Extinct or Not? Confirming the “Extinct” Status of Hieracium tolstoii (Asteraceae) with Integrated Taxonomic Investigation
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphometric Analysis
2.2. Statistical Analysis
2.3. DNA Extraction
2.4. Markers Amplification
2.5. Phylogenetic Analysis
3. Results
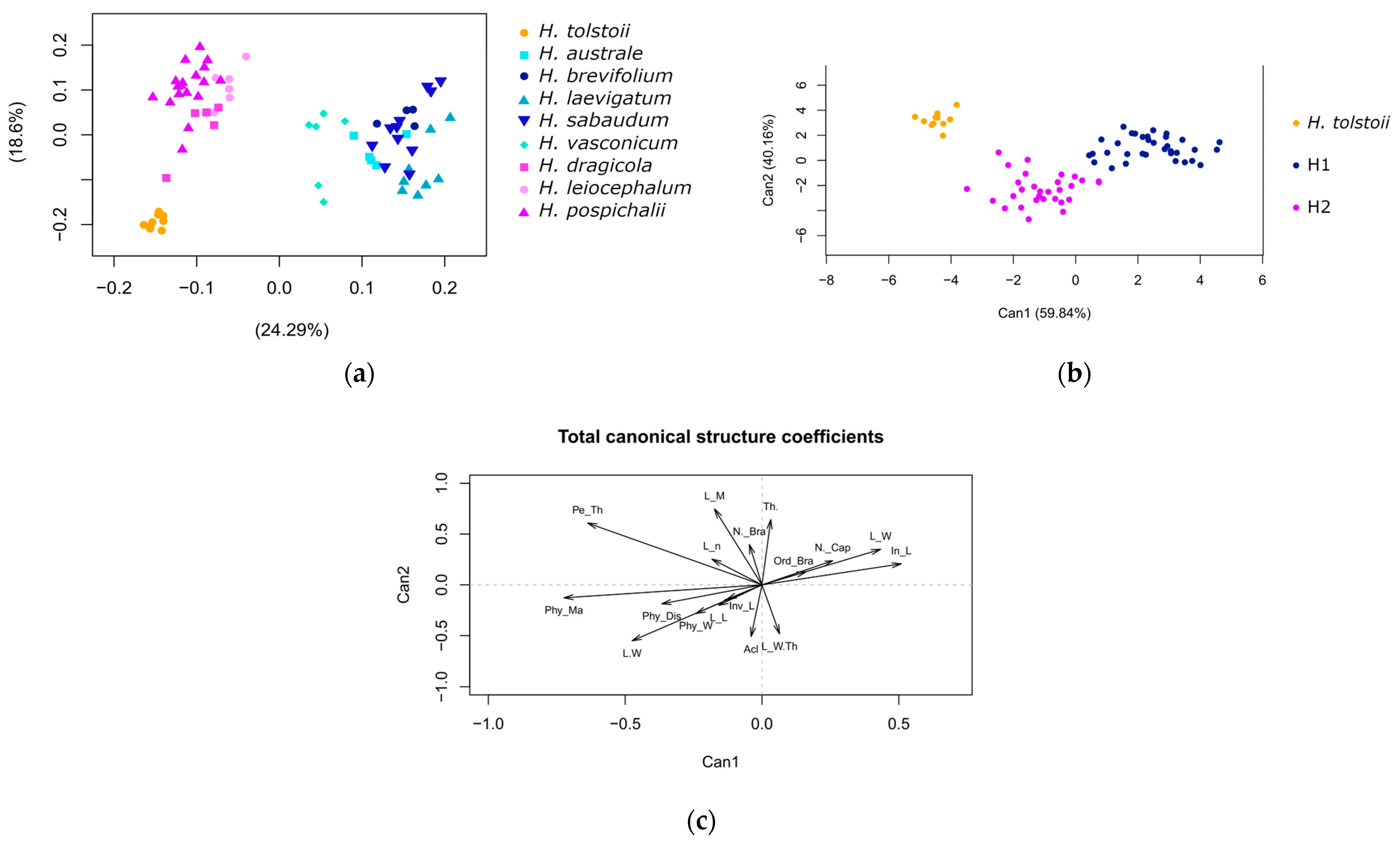
3.1. Morphometric Analysis
3.2. Molecular Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nic Lughadha, E.; Bachman, S.P.; Leão, T.C.C.; Forest, F.; Halley, J.M.; Moat, J.; Acedo, C.; Bacon, K.L.; Brewer, R.F.A.; Gâteblé, G.; et al. Extinction risk and threats to plants and fungi. Plant People Planet 2020, 2, 389–408. [Google Scholar] [CrossRef]
- De Vos, J.M.; Joppa, L.N.; Gittleman, J.L.; Stephens, P.R.; Pimm, S.L. Estimating the normal background rate of species extinction. Conserv. Biol. 2015, 29, 452–462. [Google Scholar] [CrossRef] [PubMed]
- IUCN. IUCN 2016: International Union for Conservation of Nature Annual Report 2016; IUCN: Gland, Switzerland, 2017. [Google Scholar]
- Seddon, P.J.; King, M. Creating proxies of extinct species: The bioethics of de-extinction. Emerg. Top. Life Sci. 2019, 3, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Abeli, T.; Rocchetti, G.A.; Barina, Z.; Bazos, I.; Draper, D.; Grillas, P.; Iriondo, J.M.; Laguna, E.; Moreno-Saiz, J.C.; Bartolucci, F. Seventeen ‘extinct’ plant species back to conservation attention in Europe. Nat. Plants 2021, 7, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, A.M.; Govaerts, R.; Ficinski, S.Z.; Nic Lughadha, E.; Vorontsova, M.S. Global dataset shows geography and life form predict modern plant extinction and rediscovery. Nat. Ecol. Evol. 2019, 3, 1043–1047. [Google Scholar] [CrossRef]
- Mráz, P.; Zdvořák, P. Reproductive pathways in Hieracium s.s. (Asteraceae): Strict sexuality in diploids and apomixis in polyploids. Ann. Bot. 2019, 123, 391–403. [Google Scholar] [CrossRef]
- IPNI—International Plant Names Index. The Royal Botanic Gardens, Kew, Harvard University Herbaria & Libraries and Australian National Herbarium. Available online: http://www.ipni.org (accessed on 2 July 2024).
- Zahn, K.H. Compositae-Hieracium. In Das Pflanzenreich IV; Engler, A., Ed.; W. Engelmann: Leipzig, Germany, 1921; pp. 1965–1966. [Google Scholar]
- Schuhwerk, F. Some thoughts on the taxonomy of Hieracium. Ber. Bayer. Bot. Ges. 2002, 72, 193–198. [Google Scholar]
- Krak, K.; Caklová, P.; Chrtek, J.; Fehrer, J. Reconstruction of phylogenetic relationships in a highly reticulate group with deep coalescence and recent speciation (Hieracium, Asteraceae). Heredity 2013, 110, 138–151. [Google Scholar] [CrossRef]
- Chrtek, J.; Mráz, P.; Belyayev, A.; Paštová, L.; Mrázová, V.; Caklová, P.; Josefiová, J.; Zagorski, D.; Hartmann, M.; Pinc, J.; et al. Evolutionary history and genetic diversity of apomictic allopolyploids in Hieracium s.str: Morphological versus genomic features. Am. J. Bot. 2020, 107, 66–90. [Google Scholar] [CrossRef]
- Stace, C.A. Species recognition in agamosperms—The need for a pragmatic approach. Folia Geobot. 1998, 33, 319–326. [Google Scholar] [CrossRef]
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Alessandrini, A.; Ardenghi, N.M.G.; Bacchetta, G.; Banfi, E.; Barberis, G.; Bernardo, L.; Bouvet, D.; et al. A second update to the checklist of the vascular flora native to Italy. Plant Biosyst. 2024, 158, 219–296. [Google Scholar] [CrossRef]
- Gottschlich, G. Hieracium. In Flora d’Italia, 2nd ed.; Pignatti, S., Guarino, R., La Rosa, M., Eds.; Edagricole: Milano, Italia, 2018; Volume 3, pp. 1138–1195. [Google Scholar]
- Di Gristina, E.; Domina, G.; Gottschlich, G.; Maturo, F.; Scafidi, F. Hieracium racemosum subsp. lucanum (Asteraceae), a new hawkweed from southern Italy. Phytotaxa 2019, 425, 297–300. [Google Scholar]
- Gottschlich, G.; Villa, M. Hieracium racemosum subsp. spinidentatum (Asteraceae), a new hawkweed from Lombardy, Italy. Phytotaxa 2022, 531, 78–82. [Google Scholar] [CrossRef]
- Gottschlich, G.; Orsenigo, S. New taxa of Hieracium (Asteraceae) from Mount Lesima and adjacent regions (Northern Apennine, Italy). Phytotaxa 2021, 505, 39–55. [Google Scholar] [CrossRef]
- Orsenigo, S.; Montagnani, C.; Fenu, G.; Gargano, D.; Peruzzi, L.; Abeli, T.; Alessandrini, A.; Bacchetta, G.; Bartolucci, F.; Bovio, M.; et al. Red Listing plants under full national responsibility: Extinction risk and threats in the vascular flora endemic to Italy. Biol. Conserv. 2018, 224, 213–222. [Google Scholar] [CrossRef]
- Bartolucci, F.; Domina, G.; Alessandrini, A.; Angiolini, C.; Ardenghi, N.M.; Bacchetta, G.; Banfi, E.; Bolpagni, R.; Bonari, G.; Bräuchler, C.; et al. Notulae to the Italian native vascular flora: 7. Ital. Bot. 2019, 7, 125–148. [Google Scholar] [CrossRef]
- Orsenigo, S.; Fenu, G.; Gargano, D.; Montagnani, C.; Abeli, T.; Alessandrini, A.; Bacchetta, G.; Bartolucci, F.; Carta, A.; Castello, M.; et al. Red list of threatened vascular plants in Italy. Plant Biosyst. 2021, 155, 310–335. [Google Scholar] [CrossRef]
- Fenaroli, L.; Zahn, K.H. Hieracia nova Italiae borealis (avec remarques sur H. australe Fr.). Bot. Jahrbücher Fur Syst. 1927, 61, 22–30. [Google Scholar]
- Fiori, A.; Béguinot, A. Flora Italica Exsiccata Series II. Centuriae XI–XII (continuatio). Nuovo G. Bot. Ital. Nuova Ser. 1910, 17, 62–120. [Google Scholar]
- Orsenigo, S.; Gottschlich, G.; Galasso, G. The typification and identity of Hieracium australe Fr. (Asteraceae). Phytotaxa 2019, 388, 207–211. [Google Scholar] [CrossRef]
- Fenu, G.; Bacchetta, G.; Bernardo, L.; Calvia, G.; Citterio, S.; Foggi, B.; Fois, M.; Gangale, C.; Galasso, G.; Gargano, D.; et al. Global and Regional IUCN Red List Assessments: 2. Ital. Bot. 2016, 2, 93–115. [Google Scholar] [CrossRef][Green Version]
- Thiers, B.M. Index Herbariorum, A Global Directory of Public Herbaria and Associated Staff; New York Botanical Garden: Bronx, NY, USA, 2021. [Google Scholar]
- Tyler, T. Reevaluation of the species of Hieracium sect. Hieracium that were described by Hylander from introduced populations in Scandinavian parks. Ann. Bot. Fenn. 2004, 41, 103–131. Available online: https://www.jstor.org/stable/23726952 (accessed on 25 June 2024).
- Tyler, T. Patterns of morphometric variation and a new supraspecific classification of apomictic taxa of Hieracium (Asteraceae) from Denmark and southern Sweden. Plant Syst. Evol. 2006, 261, 39–88. [Google Scholar] [CrossRef]
- Tyler, T. Hieracium sect. Oreadea (Asteraceae) in Sweden–from a complete mess to a preliminary taxonomic classification. Nord. J. Bot. 2011, 29, 538–589. [Google Scholar] [CrossRef]
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software, PBC: Boston, MA, USA, 2024. [Google Scholar]
- Šlenker, M.; Koutecký, P.; Marhold, K. MorphoTools2: An R package for multivariate morphometric analysis. Bioinformatics 2022, 38, 2954–2955. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium, and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef]
- Fehrer, J.; Gemeinholzer, B.; Chrtek, J.; Bräutigam, S. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol. Phylogenet. Evol. 2007, 42, 347–361. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP. Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.; Ronquist, F.; Huelsenbeck, J.P.; Nieves-Aldrey, J. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004, 53, 47–67. [Google Scholar] [CrossRef] [PubMed]
- Klecka, W.R. Discriminant Analysis; Sage Publications: Thousand Oaks, CA, USA, 1980. [Google Scholar]
- Fehrer, J.; Krak, K.; Chrtek, J. Intra-individual polymorphism in diploid and apomictic polyploid hawkweeds (Hieracium, Lactuceae, Asteraceae): Disentangling phylogenetic signal, reticulation, and noise. BMC Evol. Biol. 2009, 9, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Chrtek, J.; Zahradníček, J.; Krak, K.; Fehrer, J. Genome size in Hieracium subgenus Hieracium (Asteraceae) is strongly correlated with major phylogenetic groups. Ann. Bot. 2009, 104, 161–178. [Google Scholar] [CrossRef][Green Version]
- Albani Rocchetti, G.; Davis, C.; Caneva, G.; Bacchetta, G.; Fabrini, G.; Fenu, G.; Foggi, B.; Galasso, G.; Gargano, D.; Giusso del Galdo, G.; et al. A pragmatic and prudent consensus on the resurrection of extinct plant species using herbarium specimens. Taxon 2021, 71, 168–177. [Google Scholar] [CrossRef]
- Fainelli, F.; Gottschlich, G.; Orsenigo, S. Nomenclatural notes on some names in the Hieracium tenuiflorum group (Asteraceae) of the Alps. Phytotaxa 2024, 645, 131–148. [Google Scholar] [CrossRef]
- Guarino, S.; Basile, S.; Caimi, M.; Carratello, A.; Manachini, B.; Peri, E. Insect pests of the Herbarium of the Palermo botanical garden and evaluation of semiochemicals for the control of the key pest Lasioderma serricorne F. (Coleoptera: Anobiidae). J. Cult. Herit. 2020, 43, 37–44. [Google Scholar] [CrossRef]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Després, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic Analyses from Ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef]
- Yeates, D.K.; Mikheyev, A.S. Museums are biobanks: Unlocking the genetic potential of the three billion specimens in the world’s biological collections. Curr. Opin. Insect. Sci. 2016, 18, 83–88. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G.J.M.E. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Verkley, G.J.; Rossman, A.; Crouch, J.A. 8 The Role of Herbaria and Culture Collections. In Systematics and Evolution; The Mycota, vol 7B; McLaughlin, D., Spatafora, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 205–225. [Google Scholar]
- Gottschlich, G. Hieracia nova alpinum V. Stapfia 2011, 95, 33–45. [Google Scholar]
- Palmer, J.D.; Jansen, R.K.; Michaels, H.J.; Chase, M.W.; Manhart, J.R. Chloroplast DNA Variation and Plant Phylogeny. Ann. Mo. Bot. Gard. 1988, 75, 1180–1206. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA Barcodes to Identify Flowering Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef]
- Logacheva, M.D.; Valiejo-Roman, C.M.; Degtjareva, G.V.; Stratton, J.M.; Downie, S.R.; Samigullin, T.H.; Pimenov, M.G. A comparison of nrDNA ITS and ETS loci for phylogenetic inference in the Umbelliferae: An example from tribe Tordylieae. Mol. Phylogenet. Evol. 2010, 57, 471–476. [Google Scholar] [CrossRef]

| Description of the Character | Character Name | Type |
|---|---|---|
| Number of cauline leaves | L_n | QD |
| Leaves distribution along stem | L_Dis | CO |
| Stem indumentum low simple | St_Low_Si | CO |
| Stem inumentum mid stellate | St_Mid_St | CO |
| Stem indumentum mid glandular | St_Mid | CO |
| Base of leaves | L_B | CO |
| Leaf colour | L_C | CO |
| Leaf color (upper/lower side) | L_Cc | BI |
| Shape of lowermost cauline leaves | L_Sh | CO |
| Leaf indumentum lower side | L_In_Low | CO |
| Upper leaf surface | L_In_Up | CO |
| Leaves 1/3 up | L_1/3_Up | CO |
| Leaf margins upper leaves | L_In_Mar_Up | CO |
| Glandular hairs on leaf margins | G_L | CO |
| Hairs on leaves/lower stem | H_L | CO |
| Leaf length (lowermost well-developed) | L_L | QC |
| Leaf width (lowermost well-developed) | L_W | QC |
| Leaf length/Leaf width | L/W | QC |
| Position of the largest teeth | L_Th | CO |
| Shape/direction of teeth | Th_Sh | CO |
| Largest teeth (cm) | Th | QC |
| Width of leaf (at largest teeth, excluding teeth) | Th_L_W | QC |
| Width of leaf/largest teeth | L_W/Th | QC |
| Pattern of leaf dentation | Th_Pa | CO |
| Dentation along stem leaves | Th_St | CO |
| Internode length (mean of lowermost three nodes) | In_L | QC |
| Dentation on petiole (pseudopetiole) | Pe_Th | CO |
| Leaf margin | L_M | CO |
| Phyllaries disposition | Phy_Dis | CO |
| Phyllaries margin | Phy_Ma | CO |
| Involucre length (from the base of the secondary capitulum to the apex of the longest) | Inv_L | QC |
| Phyllaries width max | Phy_W | QC |
| Apex of phyllaries | Phy_Ap | CO |
| Simple hairs on phyllaries | S_H | CO |
| Simple hair length | S_H_L | CO |
| Glandular hairs on phyllaries | G_H | CO |
| Glandular hairs length | G_H_L | CO |
| Stellate hairs on phyllaries and distribution | St_H_D | CO |
| Abundance of stellate hairs | St_H | CO |
| Peduncle indumentum | Ped_In | CO |
| Bracts below capitulum | B_Cap | CO |
| Bracts on peduncle | B_Ped | CO |
| Style pigmentation (in exsiccata) | Sty_Pig | CO |
| Orders of branching | Ord_Bra | CO |
| Number of branches | N_Bra | QD |
| Number of well-developed capitula | N_Cap | QD |
| Acladium length | Acl | QC |
| Presence of aborted capitula | Cap_Ab | CO |
| Region | 5′ Sequences 3′ | Ta (°C) | Source |
|---|---|---|---|
| trnH-psbA | pbAF: GTTATGCATGAACGTAATGCTC trnHR: CGCGCATGGTGGATTCACAAATC | 56 | Sang et al., 1997 [36] |
| trnT-trnL | a: ATTACAAATGCGATGCTCT b: TCTACCGATTTCGCCATATC | 46 | Taberlet et al., 1991 [37] |
| trnT-trnL portion1 | a: ATTACAAATGCGATGCTCT b+: TATACATCTGTCTCTCTTCC | 46 | Taberlet et al., 1991 [37] Fehrer et al., 2007 [35] |
| trnT-trnL portion2 | a+: AAGAGAGACAGATGTATAGC b: TCTACCGATTTCGCCATATC | 46 | Fehrer et al., 2007 [35] Taberlet et al., 1991 [37] |
| trnV-ndhC | trnV-a: GAAGGTCTACGGTTCGAGTC ndhC-a: AGAAATGCCCAAAAAATATCATATTC | 52 | Krak et al., 2013 [11] |
| trnV-ndhC portion 1 | trnV-a: GAAGGTCTACGGTTCGAGTC V.C-R1: CCTCCATCGGGATTGGATTC | 57 | Krak et al., 2013 [11] New design |
| trnV-ndhC portion 2 | V.C-F2: CGCAGGAAAATTTATATGGA V.C-R2: CCAAATTCTCTTGTTTTCAT | 50 | New design |
| trnV-ndhC portion 3 | V.C-F3: CTCGGTAAGATTGAGATGAAAACA ndhC-a: AGAAATGCCCAAAAAATATCATATTC | 55 | New design Krak et al., 2013 [11] |
| Group | N. Samples | As H. tolstoii | As H1 | As H2 | Correct | Correct (%) |
|---|---|---|---|---|---|---|
| H. tolstoii | 11 | 11 | 0 | 0 | 11 | 100 |
| H1 | 34 | 0 | 29 | 5 | 29 | 85.29 |
| H2 | 30 | 1 | 1 | 28 | 28 | 93.33 |
| Total | 75 | 12 | 30 | 33 | 68 | 90.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fainelli, F.; Baldesi, G.; Pallanza, M.; Orsenigo, S. Extinct or Not? Confirming the “Extinct” Status of Hieracium tolstoii (Asteraceae) with Integrated Taxonomic Investigation. Diversity 2024, 16, 591. https://doi.org/10.3390/d16090591
Fainelli F, Baldesi G, Pallanza M, Orsenigo S. Extinct or Not? Confirming the “Extinct” Status of Hieracium tolstoii (Asteraceae) with Integrated Taxonomic Investigation. Diversity. 2024; 16(9):591. https://doi.org/10.3390/d16090591
Chicago/Turabian StyleFainelli, Federico, Giacomo Baldesi, Mattia Pallanza, and Simone Orsenigo. 2024. "Extinct or Not? Confirming the “Extinct” Status of Hieracium tolstoii (Asteraceae) with Integrated Taxonomic Investigation" Diversity 16, no. 9: 591. https://doi.org/10.3390/d16090591
APA StyleFainelli, F., Baldesi, G., Pallanza, M., & Orsenigo, S. (2024). Extinct or Not? Confirming the “Extinct” Status of Hieracium tolstoii (Asteraceae) with Integrated Taxonomic Investigation. Diversity, 16(9), 591. https://doi.org/10.3390/d16090591






