysis Using Parsimony (and Other Methods); Version 4.10; Illinois Natural History Survey: Champaigne, IL, USA, 2002. [Google Scholar]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Res. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A.A. general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Reid, N.M.; Carstens, B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed yule-coalescent model. BMC Evol. Biol. 2012, 12, 196. [Google Scholar] [CrossRef]
- Jacobs, S.J.; Kristofferson, C.; Uribe-Convers, S.; Latvis, M.; Tank, D.C. Incongruence in molecular species delimitation schemes: What to do when adding more data is difficult. Mol. Ecol. 2018, 27, 2397–2413. [Google Scholar] [CrossRef]
- Tomasello, S. How many names for a beloved genus?—Coalescent-based species delimitation in Xanthium L. (Ambrosiinae, Asteraceae). Mol. Phylogenet. Evol. 2018, 127, 135–145. [Google Scholar] [CrossRef]
- Busschau, T.; Conradie, W.; Daniels, S.R. Evidence for cryptic diversification in a rupicolous forest-dwelling gecko (Gekkonidae: Afroedura pondolia) from a biodiversity hotspot. Mol. Phylogenet. Evol. 2019, 139, 106549. [Google Scholar] [CrossRef]
- Klimov, P.B.; Skoracki, M.; Bochkov, A.V. Cox 1 barcoding versus multilocus species delimitation: Validation of two mite species with contrasting effective population sizes. Paras. Vect. 2019, 12, 8. [Google Scholar] [CrossRef]
- Jones, G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J. Math. Biol. 2017, 74, 447–467. [Google Scholar] [CrossRef]
- Vitecek, S.; Kucinic, M.; Previšić, A.; Živic, I.; Stojanovic, K.; Keresztes, L.; Bálint, M.; Hoppeler, F.; Waringer, J.; Graf, W.; et al. Integrative taxonomy by molecular species delimitation: Multi-locus data corroborate a new species of Balkan Drusinae micro-endemics. BMC Evol. Biol. 2017, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Aydin, Z.; Oxelman, B. DISSECT: An assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 2015, 31, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Solís-Lemus, C.; Knowles, L.L.; Ané, C. Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution 2015, 69, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Rannala, B. Bayesian species delimitation using multilocus sequence data. Proc. Nat. Acad. Sci. USA 2010, 107, 9264–9269. [Google Scholar] [CrossRef]
- Barley, A.J.; Brown, J.M.; Thomson, R.C. Impact of model violations on the inference of species boundaries under the multispecies coalescent. Syst. Biol. 2018, 67, 269–284. [Google Scholar] [CrossRef]
- Parmakelis, A.; Kotsakiozi, P.; Stathi, I.; Poulikarakou, S.; Fet, V. Hidden diversity of Euscorpius (Scorpiones: Euscorpiidae) in Greece revealed by multilocus species- delimitation approaches. Biol. J. Linn. Soc. 2013, 110, 728–748. [Google Scholar] [CrossRef][Green Version]
- Ruhberg, H. Die Peripatopsidae (Onychophora). Systematik, Ökologie, Chorologie und phylogenetische Aspekte. Zoologica 1985, 46, 1–183. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).