Uncovering the Evolutionary History in Lineage of Caribbean Octocorals: Phylogenomics Reveals Unrecognized Diversity in Eunicea †
Abstract
:1. Introduction
2. Materials and Methods
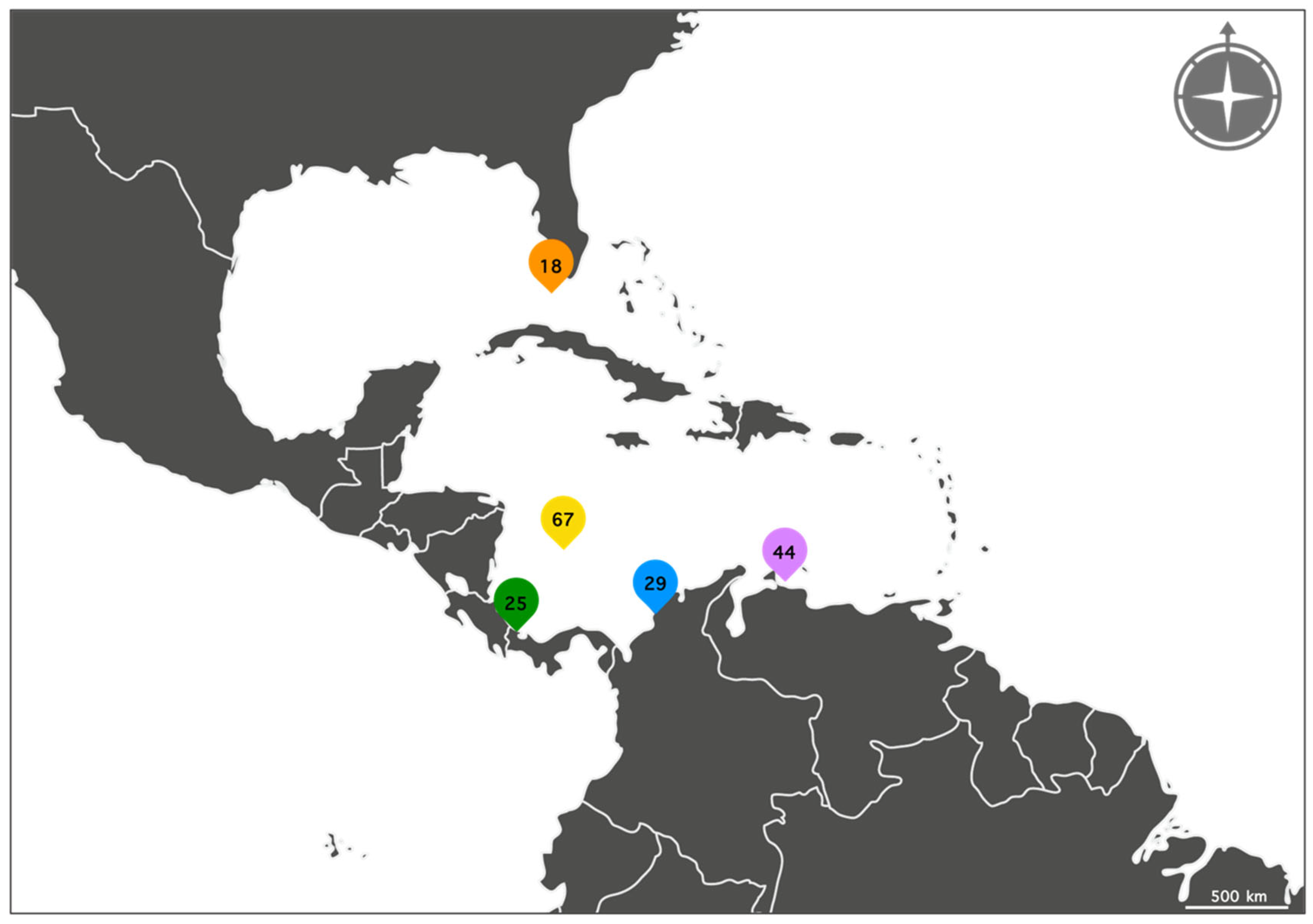
2.1. Sample Collection and Morphospecies Identifications
2.2. DNA Extraction, Library Construction and Sequencing
2.3. Data Cleaning and De Novo Assembly
2.4. Phylogenomic Analysis and Species Delimitation
2.5. Morphological Analysis from New Eunicea Species
3. Results
3.1. Data Cleaning and De Novo Assembly
3.2. Variant Calling and Filtering
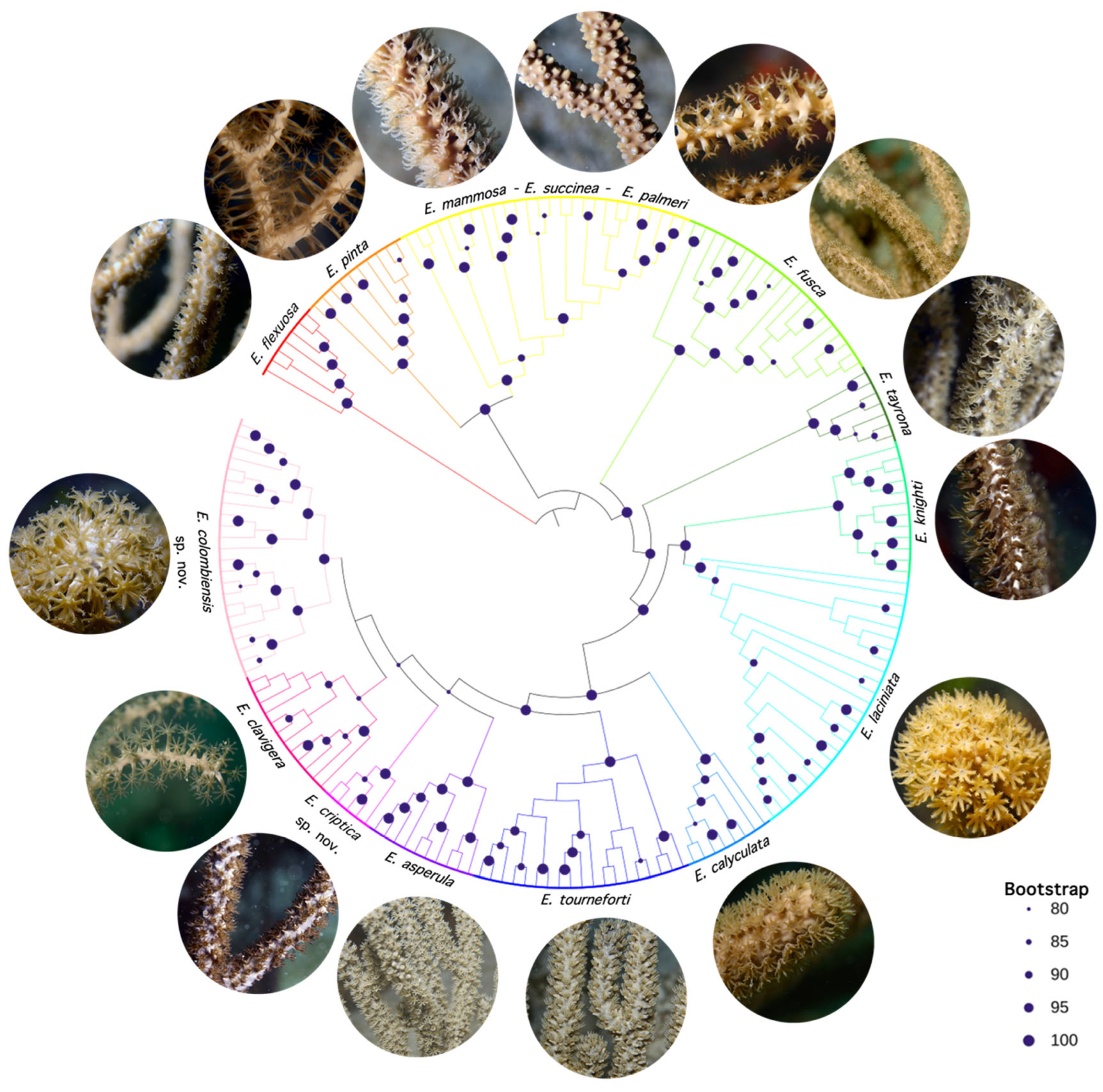
3.3. Phylogenomic Inference and Species Delimitation
- Clade A
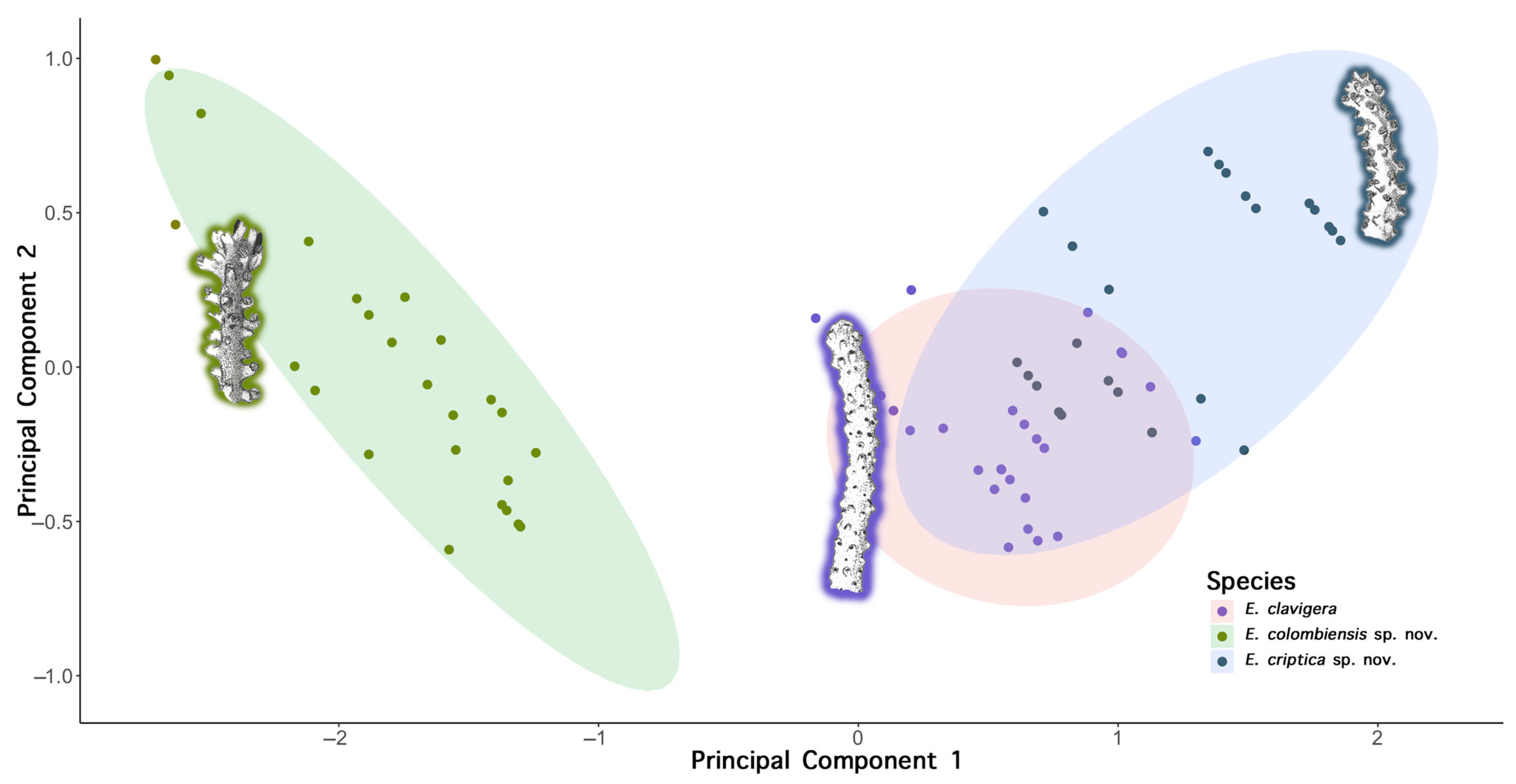
3.4. Systematics
- Examined Material:
- Diagnosis:

- Eunicea criptica sp. nov.
- https://zoobank.org/NomenclaturalActs/262AFAA2-62AB-46FD-B8E8-14159E4AFE70 (accessed on 25 February 2025).
- Material examined:
- Type locality:
- Diagnosis:
- Description:

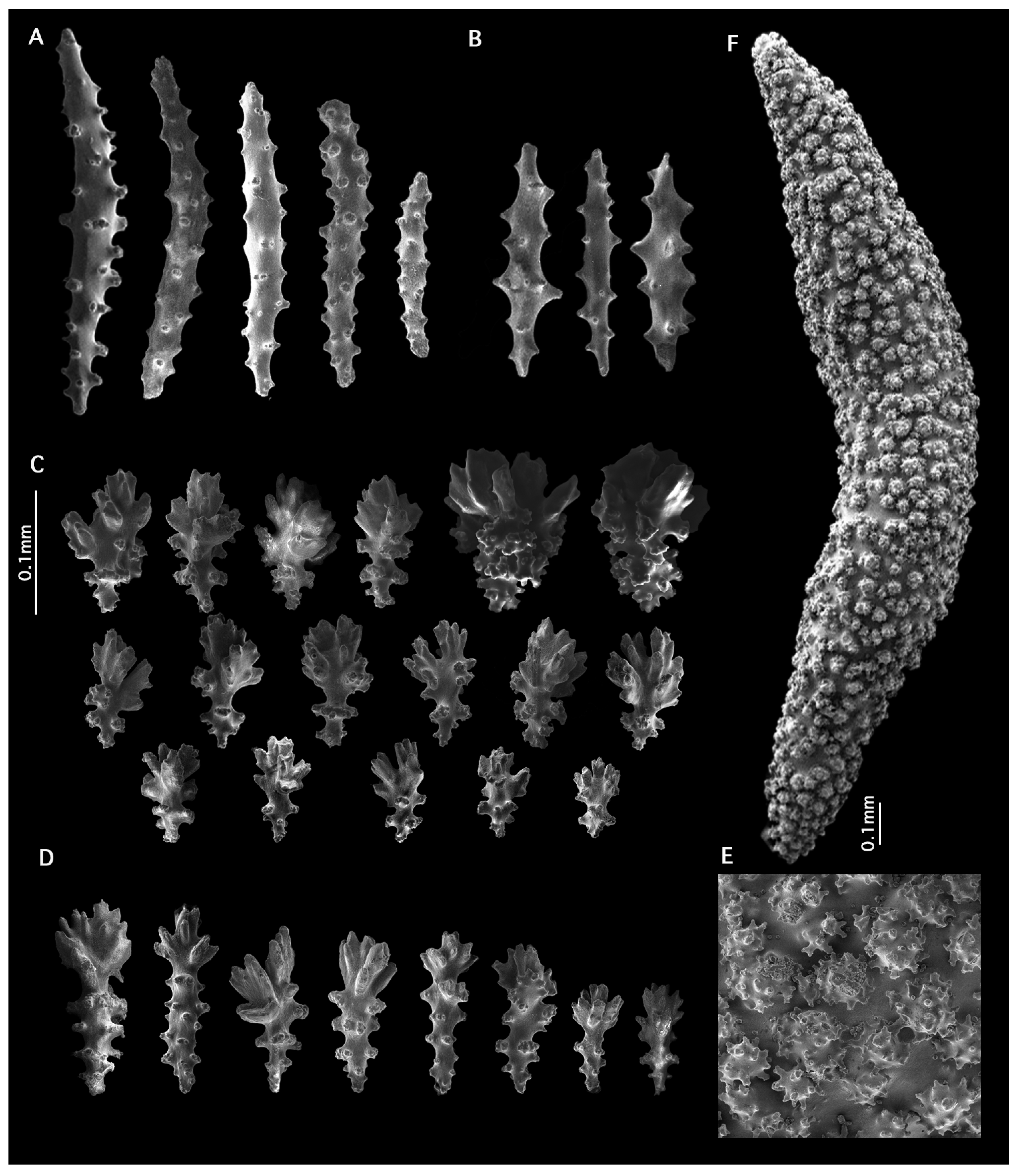
- Etymology:
- Distribution:
- Species comparisons:
- Eunicea colombiensis sp. nov.
- https://zoobank.org/NomenclaturalActs/F59458C7-12A8-41AA-AB4A-F49C86FE51E6 (accessed on 25 February 2025).
- Material examined:
- Type locality:
- Diagnosis:
- Description:

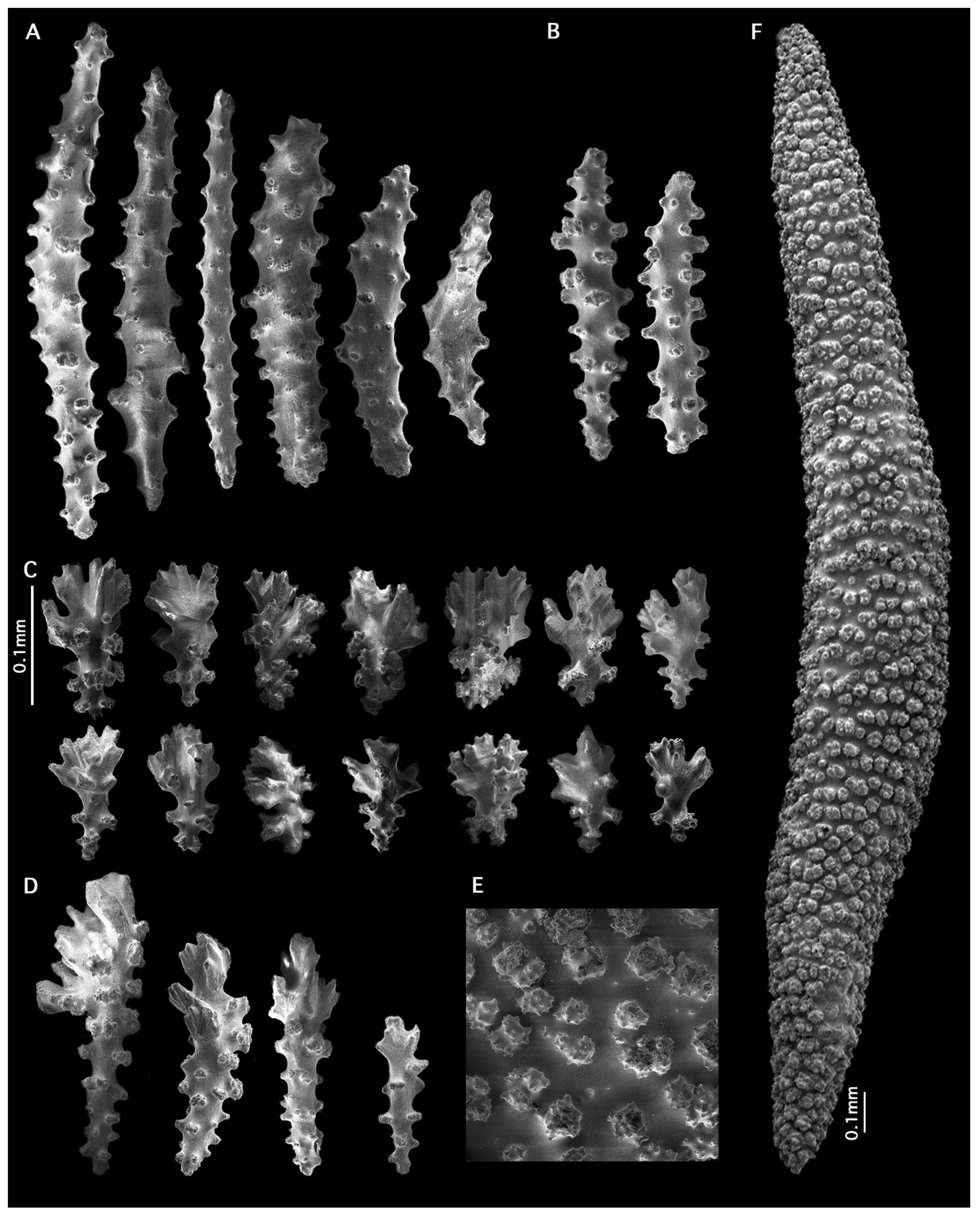
- Etymology:
- Distribution:
- Species comparisons:
- Additional taxonomic remarks:
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Hausdorf, B. Progress Toward a General Species Concept. Evolution 2011, 65, 923–931. [Google Scholar] [CrossRef] [PubMed]
- De Queiroz, K. Species Concepts and Species Delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.L.; Knowles, L.L. Species Detection and Individual Assignment in Species Delimitation: Can Integrative Data Increase Efficacy? Proc. R. Soc. B Biol. Sci. 2014, 281, 20132765. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.A. Diversity and Evolution of Octocoral Animal Forests at Both Sides of Tropical America. In Marine Animal Forests: The Ecology of Benthic Biodiversity Hotspots; Rossi, S., Bramanti, L., Gori, A., Orejas, C., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 111–143. ISBN 978-3-319-21012-4. [Google Scholar]
- Potkamp, G.; Fransen, C.H.J.M. Speciation with Gene Flow in Marine Systems. Contrib. Zool. 2019, 88, 133–172. [Google Scholar] [CrossRef]
- Ruzicka, R.; Colella, M.; Porter, J.; Morrison, J.; Kidney, J.; Brinkhuis, V.; Lunz, K.; Macaulay, K.; Bartlett, L.; Meyers, M.; et al. Temporal Changes in Benthic Assemblages on Florida Keys Reefs 11 Years after the 1997/1998 El Niño. Mar. Ecol. Prog. Ser. 2013, 489, 125–141. [Google Scholar] [CrossRef]
- Edmunds, P.J.; Lasker, H.R. Cryptic Regime Shift in Benthic Community Structure on Shallow Reefs in St. John, US Virgin Islands. Mar. Ecol. Prog. Ser. 2016, 559, 1–12. [Google Scholar] [CrossRef]
- Tsounis, G.; Edmunds, P.J. Three Decades of Coral Reef Community Dynamics in St. John, USVI: A Contrast of Scleractinians and Octocorals. Ecosphere 2017, 8, e01646. [Google Scholar] [CrossRef]
- Sánchez, J.A.; Diaz, J.; Zea, S. Gorgonian Communities in Two Contrasting Environments on Oceanic Atolls of the Southwestern Caribbean. Bull. Mar. Sci. 1997, 61, 453–465. [Google Scholar]
- Sánchez, J.A. Systematics of the Candelabrum Gorgonian Corals (Eunicea Lamouroux; Plexauridae; Octocorallia; Cnidaria). Zool. J. Linn. Soc. 2009, 157, 237–263. [Google Scholar] [CrossRef]
- Sánchez, J.A.; González-Zapata, F.L.; Dueñas, L.F.; Andrade, J.; Pico-Vargas, A.L.; Vergara, D.C.; Sarmiento, A.; Bolaños, N. Corals in the Mesophotic Zone (40–115 m) at the Barrier Reef Complex From San Andrés Island (Southwestern Caribbean). Front. Mar. Sci. 2019, 6, 536. [Google Scholar] [CrossRef]
- Gerhart, D.J. The Chemical Systematics of Colonial Marine Animals: An Estimated Phylogeny of the Order Gorgonacea Based on Terpenoid Characters. Biol. Bull. 1983, 164, 71–81. [Google Scholar] [CrossRef]
- Wirshing, H.H.; Messing, C.G.; Douady, C.J.; Reed, J.; Stanhope, M.J.; Shivji, M.S. Molecular Evidence for Multiple Lineages in the Gorgonian Family Plexauridae (Anthozoa: Octocorallia). Mar. Biol. 2005, 147, 497–508. [Google Scholar] [CrossRef]
- Aguilar, C. Phylogenetic Hypotheses of Octocorals Species Using Predicted RNA Secondary Structures of the Internal Transcribed Spacer 2 (ITS2). Master’s Thesis, Universidad de los Andes, Bogotá, Colombia, 2006. [Google Scholar]
- Grajales, A.; Aguilar, C.; Sánchez, J.A. Phylogenetic Reconstruction Using Secondary Structures of Internal Transcribed Spacer 2 (ITS2, rDNA): Finding the Molecular and Morphological Gap in Caribbean Gorgonian Corals. BMC Evol. Biol. 2007, 7, 90. [Google Scholar] [CrossRef]
- Sarmiento, A.; Sánchez, J.A. Phylogenetic Reconstruction of the Eunicea Caribbean Octocorals Using Secondary Structures of Internal Transcribed Spacer 2 (ITS2). Master’s Thesis, Universidad de los Andes, Bogotá, Colombia, 2015. [Google Scholar]
- Bayer, F.M. The Shallow-Water Octocorallia of the West Indian Region. Stud. Fauna Curaçao Caribb. Isl. 1961, 12, 1–373. [Google Scholar]
- Coffroth, M.A.; Lasker, H.R.; Diamond, M.E.; Bruenn, J.A.; Bermingham, E. DNA Fingerprints of a Gorgonian Coral: A Method for Detecting Clonal Structure in a Vegetative Species. Mar. Biol. 1992, 114, 317–325. [Google Scholar] [CrossRef]
- Russello, M.A.; Waterhouse, M.D.; Etter, P.D.; Johnson, E.A. From Promise to Practice: Pairing Non-Invasive Sampling with Genomics in Conservation. PeerJ 2015, 3, e1106. [Google Scholar] [CrossRef]
- Titus, B.M.; Daly, M. Reduced Representation Sequencing for Symbiotic Anthozoans: Are Reference Genomes Necessary to Eliminate Endosymbiont Contamination and Make Robust Phylogeographic Inference? bioRxiv 2018. bioRxiv:440289. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Eaton, D.A.R.; Overcast, I. Ipyrad: Interactive Assembly and Analysis of RADseq Datasets. Bioinformatics 2020, 36, 2592–2594. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 March 2024).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An Automated Data Conversion Tool for Connecting Population Genetics and Genomics Programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A Cluster Matching and Permutation Program for Dealing with Label Switching and Multimodality in Analysis of Population Structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A Program for the Graphical Display of Population Structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Wang, X.; Liew, Y.J.; Li, Y.; Zoccola, D.; Tambutte, S.; Aranda, M. Draft Genomes of the Corallimorpharians Amplexidiscus Fenestrafer and Discosoma sp. Mol. Ecol. Resour. 2017, 17, e187–e195. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. Hierfstat, a Package for r to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Bayer, F.M.; Grasshoff, M.; Verseveldt, J. Illustrated Trilingual Glossary of Morphological and Anatomical Terms Applied to Octocorallia; Brill: Leiden, The Netherlands, 1983. [Google Scholar]
- Ehrenberg, C.G. Beiträge Zur Physiologischen Kenntniss Der Corallenthiere Im Allgemeinen, Und Besonders Des Rothen Meeres, Nebst Einem Versuche Zur Physiologischen Systematik Derselben. Abh. K. Akad. Wiss. Berl. 1834, 1, 225–380. [Google Scholar]
- Haeckel, E. Generelle Morphologie der Organismen; Reimer, G., Ed.; De Gruyter: Berlin, Germany,, 1866; ISBN 978-3-11-084828-1. [Google Scholar]
- McFadden, C.S.; van Ofwegen, L.P.; Quattrini, A.M. Revisionary Systematics of Octocorallia (Cnidaria: Anthozoa) Guided by Phylogenomics. Bull. Soc. Syst. Biol. 2022, 1. [Google Scholar] [CrossRef]
- Gray, J.E. XLV.—On the Arrangement of Zoophytes with Pinnated Tentacles. Ann. Mag. Nat. Hist. 1859, 4, 439–444. [Google Scholar] [CrossRef]
- Lamouroux, M. Histoire Des Polypiers Coralligènes Flexibles, Vulgairement Nommés Zoophytes; Impr. de F. Poisson: Caen, France, 1816. [Google Scholar]
- Verrill, A. List of the Polyps and Corals Sent by the Museum of Comparative Zoology to Other Institutions in Exchange, with Annotations. Bull. Mus. Comp. Zool. Harv. Coll. 1864, 1, 29–60. [Google Scholar]
- McFadden, C.S.; Sanchez, J.A.; France, S.C. Molecular Phylogenetic Insights into the Evolution of Octocorallia: A Review. Integr. Comp. Biol. 2010, 50, 389–410. [Google Scholar] [CrossRef]
- Pante, E.; Abdelkrim, J.; Viricel, A.; Gey, D.; France, S.C.; Boisselier, M.C.; Samadi, S. Use of RAD Sequencing for Delimiting Species. Heredity 2015, 114, 450–459. [Google Scholar] [CrossRef]
- Herrera, S.; Shank, T.M. RAD Sequencing Enables Unprecedented Phylogenetic Resolution and Objective Species Delimitation in Recalcitrant Divergent Taxa. Mol. Phylogenet. Evol. 2016, 100, 70–79. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Wu, T.; Soong, K.; Jeng, M.-S.; Benayahu, Y.; McFadden, C.S. A next Generation Approach to Species Delimitation Reveals the Role of Hybridization in a Cryptic Species Complex of Corals. BMC Evol. Biol. 2019, 19, 116. [Google Scholar] [CrossRef]
- Richards, Z.T.; Berry, O.; van Oppen, M.J.H. Cryptic Genetic Divergence within Threatened Species of Acropora Coral from the Indian and Pacific Oceans. Conserv. Genet. 2016, 17, 577–591. [Google Scholar] [CrossRef]
- Bongaerts, P.; Riginos, C.; Ridgway, T.; Sampayo, E.M.; van Oppen, M.J.H.; Englebert, N.; Vermeulen, F.; Hoegh-Guldberg, O. Genetic Divergence across Habitats in the Widespread Coral Seriatopora Hystrix and Its Associated Symbiodinium. PLoS ONE 2010, 5, e10871. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, A.; Breedy, O.; Guzman, H.M.; Vargas, S. Hybridization and Cryptic Speciation in the Tropical Eastern Pacific Octocoral Genus Pacifigorgia. bioRxiv 2021. bioRxiv:2021.04.29.442007. [Google Scholar] [CrossRef]
- Rippe, J.P.; Dixon, G.; Fuller, Z.L.; Liao, Y.; Matz, M. Environmental Specialization and Cryptic Genetic Divergence in Two Massive Coral Species from the Florida Keys Reef Tract. Mol. Ecol. 2021, 30, 3468–3484. [Google Scholar] [CrossRef]
- Matias, A.M.A.; Popovic, I.; Thia, J.A.; Cooke, I.R.; Torda, G.; Lukoschek, V.; Bay, L.K.; Kim, S.W.; Riginos, C. Cryptic Diversity and Spatial Genetic Variation in the Coral Acropora Tenuis and Its Endosymbionts across the Great Barrier Reef. Evol. Appl. 2022, 16, 293–310. [Google Scholar] [CrossRef]
- Faria, R.; Johannesson, K.; Stankowski, S. Speciation in Marine Environments: Diving under the Surface. J. Evol. Biol. 2021, 34, 4–15. [Google Scholar] [CrossRef]
- Meziere, Z.; Popovic, I.; Prata, K.; Ryan, I.; Pandolfi, J.; Riginos, C. Exploring Coral Speciation: Multiple Sympatric Stylophora Pistillata Taxa along a Divergence Continuum on the Great Barrier Reef. Evol. Appl. 2024, 17, e13644. [Google Scholar] [CrossRef]
- Prada, C.; Hellberg, M.E. Long Prereproductive Selection and Divergence by Depth in a Caribbean Candelabrum Coral. Proc. Natl. Acad. Sci. USA 2013, 110, 3961–3966. [Google Scholar] [CrossRef]
- Prada, C.; Hellberg, M.E. Speciation-by-Depth on Coral Reefs: Sympatric Divergence with Gene Flow or Cryptic Transient Isolation? J. Evol. Biol. 2021, 34, 128–137. [Google Scholar] [CrossRef]
- Rasband, W.S. ImageJ; U.S. National Institutes of Health: Bethesda, MD, USA, 1997–2018; Available online: https://imagej.net/ij/ (accessed on 8 May 2024).
- R Core Team. R: A Language and Environment for Statistical Computing; Foundation Statistics and Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]


| Species | N | Sites Collected | Depth Range (m) |
|---|---|---|---|
| E. flexuosa | 7 | FLO, SAI | 5–15 |
| E. pinta | 10 | SAI | 35–45 |
| E. mammosa | 14 | BDT, CUR, FLO, SAI | 3–15 |
| E. succinea | 7 | SAI | 10–20 |
| E. palmeri | 5 | SAI | 7–20 |
| E. fusca | 19 | CAR, CUR, SAI | 8–34 |
| E. asperula | 10 | CAR, CUR, SAI | 10–26 |
| E. tayrona | 7 | BDT, SAI | 3–17 |
| E. knighti | 12 | BDT, SAI | 3–41 |
| E. laciniata | 25 | CAR, CUR, FLO, SAI | 5–26 |
| E. calyculata | 8 | CAR, CUR, FLO, SAI | 5–26 |
| E. tourneforti | 19 | BDT, CAR, CUR, FLO, SAI | 3–26 |
| E. clavigera | 12 | BDT, CAR, SAI | 3–26 |
| E. criptica sp. nov. | 5 | CUR | 15–27 |
| E. colombiensis sp. nov. | 23 | BDT, CAR, CUR, SAI | 3–41 |
| Clustering Threshold | Taxon Occupancy (%) | Shared Polymorphic Sites | Total Filtered Loci | Sequence Matrix Size | Missing Sites (%) | SNPs Matrix size | Missing Sites (%) |
|---|---|---|---|---|---|---|---|
| 0.8 | 10 | 0.25 | 17,600 | 2,455,879 | 81.12 | 241,830 | 79.20 |
| 0.5 | 18,777 | 2,620,265 | 81.12 | 258,564 | 79.37 | ||
| 25 | 0.25 | 3303 | 448,561 | 58.73 | 57,729 | 57.23 | |
| 0.5 | 3540 | 482,734 | 59.15 | 61,614 | 57.73 | ||
| 50 | 0.25 | 662 | 96,955 | 31.25 | 14,338 | 30.98 | |
| 0.5 | 676 | 99,082 | 31.40 | 14,611 | 31.12 | ||
| 0.85 | 10 | 0.25 | 21,557 | 3,049,774 | 81.45 | 305,958 | 79.83 |
| 0.5 | 23,108 | 3,270,269 | 81.46 | 328,314 | 79.92 | ||
| 25 | 0.25 | 3877 | 534,595 | 59.35 | 69,203 | 57.87 | |
| 0.5 | 4156 | 575,576 | 59.72 | 73,832 | 58.28 | ||
| 50 | 0.25 | 728 | 107,010 | 32.11 | 16,000 | 31.78 | |
| 0.5 | 749 | 110,192 | 32.32 | 16,432 | 31.98 |
| E. clavigera | E. asperula | E. tourneforti | E. criptica sp. nov. | |
|---|---|---|---|---|
| E. asperula | 0.141 | NA | ||
| E. tourneforti | 0.222 | 0.221 | NA | |
| E. criptica sp. nov. | 0.095 | 0.129 | 0.176 | NA |
| E. colombiensis sp. nov. | 0.103 | 0.198 | 0.207 | 0.091 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmiento, A.; Calixto-Botía, I.; Julio-Rodríguez, T.; Quattrini, A.M.; Sánchez, J.A. Uncovering the Evolutionary History in Lineage of Caribbean Octocorals: Phylogenomics Reveals Unrecognized Diversity in Eunicea. Diversity 2025, 17, 173. https://doi.org/10.3390/d17030173
Sarmiento A, Calixto-Botía I, Julio-Rodríguez T, Quattrini AM, Sánchez JA. Uncovering the Evolutionary History in Lineage of Caribbean Octocorals: Phylogenomics Reveals Unrecognized Diversity in Eunicea. Diversity. 2025; 17(3):173. https://doi.org/10.3390/d17030173
Chicago/Turabian StyleSarmiento, Adriana, Iván Calixto-Botía, Tatiana Julio-Rodríguez, Andrea M. Quattrini, and Juan A. Sánchez. 2025. "Uncovering the Evolutionary History in Lineage of Caribbean Octocorals: Phylogenomics Reveals Unrecognized Diversity in Eunicea" Diversity 17, no. 3: 173. https://doi.org/10.3390/d17030173
APA StyleSarmiento, A., Calixto-Botía, I., Julio-Rodríguez, T., Quattrini, A. M., & Sánchez, J. A. (2025). Uncovering the Evolutionary History in Lineage of Caribbean Octocorals: Phylogenomics Reveals Unrecognized Diversity in Eunicea. Diversity, 17(3), 173. https://doi.org/10.3390/d17030173









