Mixed Mating System, Dispersal Limitation Shape, and Spatial Genetic Structure of Tamarix chinensis on Isolated Wudi Seashell Island
Abstract
:1. Introduction
2. Materials and Methods
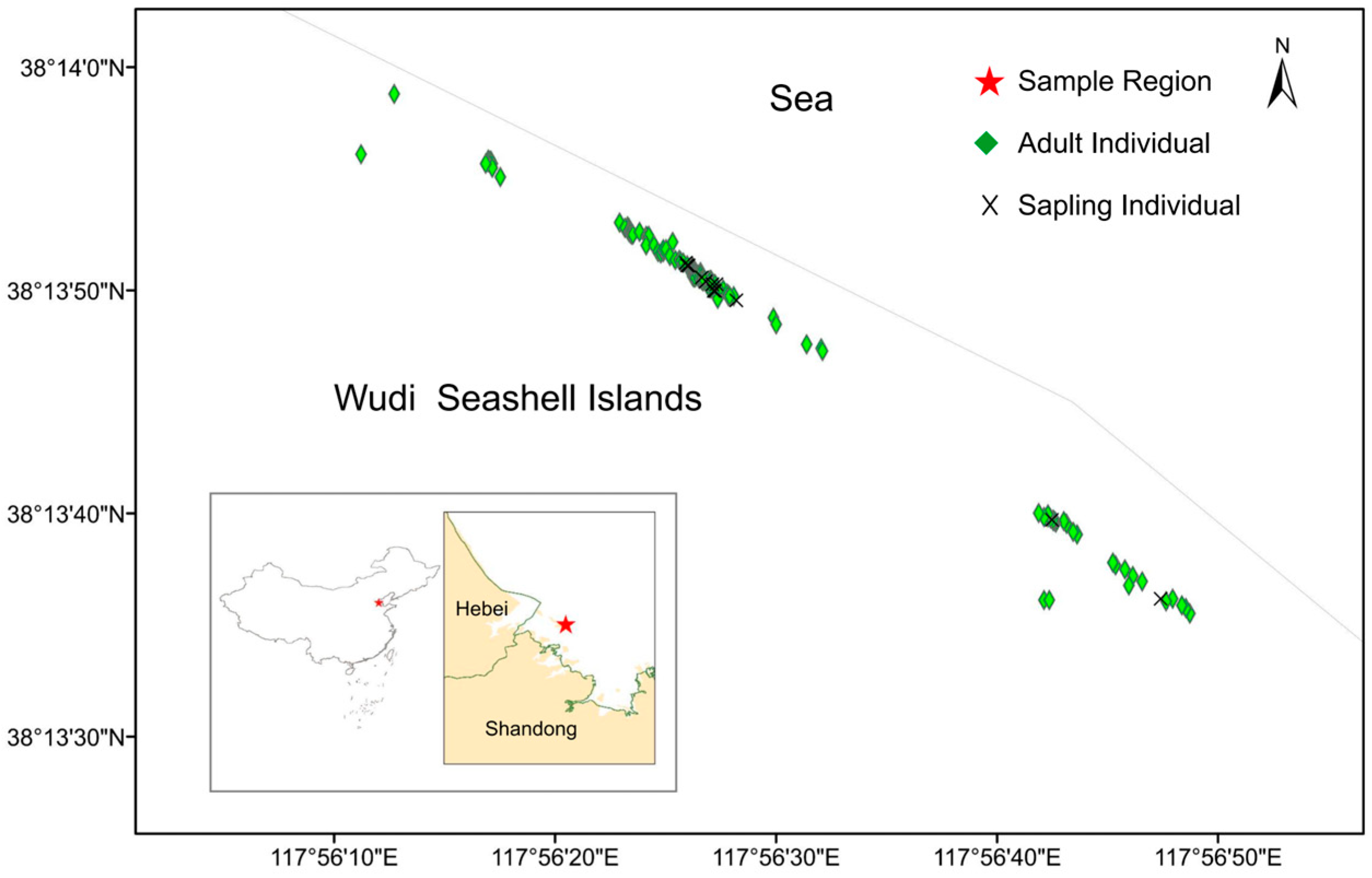
2.1. Study Area and Sampling
2.2. DNA Extraction and PCR Amplification
2.3. Genetic Parameters
2.4. Paternity Analysis and Mating System
2.5. Spatial Genetic Structure
2.6. Bottleneck Analysis and Estimation of Effective Population Size
3. Results
3.1. Genetic Diversity
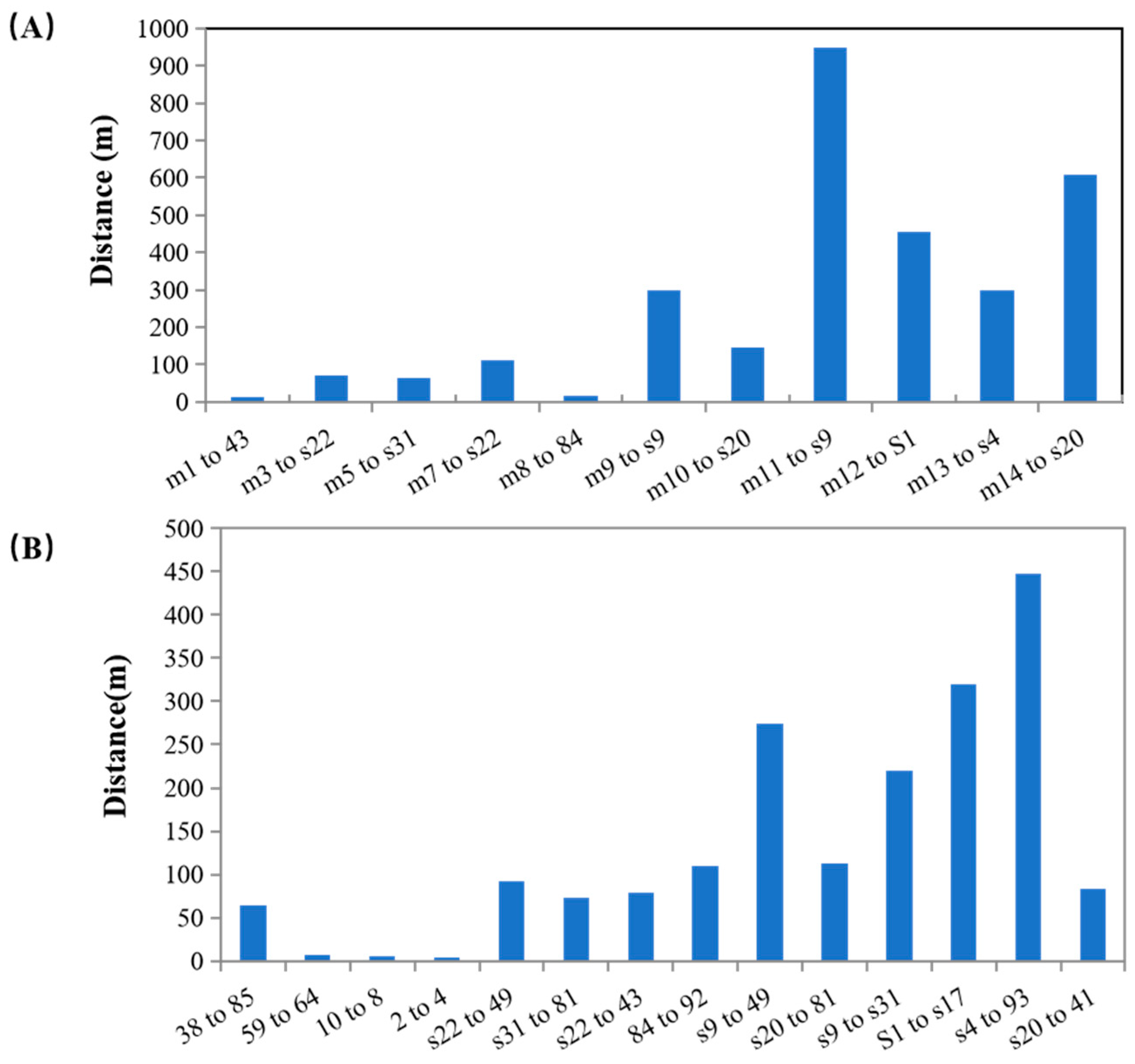
3.2. Paternity Analysis and Mating System
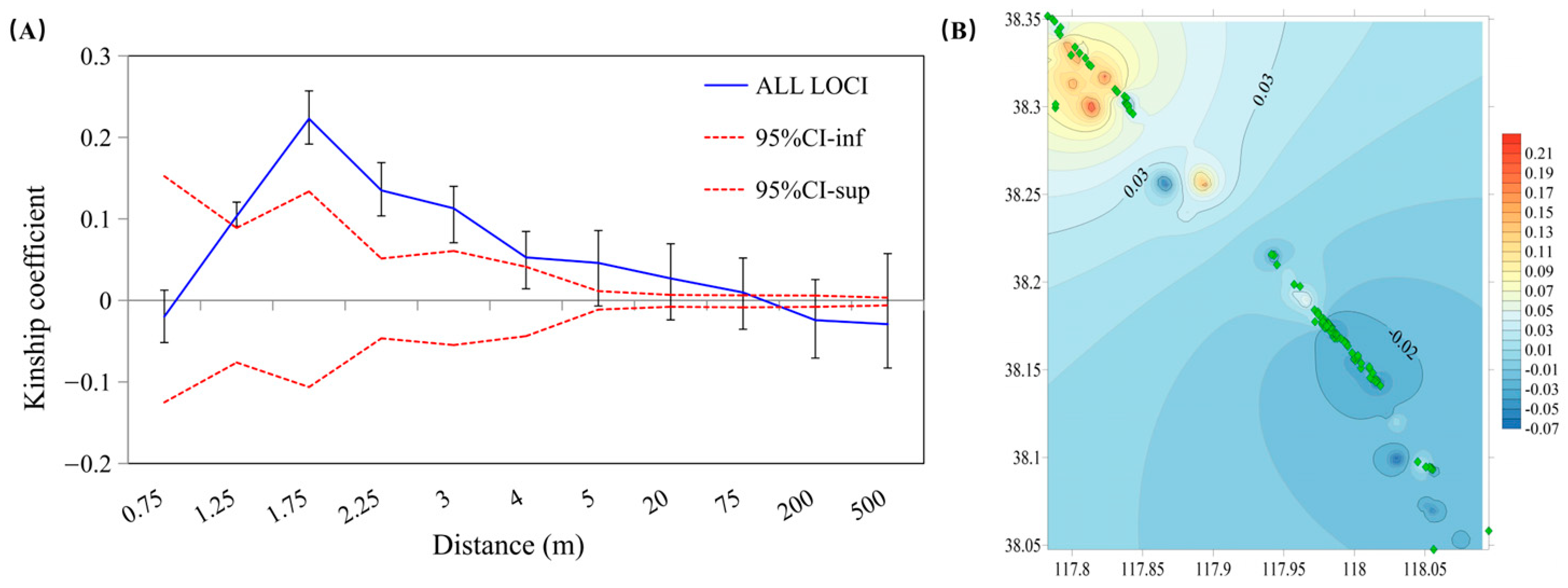
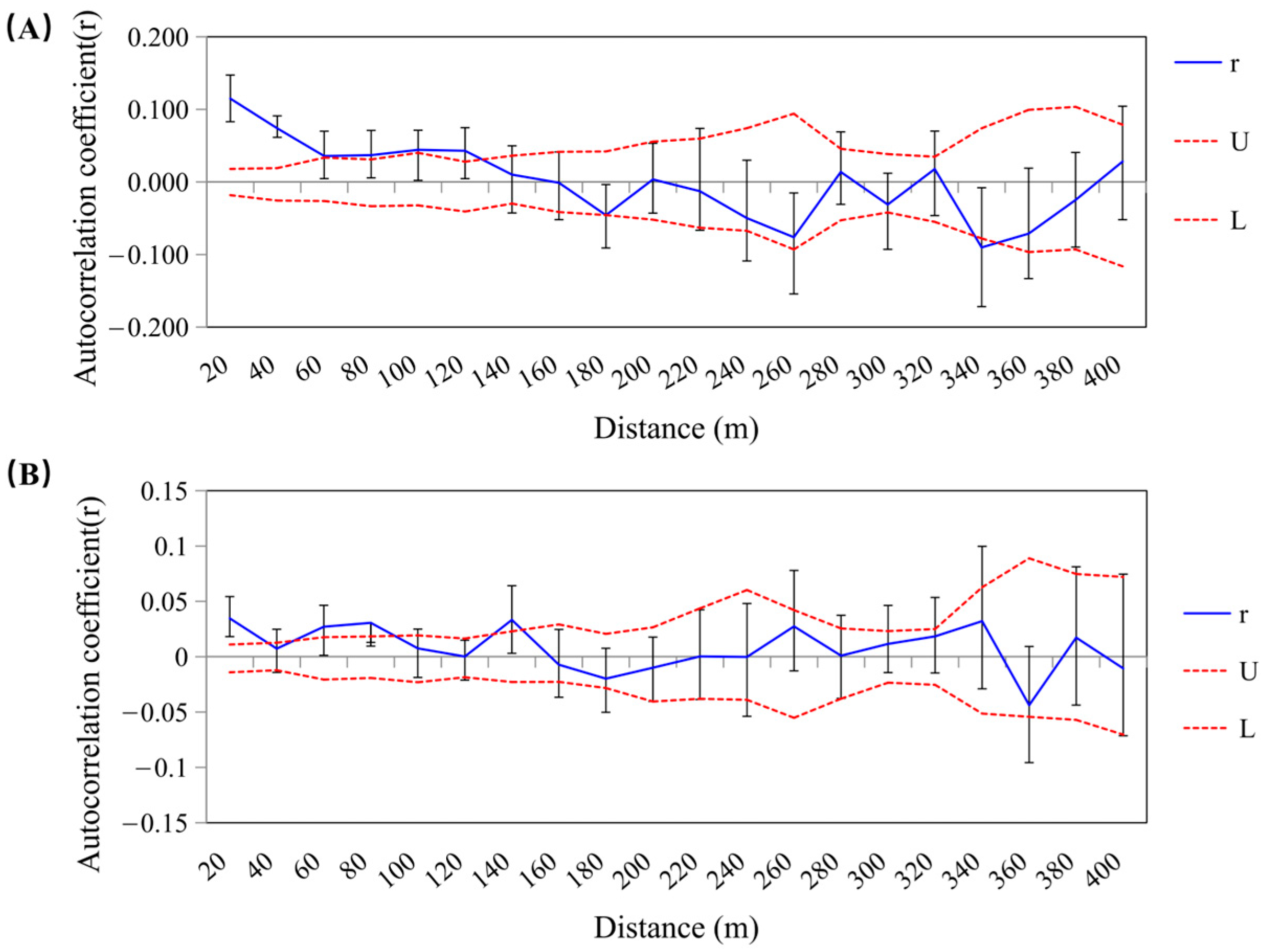
3.3. Spatial Genetic Structure Analysis
3.4. Bottleneck and Effective Population Size
4. Discussion
4.1. Genetic Diversity
4.2. Paternity Analysis and Mating System
4.3. Spatial Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Z.Y.; Yang, A.-A.; Zhang, H.-G.; Wang, W.-B.; Zhang, R.-H. Population structure and genetic diversity of Tamarix chinensis as revealed with microsatellite markers in two estuarine flats. Peerj 2023, 11, e15882. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xu, L.; Xie, H.; Zhao, D.; Huang, M. RAPD analysis of population genetic diversity of Tamarix chinensis in Yellow River delta. J. Nanjing For. Univ. 2008, 32, 56–60. [Google Scholar] [CrossRef]
- Xu, Z.; Li, R.; Dou, W.; Wen, H.; Yu, S.; Wang, P.; Ning, L.; Duan, J.; Wang, J. Plant Diversity Response to Environmental Factors in Yellow River Delta, China. Land 2024, 13, 264. [Google Scholar] [CrossRef]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Liang, H.; Liu, C.; Li, Y.; Wang, Y.; Kong, Y.; Quan, J.; Yang, X. Low population genetic differentiation in two Tamarix species (Tamarix austromongolica and Tamarix chinensis) along the Yellow River. Genetica 2019, 147, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Chen, Y.; Bao, Y. Population genetic structure of Tamarix chinensis in the Yellow River Delta, China. Plant Syst. Evol. 2012, 298, 147–153. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, L.; Gao, L.; Tang, S.; Zhao, Y.; Yang, J. Local habitat condition rather than geographic distance determines the genetic structure of Tamarix chinensis populations in Yellow River Delta, China. Tree Genet. Genomes 2016, 12, 14. [Google Scholar] [CrossRef]
- Sun, L.K.; Liu, G.X.; Lu, Y.L.; Zhang, B.G.; Zhang, G.S. Molecular data and ecological niche modelling reveal the phylogeographic pattern of the widespread shrub Tamarix chinensis Lour. (Tamaricaceae) in China. Kew Bull. 2020, 75, 41. [Google Scholar] [CrossRef]
- Bacles, C.F.E.; Ennos, R.A. Paternity analysis of pollen-mediated gene flow for Fraxinus excelsior L. in a chronically fragmented landscape. Heredity 2008, 101, 368–380. [Google Scholar] [CrossRef]
- Geng, Q.; Lian, C.; Goto, S.; Tao, J.; Kimura, M.; Islam, S.; Hogetsu, T. Mating system, pollen and propagule dispersal, and spatial genetic structure in a high-density population of the mangrove tree Kandelia candel. Mol. Ecol. 2008, 17, 4724–4739. [Google Scholar] [CrossRef]
- Yue, X.Y.; Yue, W. Ndvi Dynamics and Its Relationship with Climatic Factors in the Shell Ridge Island of the Yellow River Delta. Appl. Ecol. Environ. Res. 2024, 22, 2809–2819. [Google Scholar] [CrossRef]
- Jiang, Z.-M.; Chen, Y.-X.; Bao, Y. Genetic Structure and Population Differentiation of Tamarix chinensis in Yellow Rive Delta, China. Plant Divers. 2011, 33, 403–408. [Google Scholar]
- Gaskin, J.F.; Pepper, A.E.; Manhart, J.R. Isolation and characterization of 10 polymorphic microsatellites in saltcedars (Tamarix chinensis and Tamarix ramosissima). Mol. Ecol. Notes 2006, 6, 1147–1149. [Google Scholar] [CrossRef]
- Zhang, R.; Wen, Q.; Ku, L.-A. Development and characterization of genomic SSR markers for Tamarix chinensis (Tamaricaceae). Appl. Plant Sci. 2019, 7, e1219. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of codominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Ritland, K. Extensions of models for the estimation of mating systems using n independent loci. Heredity 2002, 88, 221–228. [Google Scholar] [CrossRef]
- Hardy, O.; Vekemans, X. SPAGeDI: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Miller, M. ALLELES IN SPACE (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. J. Hered. 2005, 96, 722–724. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Md, S.I.; Chunlan, L.; Norikazu, K.; Taizo, H. Analysis of the mating system, reproductive characteristics, and spatial genetic structure in a natural mangrove tree (Bruguiera gymnorrhiza) population at its northern biogeographic limit in the southern Japanese archipelago. J. For. Res. 2015, 20, 293–300. [Google Scholar] [CrossRef]
- Luikart, G.; Allendorf, F.W.; Cornuet, J.M.; Sherwin, W.B. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998, 89, 238–247. [Google Scholar] [CrossRef]
- Pinto, A.V.; Hansson, B.; Patramanis, I.; Morales, H.E.; van Oosterhout, C. The impact of habitat loss and population fragmentation on genomic erosion. Conserv. Genet. 2024, 25, 49–57. [Google Scholar] [CrossRef]
- Baldauf, C.; Ciampi-Guillardi, M.; Aguirra, T.J.; Corrêa, C.E.; dos Santos, F.A.M.; de Souza, A.P.; Sebbenn, A.M. Genetic diversity, spatial genetic structure and realised seed and pollen dispersal of Himatanthus drasticus (Apocynaceae) in the Brazilian savanna. Conserv. Genet. 2014, 15, 1073–1083. [Google Scholar] [CrossRef]
- Bittencourt, J.V.M.; Sebbenn, A.M. Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 2007, 99, 580–591. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Tambarussi, E.V.; Boshier, D.; Vencovsky, R.; Freitas, M.L.M.; Sebbenn, A.M. Paternity analysis reveals significant isolation and near neighbor pollen dispersal in small Cariniana legalis Mart. Kuntze populations in the Brazilian Atlantic Forest. Ecol. Evol. 2015, 5, 5588–5600. [Google Scholar] [CrossRef]
- Cui, B.; Yang, Q.; Zhang, K.; Zhao, X.; You, Z. Responses of saltcedar (Tamarix chinensis) to water table depth and soil salinity in the Yellow River Delta, China. Plant Ecol. 2010, 209, 279–290. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, L.; Fang, Y. Study on the Flowering Characteristics and Pollination Ecology of Tamarix chinensis in the Yellow River Delta. J. Trop. Subtrop. Bot. 2005, 13, 353–357. [Google Scholar]
- Sebbenn, A.M.; Carvalho, A.C.M.; Freitas, M.L.M.; Moraes, S.M.B.; Gaino, A.P.S.C.; da Silva, J.M.; Jolivet, C.; Moraes, M.L.T. Low levels of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf. Heredity 2011, 106, 134–145. [Google Scholar] [CrossRef]
| Locus | N | Na | Ne | I | He | uHe | PPL |
|---|---|---|---|---|---|---|---|
| 823 | 107 | 1.909 | 1.704 | 0.566 | 0.393 | 0.395 | - |
| 824 | 107 | 1.857 | 1.625 | 0.516 | 0.354 | 0.356 | - |
| 834 | 107 | 2.000 | 1.679 | 0.583 | 0.396 | 0.398 | - |
| 835 | 107 | 2.000 | 1.782 | 0.612 | 0.425 | 0.427 | - |
| 840 | 107 | 2.000 | 1.869 | 0.655 | 0.462 | 0.465 | - |
| 841 | 107 | 2.000 | 1.592 | 0.529 | 0.352 | 0.353 | - |
| 844 | 107 | 2.000 | 1.695 | 0.580 | 0.395 | 0.397 | - |
| 873 | 107 | 2.000 | 1.753 | 0.613 | 0.424 | 0.426 | - |
| 880 | 107 | 2.000 | 1.761 | 0.611 | 0.423 | 0.425 | - |
| 888 | 107 | 2.000 | 1.725 | 0.593 | 0.407 | 0.409 | - |
| mean | 107 | 1.979 | 1.715 | 0.585 | 0.402 | 0.404 | 97.87% |
| SE | - | 0.015 | 0.026 | 0.013 | 0.011 | 0.011 | - |
| Locus | N | Na | Ne | I | He | Ho | Fi | Fis |
|---|---|---|---|---|---|---|---|---|
| E1 | 109 | 12 | 5.48 | 1.951 | 0.8177 | 0.881 | −0.078 | 0.137 |
| E2 | 109 | 8 | 2.99 | 1.382 | 0.6655 | 0.514 | 0.229 | 0.037 |
| E3 | 109 | 12 | 3.42 | 1.590 | 0.7077 | 0.679 | 0.041 | 0.091 |
| E4 | 109 | 14 | 6.13 | 2.129 | 0.8369 | 0.716 | 0.146 | 0.054 |
| mean | 109 | 11.5 | 4.51 | 1.763 | 0.7569 | 0.697 | 0.079 | 0.078 |
| Locus | N | PIC | NE-1P | NE-2P | NE-PP | F(Null) |
|---|---|---|---|---|---|---|
| E1 | 109 | 0.792 | 0.529 | 0.355 | 0.171 | −0.046 |
| E2 | 109 | 0.618 | 0.742 | 0.570 | 0.380 | 0.134 |
| E3 | 109 | 0.663 | 0.691 | 0.517 | 0.321 | 0.016 |
| E4 | 109 | 0.815 | 0.486 | 0.319 | 0.141 | 0.077 |
| Mean | 109 | 0.722 | 0.612 | 0.440 | 0.253 | 0.045 |
| Combined | - | - | 0.868 | 0.967 | 0.997 | - |
| Group | ts | tm | tm − ts |
|---|---|---|---|
| Saplings | 0.455(0.525) | 0.492(0.785) | 0.037 |
| Adult individuals | 0.369(0.439) | 0.583(0.553) | 0.216 |
| All | 0.210(0.945) | 0.554(1.468) | 0.343 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Lan, X.; Yang, S.; Hui, M. Mixed Mating System, Dispersal Limitation Shape, and Spatial Genetic Structure of Tamarix chinensis on Isolated Wudi Seashell Island. Diversity 2025, 17, 285. https://doi.org/10.3390/d17040285
Zhang B, Lan X, Yang S, Hui M. Mixed Mating System, Dispersal Limitation Shape, and Spatial Genetic Structure of Tamarix chinensis on Isolated Wudi Seashell Island. Diversity. 2025; 17(4):285. https://doi.org/10.3390/d17040285
Chicago/Turabian StyleZhang, Binghuang, Xiao Lan, Shengchang Yang, and Ma Hui. 2025. "Mixed Mating System, Dispersal Limitation Shape, and Spatial Genetic Structure of Tamarix chinensis on Isolated Wudi Seashell Island" Diversity 17, no. 4: 285. https://doi.org/10.3390/d17040285
APA StyleZhang, B., Lan, X., Yang, S., & Hui, M. (2025). Mixed Mating System, Dispersal Limitation Shape, and Spatial Genetic Structure of Tamarix chinensis on Isolated Wudi Seashell Island. Diversity, 17(4), 285. https://doi.org/10.3390/d17040285






