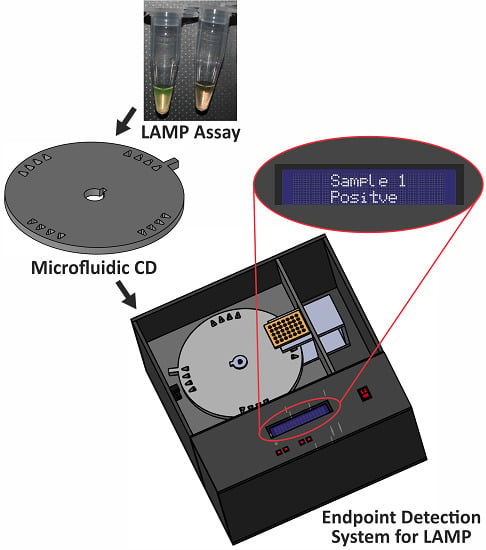
A Portable Automatic Endpoint Detection System for Amplicons of Loop Mediated Isothermal Amplification on Microfluidic Compact Disk Platform
Abstract
:1. Introduction
2. Methodology
2.1. Sample Preparation
2.2. Disc Fabrication

2.3. Development of the Endpoint Detection System for LAMP
2.3.1. Design of Detection System



2.3.2. Operation of Detection System
3. Results and Discussion


| Sample ID | Concentration of DNA Template (ng/µL) for LAMP Reaction | Color Changes of Resulting Dye ** | Visual Identification of Resulting Dye ** | Sensor Reading (Normalized Pulse Duration) Mean ± SD | Automatic Detection System Interpretation of Resulting Dye ** |
|---|---|---|---|---|---|
| 1 | 2.5E+00 | Yellowish Green | Positive | 0.067 ± 0.038 | Positive |
| 2 | 2.5E-01 | Yellowish Green | Positive | 0.025 ± 0.035 | Positive |
| 3 | 2.5E-02 | Yellowish Green | Positive | 0.045 ± 0.043 | Positive |
| 4 | 2.5E-03 | Yellowish Green | Positive | 0.065 ± 0.033 | Positive |
| 5 | 2.5E-04 | Faded Orange | Negative | 0.834 ± 0.042 | Negative |
| 6 | 2.5E-05 | Faded Orange | Negative | 0.832 ± 0.035 | Negative |
| 7 | 2.5E-06 | Faded Orange | Negative | 0.820 ± 0.037 | Negative |
| 8 | 0 * | Unchanged | Negative | 0.971 ± 0.037 | Negative |

4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Crim, S.M.; Iwamoto, M.; Huang, J.Y.; Griffin, P.M.; Gilliss, D.; Cronquist, A.B.; Cartter, M.; Tobin-D’Angelo, M.; Blythe, D.; Smith, K. Incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 us sites, 2006–2013. Morb. Mortal. Wkly. Rep. 2014, 63, 328–332. [Google Scholar]
- Mandal, P.; Biswas, A.; Choi, K.; Pal, U. Methods for rapid detection of foodborne pathogens: An overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Jofré, A.; Martin, B.; Garriga, M.; Hugas, M.; Pla, M.; Rodrı́guez-Lázaro, D.; Aymerich, T. Simultaneous detection of Listeria monocytogenes and Salmonella by multiplex PCR in cooked ham. Food Microbiol. 2005, 22, 109–115. [Google Scholar] [CrossRef]
- Ko, S.; Grant, S.A. A novel fret-based optical fiber biosensor for rapid detection of Salmonella Typhimurium. Biosens. Bioelectron. 2006, 21, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.K. Biosensors and bio‐based methods for the separation and detection of foodborne pathogens. Adv. Food Nutr. Res. 2008, 54, 1–44. [Google Scholar] [PubMed]
- Rasooly, A.; Herold, K.E. Biosensors for the analysis of food-and waterborne pathogens and their toxins. J. AOAC Int. 2006, 89, 873–883. [Google Scholar] [PubMed]
- Leonard, P.; Hearty, S.; Quinn, J.; O’Kennedy, R. A generic approach for the detection of whole Listeria Monocytogenes cells in contaminated samples using surface plasmon resonance. Biosens. Bioelectron. 2004, 19, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Warsinke, A.; Benkert, A.; Scheller, F. Electrochemical immunoassays. Fresenius J. Anal. Chem. 2000, 366, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Grant, S.A. Development of a novel fret method for detection of Listeria or Salmonella. Sens. Actuators B Chem. 2003, 96, 372–378. [Google Scholar] [CrossRef]
- Majoul, I. Analysing the action of bacterial toxins in living cells with fluorescence resonance energy transfer (fret). Int. J. Med. Microbiol. 2004, 293, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Ulvick, S.J.; Uzzell, G.L.; Tabb, J.S.; Valdes, E.R.; Batt, C.A. Novel immuno-fret assay method for Bacillus spores and Escherichia coli O157: H7. Biochem. Biophys. Res. Commun. 2001, 287, 875–880. [Google Scholar] [CrossRef] [PubMed]
- O’sullivan, C.; Vaughan, R.; Guilbault, G. Piezoelectric Immunosensors-Theory and Applications; Taylor & Francis: Cork, Ireland, 1999. [Google Scholar]
- Pancrazio, J.; Whelan, J.; Borkholder, D.; Ma, W.; Stenger, D. Development and application of cell-based biosensors. Ann. Biomed. Eng. 1999, 27, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.K.; Wampler, J.L. Animal and cell culture models for foodborne bacterial pathogens. Foodborne Pathog. Microbiol. Mol. Biol. 2005, 15–32. [Google Scholar]
- Ziegler, C. Cell-based biosensors. Fresenius’ J. Anal. Chem. 2000, 366, 552–559. [Google Scholar] [CrossRef]
- Wang, R.F.; Cao, W.W.; Cerniglia, C. A universal protocol for PCR detection of 13 species of foodborne pathogens in foods. J. Appl. Microbiol. 1997, 83, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Jeníková, G.; Pazlarová, J.; Demnerová, K. Detection of Salmonella in food samples by the combination of immunomagnetic separation and pcr assay. Int. Microbiol. 2010, 3, 225–229. [Google Scholar]
- Oliveira, S.; Santos, L.; Schuch, D.; Silva, A.; Salle, C.; Canal, C. Detection and identification of Salmonellas from poultry-related samples by PCR. Vet. Microbiol. 2002, 87, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Paccassoni, E.; Fach, P.; Bunge, C.; Martin, A.; Helmuth, R. Diagnostic real-time pcr for detection of Salmonella in food. Appl. Environ. Microbiol. 2004, 70, 7046–7052. [Google Scholar] [CrossRef] [PubMed]
- Beckers, H.; Tips, P.; Soentoro, P.; Delfgou-Van Asch, E.; Peters, R. The efficacy of enzyme immunoassays for the detection of Salmonellas. Food Microbiol. 1988, 5, 147–156. [Google Scholar] [CrossRef]
- Magliulo, M.; Simoni, P.; Guardigli, M.; Michelini, E.; Luciani, M.; Lelli, R.; Roda, A. A rapid multiplexed chemiluminescent immunoassay for the detection of Escherichia coli O157:H7, Yersinia Enterocolitica, Salmonella Typhimurium, and Listeria Monocytogenes pathogen bacteria. J. Agric. Food Chem. 2007, 55, 4933–4939. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.; Malo, A.; Siddons, C.; Harkin, M. Use of commercial enzyme immunoassays and immunomagnetic separation systems for detecting Escherichia coli O157 in bovine fecal samples. Appl. Environ. Microbiol. 1997, 63, 2549–2553. [Google Scholar] [PubMed]
- Rasooly, A.; Herold, K.E. Food microbial pathogen detection and analysis using DNA microarray technologies. Foodborne Pathog. Dis. 2008, 5, 531–550. [Google Scholar] [CrossRef] [PubMed]
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.K.; Kricka, L.J. Microfluidics and point-of-care testing. Lab Chip 2008, 8, 1982–1983. [Google Scholar] [CrossRef] [PubMed]
- Mairhofer, J.; Roppert, K.; Ertl, P. Microfluidic systems for pathogen sensing: A review. Sensors 2009, 9, 4804–4823. [Google Scholar] [CrossRef] [PubMed]
- Lagally, E.; Scherer, J.; Blazej, R.; Toriello, N.; Diep, B.; Ramchandani, M.; Sensabaugh, G.; Riley, L.; Mathies, R. Integrated portable genetic analysis microsystem for pathogen/infectious disease detection. Anal. Chem. 2004, 76, 3162–3170. [Google Scholar] [CrossRef]
- Easley, C.J.; Karlinsey, J.M.; Bienvenue, J.M.; Legendre, L.A.; Roper, M.G.; Feldman, S.H.; Hughes, M.A.; Hewlett, E.L.; Merkel, T.J.; Ferrance, J.P. A fully integrated microfluidic genetic analysis system with sample-in–answer-out capability. Proc. Natl. Acad. Sci. USA 2006, 103, 19272–19277. [Google Scholar] [CrossRef] [PubMed]
- Beyor, N.; Yi, L.; Seo, T.S.; Mathies, R.A. Integrated capture, concentration, polymerase chain reaction, and capillary electrophoretic analysis of pathogens on a chip. Anal. Chem. 2009, 81, 3523–3528. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, U.; Witek, M.A.; Adams, A.A.; Osiri, J.K.; Hupert, M.L.; Bianchi, T.S.; Roelke, D.L.; Soper, S.A. Enrichment and detection of Escherichia coli O157:H7 from water samples using an antibody modified microfluidic chip. Anal. Chem. 2010, 82, 2844–2849. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Wang, H.; Hupert, M.; Witek, M.; Dharmasiri, U.; Pingle, M.R.; Barany, F.; Soper, S.A. Modular microfluidic system fabricated in thermoplastics for the strain-specific detection of bacterial pathogens. Lab Chip 2012, 12, 3348–3355. [Google Scholar] [CrossRef] [PubMed]
- Madou, M.; Zoval, J.; Jia, G.; Kido, H.; Kim, J.; Kim, N. Lab on a cd. Annu. Rev. Biomed. Eng. 2006, 8, 601–628. [Google Scholar] [CrossRef] [PubMed]
- Gorkin, R.; Park, J.; Siegrist, J.; Amasia, M.; Lee, B.S.; Park, J.-M.; Kim, J.; Kim, H.; Madou, M.; Cho, Y.-K. Centrifugal microfluidics for biomedical applications. Lab Chip 2010, 10, 1758–1773. [Google Scholar] [CrossRef] [PubMed]
- Zoval, J.V.; Madou, M.J. Centrifuge-based fluidic platforms. IEEE Proc. 2004, 92, 140–153. [Google Scholar] [CrossRef]
- Grumann, M.; Geipel, A.; Riegger, L.; Zengerle, R.; Ducrée, J. Batch-mode mixing on centrifugal microfluidic platforms. Lab Chip 2005, 5, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Haeberle, S.; Brenner, T.; Schlosser, H.P.; Zengerle, R.; Ducrée, J. Centrifugal micromixery. Chem. Eng. Technol. 2005, 28, 613–616. [Google Scholar] [CrossRef]
- Park, J.-M.; Cho, Y.-K.; Lee, B.-S.; Lee, J.-G.; Ko, C. Multifunctional microvalves control by optical illumination on nanoheaters and its application in centrifugal microfluidic devices. Lab Chip 2007, 7, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Steigert, J.; Grumann, M.; Brenner, T.; Riegger, L.; Harter, J.; Zengerle, R.; Ducrée, J. Fully integrated whole blood testing by real-time absorption measurement on a centrifugal platform. Lab Chip 2006, 6, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Duarte, R.; Gorkin, R.A., III; Abi-Samra, K.; Madou, M.J. The integration of 3d carbon-electrode dielectrophoresis on a cd-like centrifugal microfluidic platform. Lab Chip 2010, 10, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Ducrée, J.; Haeberle, S.; Lutz, S.; Pausch, S.; Von Stetten, F.; Zengerle, R. The centrifugal microfluidic bio-disk platform. J. Micromech. Microeng. 2007, 17, S103–S115. [Google Scholar] [CrossRef]
- Lee, B.S.; Lee, Y.U.; Kim, H.-S.; Kim, T.-H.; Park, J.; Lee, J.-G.; Kim, J.; Kim, H.; Lee, W.G.; Cho, Y.-K. Fully integrated lab-on-a-disc for simultaneous analysis of biochemistry and immunoassay from whole blood. Lab Chip 2011, 11, 70–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Sunkara, V.; Kim, T.-H.; Hwang, H.; Cho, Y.-K. Lab-on-a-disc for fully integrated multiplex immunoassays. Anal. Chem. 2012, 84, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Riegger, L.; Grumann, M.; Nann, T.; Riegler, J.; Ehlert, O.; Bessler, W.; Mittenbuehler, K.; Urban, G.; Pastewka, L.; Brenner, T. Read-out concepts for multiplexed bead-based fluorescence immunoassays on centrifugal microfluidic platforms. Sens. Actuators A Phys. 2006, 126, 455–462. [Google Scholar] [CrossRef]
- Cho, Y.-K.; Lee, J.-G.; Park, J.-M.; Lee, B.-S.; Lee, Y.; Ko, C. One-step pathogen specific DNA extraction from whole blood on a centrifugal microfluidic device. Lab Chip 2007, 7, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Kim, Y.; Cho, J.; Lee, J.-Y.; Choi, M.-S.; Cho, Y.-K. Lab-on-a-disc for simultaneous determination of nutrients in water. Anal. Chem. 2013, 85, 2954–2960. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.S.; Lee, J.-N.; Park, J.-M.; Lee, J.-G.; Kim, S.; Cho, Y.-K.; Ko, C. A fully automated immunoassay from whole blood on a disc. Lab Chip 2009, 9, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Wang, S.; Luo, J.; Lee, L.J.; Yang, S.-T.; Madou, M.J. Design of a compact disk-like microfluidic platform for enzyme-linked immunosorbent assay. Anal. Chem. 2004, 76, 1832–1837. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Park, J.; Kim, C.-J.; Cho, Y.-K. Fully integrated lab-on-a-disc for nucleic acid analysis of food-borne pathogens. Anal. Chem. 2014, 86, 3841–3848. [Google Scholar] [CrossRef] [PubMed]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Chen, C.-C.; Wei, S.-C.; Lu, H.-H.; Liang, Y.-H.; Lin, C.-W. Diagnostic devices for isothermal nucleic acid amplification. Sensors 2012, 12, 8319–8337. [Google Scholar] [CrossRef] [PubMed]
- Muangchuen, A.; Chaumpluk, P.; Suriyasomboon, A.; Ekgasit, S. Colorimetric detection of ehrlichia canis via nucleic acid hybridization in gold nano-colloids. Sensors 2014, 14, 14472–14487. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Jin, W.; Zhou, C.; Han, S.; Yang, W.; Zhu, Q.; Jin, Q.; Mu, Y. Integrated glass microdevice for nucleic acid purification, loop-mediated isothermal amplification, and online detection. Anal. Chem. 2011, 83, 3336–3342. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Lee, C.-N.; Mark, H.; Meldrum, D.R.; Lin, C.-W. Efficient, specific, compact Hepatitis b diagnostic device: Optical detection of the Hepatitis b virus by isothermal amplification. Sens. Actuators B Chem. 2007, 127, 598–605. [Google Scholar] [CrossRef]
- Li, F.; Yan, W.; Long, L.; Qi, X.; Li, C.; Zhang, S. Development and application of loop-mediated isothermal amplification assays for rapid visual detection of cry2Ab and cry3A genes in genetically-modified crops. Int. J. Mol. Sci. 2014, 15, 15109–15121. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Cheng, S.; Chu, Y.; Wu, H.; Zou, B.; Huang, H.; Xi, T.; Zhou, G. A closed-tube detection of loop-mediated isothermal amplification (lamp) products using a wax-sealed fluorescent intercalator. J. Nanosci. Nanotechnol. 2013, 13, 3999–4005. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Geva, E.; Mauk, M.; Qiu, X.; Abrams, W.R.; Malamud, D.; Curtis, K.; Owen, S.M.; Bau, H.H. An isothermal amplification reactor with an integrated isolation membrane for point-of-care detection of infectious diseases. Analyst 2011, 136, 2069–2076. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Liu, Y.; Kong, J.; Jiang, X. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal. Chem. 2010, 82, 3002–3006. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28. [Google Scholar] [CrossRef] [PubMed]
- Primerexplore v4. Available online: http://primerexplorer.jp/elamp4.0.0/index.html (accessed on 13 November 2014).
- Arunrut, N.; Prombun, P.; Saksmerprome, V.; Flegel, T.W.; Kiatpathomchai, W. Rapid and sensitive detection of infectious hypodermal and hematopoietic necrosis virus by loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods 2011, 171, 21–25. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, S.M.; Ibrahim, F.; Sayad, A.A.; Thiha, A.; Pei, K.X.; Mohktar, M.S.; Hashim, U.; Cho, J.; Thong, K.L. A Portable Automatic Endpoint Detection System for Amplicons of Loop Mediated Isothermal Amplification on Microfluidic Compact Disk Platform. Sensors 2015, 15, 5376-5389. https://doi.org/10.3390/s150305376
Uddin SM, Ibrahim F, Sayad AA, Thiha A, Pei KX, Mohktar MS, Hashim U, Cho J, Thong KL. A Portable Automatic Endpoint Detection System for Amplicons of Loop Mediated Isothermal Amplification on Microfluidic Compact Disk Platform. Sensors. 2015; 15(3):5376-5389. https://doi.org/10.3390/s150305376
Chicago/Turabian StyleUddin, Shah Mukim, Fatimah Ibrahim, Abkar Ahmed Sayad, Aung Thiha, Koh Xiu Pei, Mas S. Mohktar, Uda Hashim, Jongman Cho, and Kwai Lin Thong. 2015. "A Portable Automatic Endpoint Detection System for Amplicons of Loop Mediated Isothermal Amplification on Microfluidic Compact Disk Platform" Sensors 15, no. 3: 5376-5389. https://doi.org/10.3390/s150305376
APA StyleUddin, S. M., Ibrahim, F., Sayad, A. A., Thiha, A., Pei, K. X., Mohktar, M. S., Hashim, U., Cho, J., & Thong, K. L. (2015). A Portable Automatic Endpoint Detection System for Amplicons of Loop Mediated Isothermal Amplification on Microfluidic Compact Disk Platform. Sensors, 15(3), 5376-5389. https://doi.org/10.3390/s150305376