DNA-Based Identification and Chemical Characteristics of Hypnea musciformis from Coastal Sites in Ghana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Seaweed Sampling and Preparation
2.2. DNA Extractions and Barcodes
2.3. PCR Amplification of DNA Barcodes
2.4. DNA Purification and Sequencing
2.5. Phylogenetic and Bioinformatics Analysis
2.6. Chemicals and HPAEC for Monosaccharide Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. DNA Extraction of Hypnea sp. and PCR Amplification
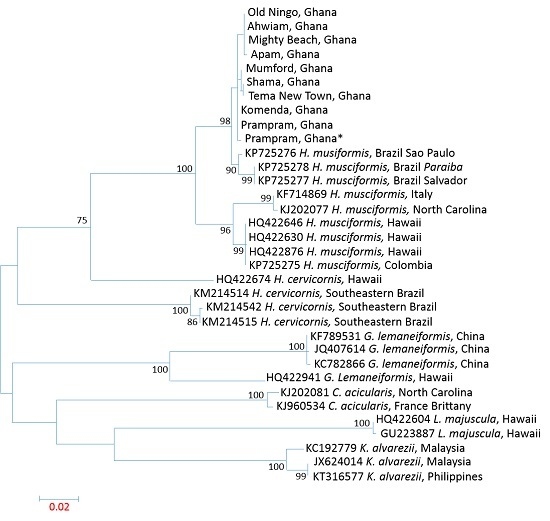
3.2. DNA barcoding by LSU, UPA, and COI
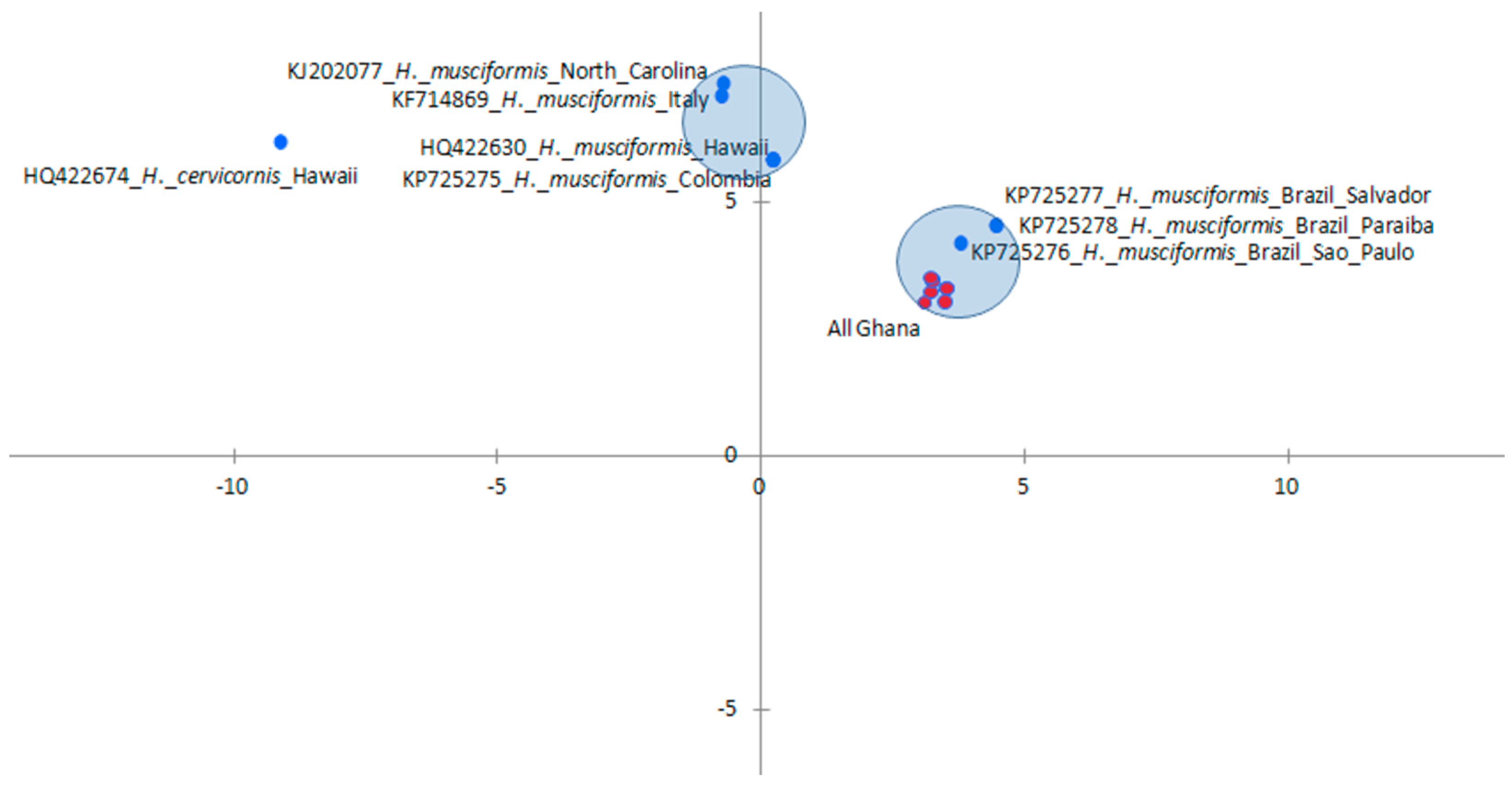
3.3. Influence of Spatial Variations on Phylogenetic Relationship
3.4. Intraspecies Divergence Analysis within COI Sequences
3.5. Chemical Characteristics of H. musciformis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sherwood, A.; Sauvage, T.; Kurihara, A.; Conklin, K.Y.; Presting, G.G. A comparative analysis of COI, LSU and UPA marker data for the Hawaiian florideophyte Rhodophyta: Implications for DNA barcoding of red algae. Cryptogamie 2010, 31, 451–465. [Google Scholar]
- De Jesus, P.B.; Silva, M.S.; de Lyra, G.M.; de Nunes, J.M.C.; Schnadelbach, A.S. Extension of the distribution range of Hypnea stellulifera (Cystocloniaceae, Rhodophyta) to the South Atlantic: Morphological and molecular evidence. Aquat. Bot. 2015, 123, 26–36. [Google Scholar] [CrossRef]
- Lawson, G.W.; Price, J.H. Seaweeds of the western coast of tropical Africa and adjacent islands: A critical assessment. I. Chlorophyta and Xanthophyta. Bot. J. Linn. Soc. 1969, 62, 279–346. [Google Scholar] [CrossRef]
- Serfor-Armah, Y.; Nyarko, B.J.B.; Osae, E.K.; Carboo, D.; Seku, F. Elemental analysis of some green and brown seaweeds from the coastal belt of Ghana. J. Radioanal. Nucl. Chem. 1999, 242, 193–197. [Google Scholar] [CrossRef]
- Mtolera, M.; Buriyo, A. Studies on Tanzanian Hypneaceae: Seasonal Variation in Content and Quality of Kappa-Carrageenan from Hypnea musciformis (Gigartinales: Rhodophyta). West. Indian Ocean J. Mar. Sci. 2005, 3, 43–49. [Google Scholar]
- Rhein-Knudsen, N.; Ale, M.T.; Meyer, A.S. Seaweed hydrocolloid production: An update on enzyme assisted extraction and modification technologies. Mar. Drugs 2015, 13, 3340–3359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durako, M.J.; Dawes, C.J. A comparative seasonal study of two populations of Hypnea musciformis from the East and West Coasts of Florida, USA. I. Growth and chemistry. Mar. Biol. 1980, 59, 151–156. [Google Scholar] [CrossRef]
- Reis, R.P.; Yoneshigue-Valentin, Y.; Pereira Dos Santos, C. Spatial and temporal variation of Hypnea musciformis carrageenan (Rhodophyta-Gigartinales) from natural beds in Rio de Janeiro State, Brazil. J. Appl. Phycol. 2008, 20, 1–8. [Google Scholar] [CrossRef]
- Saunders, G.W. Applying DNA barcoding to red macroalgae: A preliminary appraisal holds promise for future applications. Philos. Trans. R. Soc. Lond. Seri. B Biol. Sci. 2005, 360, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Saunders, G.W.; Hommersand, M.H. Assessing red algal supraordinal diversity and taxonomy in the context of contemporary systematic data. Am. J. Bot. 2004, 91, 1494–1507. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci./R. Soc. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- De Barros-Barreto, M.B.; McIvor, L.; Maggs, C.A.; Gomes Ferreira, P.C. Molecular systematics of Ceramium and Centroceras (Ceramiaceae, Rhodophyta) from Brazil. J. Phycol. 2006, 42, 905–921. [Google Scholar] [CrossRef]
- Carro, B.; Lopez, L.; Peña, V.; Bárbara, I.; Barreiro, R. DNA barcoding allows the accurate assessment of European maerl diversity: A Proof-of-Concept study. Phytotaxa 2014, 190, 176–189. [Google Scholar] [CrossRef]
- Moritz, C.; Cicero, C. DNA barcoding: Promise and pitfalls. PLoS Biol. 2004, 2, e354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radulovici, A.E.; Archambault, P.; Dufresne, F. DNA Barcodes for Marine Biodiversity: Moving Fast Forward? Diversity 2010, 2, 450–472. [Google Scholar] [CrossRef]
- Du, G.; Wu, F.; Guo, H.; Xue, H.; Mao, Y. DNA barcode assessment of Ceramiales (Rhodophyta) in the intertidal zone of the northwestern Yellow Sea. Chin. J. Oceanol. Limnol. 2015, 33, 685–695. [Google Scholar] [CrossRef]
- Freshwater, D.W.; Tudor, K.; O’Shaughnessy, K.; Wysor, B. DNA barcoding in the red algal order Gelidiales: Comparison of COI with rbcL and verification of the barcoding gap. Cryptogam. Algol. 2010, 31, 435–449. [Google Scholar]
- Conklin, K.Y.; Kurihara, A.; Sherwood, A.R. A molecular method for identification of the morphologically plastic invasive algal genera Eucheuma and Kappaphycus (Rhodophyta, Gigartinales) in Hawaii. J. Appl. Phycol. 2009, 21, 691–699. [Google Scholar] [CrossRef]
- Sherwood, A.R.; Presting, G.G. Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. J. Phycol. 2007, 43, 605–608. [Google Scholar] [CrossRef]
- Hu, Z.; Guiry, M.D.; Duan, D. Using the ribosomal internal transcribed spacer (ITS) as a complement marker for species identification of red macroalgae. Hydrobiologia 2009, 635, 279–287. [Google Scholar] [CrossRef]
- Greco, M.; Sáez, C.A.; Brown, M.T.; Bitonti, M.B. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model brown alga. PLoS ONE 2014, 9, e96470. [Google Scholar] [CrossRef] [PubMed]
- Stiger, V.; Horiguchi, T.; Yoshida, T.; Coleman, A.W.; Masuda, M. Phylogenetic relationships of Sargassum (Sargassaceae, Phaeophyceae) with reference to a taxonomic revision of the section Phyllocystae based on ITS-2 nrDNA sequences. Phycol. Res. 2000, 48, 251–260. [Google Scholar] [CrossRef]
- Yoshida, T.; Stiger, V.; Horiguchi, T. Sargassum boreale sp. nov. (Fucales, Phaeophyceae) from Hokkaido, Japan. Phycol. Res. 2000, 48, 125–131. [Google Scholar] [CrossRef]
- Du, G.; Wu, F.; Mao, Y.; Guo, S.; Xue, H.; Bi, G. DNA barcoding assessment of green macroalgae in coastal zone around Qingdao, China. J. Ocean Univ. China 2014, 13, 97–103. [Google Scholar] [CrossRef]
- Francavilla, M.; Franchi, M.; Monteleone, M.; Caroppo, C. The red seaweed Gracilaria gracilis as a multi products source. Mar. Drugs 2013, 11, 3754–3776. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Ingram, J. Tropical Soil Biology and Fertility: A Handbook of Methods. CAB International: Wallingford, UK, 1989. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Bradley, N.; Lymperopoulou, D.S.; Williams, H.N. Differences in bacterial community structure on Hydrilla verticillata and Vallisneria americana in a freshwater spring. Microbes Environ. 2014, 29, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Sluiter, J.B.; Ruiz, R.O.; Scarlata, C.J.; Sluiter, A.D.; Templeton, D.W. Compositional analysis of lignocellulosic feedstocks. 1. Review and description of methods. J. Agric. Food Chem. 2010, 58, 9043–9053. [Google Scholar] [CrossRef] [PubMed]
- Novozamsky, I.; Houba, V.J.G.; van Eck, R.; van Vark, W. A novel digestion technique for multi-element plant analysis. Commun. Soil Sci. Plant Anal. 2008, 14, 239–248. [Google Scholar] [CrossRef]
- Clesceri, L.S. Standard Methods for the Examination of Water and Wastewater, 20th ed.; APHA: Washington, DC, USA, 1998. [Google Scholar]
- Manns, D.; Deutschle, A.L.; Saake, B.; Meyer, A.S. Methodology for quantitative determination of the carbohydrate composition of brown seaweeds (Laminariaceae). RSC Adv. 2014, 4, 25736–25746. [Google Scholar] [CrossRef] [Green Version]
- Ale, M.T.; Maruyama, H.; Tamauchi, H.; Mikkelsen, J.D.; Meyer, A.S. Fucose-containing sulfated polysaccharides from brown seaweeds inhibit proliferation of melanoma cells and induce apoptosis by activation of caspase-3 in vitro. Mar. Drugs 2011, 9, 2605–2621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattio, L.; Payri, C.E.; Verlaque, M. Taxonomic revision and geographic distribution of the subgenus Sargassum (Fucales, Phaeophyceae) in the western and central Pacific Islands based n morphological and molecular analysis. J. Phycol. 2009, 45, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Saunders, G.W.; Kucena, H. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogam. Algol. 2010, 31, 487–528. [Google Scholar]
- Davalieva, K.; Efremov, G.D. Influence of salts and pcr inhibitors on the amplification capacity of three thermostable DNA polymerases. Maced. J. Chem. Chem. Eng. 2010, 29, 57–62. [Google Scholar]
- McInerney, P.; Adams, P.; Hadi, M.Z. Error Rate Comparison during Polymerase Chain Reaction by DNA Polymerase. Mol. Biol. Int. 2014, 2014, 287430. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, A.R.; Kurihara, A.; Conklin, K.Y.; Sauvage, T.; Presting, G.G. The Hawaiian Rhodophyta Biodiversity Survey (2006–2010): A summary of principal findings. BMC Plant Biol. 2010, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Nauer, F.; Cassano, V.; Oliveira, M.C. Description of Hypnea pseudomusciformis sp. nov., a new species based on molecular and morphological analyses, in the context of the H. musciformis complex (Gigartinales, Rhodophyta). J. Appl. Phycol. 2014, 27, 2405–2417. [Google Scholar] [CrossRef]
- Freshwater, D.W.; Parham, S.L.; Phillips, P.R.; Lyon, S.F.; Wilson, R. Hypnea musciformis cytochrome oxidase subunit 1 (COI) gene, partial cd. Available online: http://www.ncbi.nlm.nih.gov/nuccore/KJ202077 (accessed on 13 April 2014).
- Manghisi, A. Identifying alien macroalgae through DNA barcoding: The case of Hypnea cornuta (Cystocloniaceae, Rhodophyta). Trans. Waters Bull. 2011, 5, 42–49. [Google Scholar]
- Usov, A.I. Polysaccharides of the red algae. Adv. Carbohydr. Chem. Biochem. 2011, 65, 115–217. [Google Scholar] [PubMed]
- Arunkumar, K.; Palanivelu, A.; Darsis, A. Proximate composition, nutraceutical constituents and fatty acid profile on GCMS of seaweeds collected from Balk Bay (Thondi), India. Int. J. Curr. Sci. 2014, 12, 57–71. [Google Scholar]
- Ding, L.; Ma, Y.; Huang, B.; Chen, S. Effects of seawater salinity and temperature on growth and pigment contents in Hypnea cervicornis J. Agardh (Gigartinales, Rhodophyta). BioMed Res. Int. 2013, 2013, 594308. [Google Scholar] [CrossRef] [PubMed]
- Marinho-Soriano, E.; Fonseca, P.C.; Carneiro, M.A.A.; Moreira, W.S.C. Seasonal variation in the chemical composition of two tropical seaweeds. Bioresour. Technol. 2006, 97, 2402–2406. [Google Scholar] [CrossRef] [PubMed]
- Charles, A.L.; Chang, C.-K.; Wu, M.-L.; Huang, T.-C. Studies on the expression of liver detoxifying enzymes in rats fed seaweed (Monostroma nitidum). Food Chem. Toxicol. 2007, 45, 2390–2396. [Google Scholar] [CrossRef] [PubMed]
- De Araújo, I.W.F.; de Vanderlei, E.S.O.; Rodrigues, J.A.G.; Coura, C.O.; Quinderé, A.L.G.; Fontes, B.P.; de Queiroz, I.N.L.; Jorge, R.J.B.; Bezerra, M.M.; Rodrigues e Silva, A.A.; et al. Effects of a sulfated polysaccharide isolated from the red seaweed Solieria filiformis on models of nociception and inflammation. Carbohydr. Polym. 2011, 86, 1207–1215. [Google Scholar] [CrossRef]
- Rodrigues, J.A.G.; de Vanderlei, E.S.O.; Silva, L.M.C.M.; de Araújo, I.W.F.; de Queiroz, I.N.L.; de Paula, G.A.; Abreu, T.M.; Ribeiro, N.A.; Bezerra, M.M.; Chaves, H.V.; et al. Antinociceptive and anti-inflammatory activities of a sulfated polysaccharide isolated from the green seaweed Caulerpa cupressoides. Pharmacol. Rep. 2012, 64, 282–292. [Google Scholar] [CrossRef]
- Mohamed, S.; Hashim, S.N.; Rahman, H.A. Seaweeds: A sustainable functional food for complementary and alternative therapy. Trends Food Sci. Technol. 2012, 23, 83–96. [Google Scholar] [CrossRef]
- Andrade, L.R.; Leal, R.N.; Noseda, M.; Duarte, M.E.R.; Pereira, M.S.; Mourão, P.A.S.; Farina, M.; Amado Filho, G.M. Brown algae overproduce cell wall polysaccharides as a protection mechanism against the heavy metal toxicity. Mar. Pollut. Bull. 2010, 60, 1482–1488. [Google Scholar] [CrossRef] [PubMed]






| Country/Region | GenBank Accession Numbers (Identity) | References | |||||
|---|---|---|---|---|---|---|---|
| UPA | (ID) | LSU | (ID) | COI | (ID) | ||
| Ahwiam, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 98.9% | [2,40,41] |
| Apam, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 98.7% | [2,40,41] |
| Komenda, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 99.2% | [2,40,41] |
| Mighty Beach, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 99.0% | [2,40,41] |
| Mumford, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 98.9% | [2,40,41] |
| Old Ningo, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 98.9% | [2,40,41] |
| Shama, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 98.9% | [2,40,41] |
| Tema New Town | KP725300 | 99% | HQ421909 | 99% | KP725276 | 98.9% | [2,40,41] |
| Prampram, Ghana | KP725300 | 99% | HQ421909 | 99% | KP725276 | 99.2% | [2,40,41] |
| Sao Paulo, Brazil | KP725300 | 100% | - | KP725276 | 100% | [41] | |
| Paraiba, Brazil | KP725302 | 100% | - | KP725278 | 99% | [41] | |
| Salvador, Brazil | - | - | KP725277 | 99% | [41] | ||
| Hawaii | HQ421578 | 99% | HQ421798 | 99% | HQ422630 | 95% | [40] |
| Hawaii | HQ421520 | 99% | HQ421909 | 100% | HQ422646 | 95% | [40] |
| Hawaii | - | HQ422316 | 99% | HQ422876 | 95% | [40] | |
| Columbia | KP725299 | 99% | - | KP725275 | 95% | [2] | |
| North Carolina | - | - | KJ202077 | 94% | [42] | ||
| Italy | - | - | KF714869 | 94% | [43] | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ale, M.T.; Barrett, K.; Addico, G.N.D.; Rhein-Knudsen, N.; DeGraft-Johnson, A.A.; Meyer, A.S. DNA-Based Identification and Chemical Characteristics of Hypnea musciformis from Coastal Sites in Ghana. Diversity 2016, 8, 14. https://doi.org/10.3390/d8020014
Ale MT, Barrett K, Addico GND, Rhein-Knudsen N, DeGraft-Johnson AA, Meyer AS. DNA-Based Identification and Chemical Characteristics of Hypnea musciformis from Coastal Sites in Ghana. Diversity. 2016; 8(2):14. https://doi.org/10.3390/d8020014
Chicago/Turabian StyleAle, Marcel Tutor, Kristian Barrett, Gloria Naa Dzama Addico, Nanna Rhein-Knudsen, Amoako Atta DeGraft-Johnson, and Anne S. Meyer. 2016. "DNA-Based Identification and Chemical Characteristics of Hypnea musciformis from Coastal Sites in Ghana" Diversity 8, no. 2: 14. https://doi.org/10.3390/d8020014
APA StyleAle, M. T., Barrett, K., Addico, G. N. D., Rhein-Knudsen, N., DeGraft-Johnson, A. A., & Meyer, A. S. (2016). DNA-Based Identification and Chemical Characteristics of Hypnea musciformis from Coastal Sites in Ghana. Diversity, 8(2), 14. https://doi.org/10.3390/d8020014