2.3.1. Discrete Optimization Based on the MRF
After obtaining the similarity measure for the two different modal images, we will use the FFD as the transformation model and use Markov random field (MRF) optimization [
32] to obtain the transformation parameters in the FFD. The reason for choosing this discrete optimization method is that it does not need to calculate the gradient of the energy function in the process of optimization, which thereby facilitates producing the good registration result by avoiding falling into the local minimum. In this method, the image registration problem will be converted into the MRF based discrete optimization problem.
where
E denotes the general form of a first-order MRF, i.e., the energy function and
is a constant;
G is the set of vertices and
denotes the number of vertices in
G, where
G can be regarded as the vertex set in the FFD, because this method uses the FFD as the deformation model;
and
refer to the neighborhood of vertices
p and
q, respectively;
is the discrete labelling while
and
are the labels that are assigned to the vertices
p and
q, respectively;
denotes the data item of the energy function
, while
represents its smooth regularization and it takes the
L1-norm to encourage the neighboring nodes
p and
q to keep the displacement.
Accordingly, the MRF optimization, actually, is to seek to assign a label that is associated with the deformation to each vertex, so that the energy function in Equation (8) is minimized. In this paper, the fast primal-dual (Fast-PD) [
33] algorithm will be used for the MRF optimization to produce the registration result. More details about the Fast-PD algorithm can be found in [
33].
2.3.2. Spatial Constraint Based on the FMIND
When the above registration method is applied to three-dimensional (3D) medical images, the number of deformation field variables will be large. If all pixels’ displacement along the x, y, and z directions is considered, the number of dense deformation field variables will be 3||||, where || and || denote the number of pixels in the x, y, and z dimensions of the image I, respectively. For example, there will be 50,331,648 deformation field variables when |||| = 256. It is indeed very time-consuming to address such a high dimensional optimization problem.
In the reference and float images, sometimes only some areas involve non-rigid deformation. In addition, the non-rigid registration is unnecessary for some smooth areas. For these regions, the mask can be used to indicate that they will be excluded from the registration process. In this way, we can not only reduce the number of variables for describing the deformation field, but also focus the calculation of the energy function in the image areas that indeed involve the local non-rigid deformation. However, the shape of areas without non-rigid deformation is often irregular. Generally, manual intervention or image segmentation is needed for obtaining the appropriate mask image. However, these technologies cannot ensure that the satisfactory mask image can be produced for US and PET images due to their low image contrast, blurriness, and edge discontinuousness.
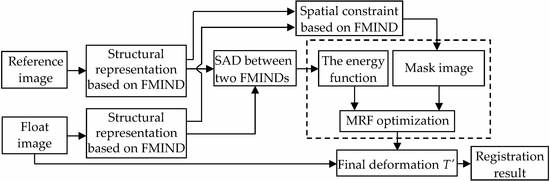
We will put forward the spatial constraint method based on the FMIND to address the above problem. From Equation (7), it can be seen that contains the corresponding relationship of the local spatial information at the pixel x in the images I and J. This information can be used to reduce the number of variables for describing the deformation field and limit the control nodes in the deformation field to move in the areas with the local non-rigid deformation. In the FMIND based spatial constraint method, the vertex set G will be divided into the set of static vertices and the set of dynamic vertices based on the local spatial information included in the FMIND. The vertices in are similar to those in the smooth areas and the areas that involve no deformation. Meanwhile, the vertices in are similar to those in the non-smooth areas involving the deformation. The calculation of the energy function can be restricted in the areas involving the non-rigid deformation through the movement of these dynamic vertices with the local non-rigid deformation. In this way, the number of deformation field variables will decrease from 3|||| to 3||||||, where ||, ||, and || denote the number of vertices in x, y, and z dimensions of . By utilizing the FMIND based spatial constraint, we can obtain the division of vertices, thereby generating the mask image without manual intervention and image segmentation.
There will be two requirements that no vertices of the whole MRF model will be omitted and no repeated division of vertices will be done to ensure the effective division of
G in the FMIND based spatial constraint method. Correspondingly, the logical relationship among
G,
, and
can be expressed as
and
, where
denotes the empty set. We have designed the following vertex partition algorithm according to the above requirements. For any vertex
of the set
G in the MRF model, i.e.,
we will check the similarity metric
for each pixel
x in the local image patch
with radius
, which takes
in
G as the center, as shown in
Figure 3. Let
con denote the number of pixels in
whose similarity
is greater than a certain threshold
. If the ratio of
con to the patch size
is greater than the static factor
,
will be regarded as a static vertex. In a similar way, we can determine other static vertices to generate the final set
. Accordingly,
will be easily computed as
.
Obviously, the performance of the vertex partition algorithm depends on three key parameters
,
, and
. Here,
will influence the decision of whether two pixels are similar and
is used to adjust the probability that
is divided into
.
Section 3.1 discusses the choice of these parameters. Algorithm 1 shows the detailed implementation of the proposed vertex partition algorithm.
| Algorithm 1. Partition of the vertex set G |
| Input: , |
| Output: |
| (1) ; |
| (2) for (i = 1; i ≤ ||; i = i++) |
| (3) for (j = 1; j ≤ ||; j = j++) |
| (4) for (k = 1; k ≤ ||; k = k++) |
| (5) con = 0; |
| (6) while ) |
| (7) if () |
| (8) con++; |
| (9) end if |
| (10) end while |
| (11) if |
| (12) ; |
| (13) end if |
| (14) end for |
| (15) end for |
| (16) end for |
| (17) = ; |
| (18) return ; |