1. Introduction
In many clinical specialties, there exists a high variety of image modalities that clinicians typically use to analyze the patient’s condition and aid in the diagnostic process. Given the importance of technology at present, namely, its inclusion in different areas to improve an expert’s work, many computational systems have been implemented to facilitate this clinical processes [
1,
2,
3]. This automatization of different clinical manual tasks provide solutions and improvements that are characterized by desirable properties that the expert cannot offer such as determinism, repeatability or objectivity. Regarding image analysis, over the years, many computational proposals have been presented based on computer vision techniques that are exploited to analyze relevant areas of the images of interest, measure relevant parameters and monitor their evolution and potential variations, which is the typical procedure that is used in many Computer Aided Diagnostic (CAD) systems.
In ophthalmology, different image modalities have increased in popularity over the last years. The color fundus retinography is the most traditional and widely extended ophthalmic image modality, that allows the direct visualization of the eye fundus and their possible pathological visible structures. Being an invasive capture technique, the Fluorescein Angiography (FA) provides an enhanced visualization of the retinal vascular structure by injecting a dye contrast into the patient. With the Optical Coherence Tomography (OCT), we can visualize and analyze volumetric scans of the inner retinal layers, non-invasively and in real time. Based on the OCT and the FA characteristics, a new image modality appeared, combining both the OCT benefits and the FA visualization: the Optical Coherence Tomography by Angiography (OCTA). OCTA [
4] imaging constitutes the new ophthalmic image modality, and it is the first that allows an exhaustive and non-invasive vascular analysis of the retina. OCTA accurately represents the vascularity of the foveal region within the retinal layers. This representation is extracted based on the blood movement in the eye microvasculature. Given that numerous systematic and eye diseases are related with the microvascular circulation, OCTA constitutes a relevant new ophthalmic image modality that better aids the diagnosis and monitoring processes of these pathologies. As reference, diseases such as diabetes can generate a Diabetic Retinopathy (DR), which provokes microvascular problems in advanced stages of the disease, eventually producing total blindness in the worst case scenario. Also, the Retinal Vein Occlusion (RVO) produces a problematic loss of vascularity and a progressive vision loss.
With these representative examples among many others, it is clear that the retinal vascular integrity is related with the Visual Acuity (VA) [
5,
6,
7,
8] of the patients. Commonly, to estimate the VA of a given patient, it is necessary to perform different rudimentary tests [
9]. In particular, these tests are normally related with the analysis of the ability of the patient to observe characteristic objects of different sizes and at different distances. Consequently, the final VA estimation is subjective whereas its process is tedious and slow. In that context, any technological contribution that avoids or facilitates this revision process is extremely desired in the ophthalmic field.
Given the potential of the OCTA image modality, over recent years, several clinical studies were published demonstrating its utility in the analysis of different vascular diseases. Hence, Balaratnasingam et al. [
5] demonstrated in their study that the VA is correlated with the Foveal Avascular Zone (FAZ) in patients with DR and RVO diseases. Other clinical studies [
6,
7,
8] related different manual biomarkers extracted from the OCTA images with the VA of the patient. Wei et al. [
10] demonstrated the adequate repeatability of the vascular density as representative biomarker in healthy subjects in both macular and optic disk regions. Posteriorly, Vivien at al. [
11], demonstrated the influence of the glaucoma disease over the vessel tree characteristics, obtaining as conclusions the absence of retinal vascularity in the pathological cases over a control group. Given its novelty, there exists a limited number of computational proposals to aid the diagnosis and monitoring of pathologies with this image modality. In fact, the existing works are mainly based on the extraction of characteristic biomarkers in the OCTA images or improving the image quality to facilitate its posterior analysis. As reference, Díaz et al. [
12] briefly introduced a method to automatically extract and measure generally the Vascular Density (VD). Also, the authors proposed a method to extract and measure the FAZ [
13] in healthy and DR OCTA images and they performed a FAZ circularity study in healthy and pathological images. Alam et al. [
14] proposed different features extracted from the OCTA images, namely, the vessel tortuosity, the mean diameter of the blood vessels, the semi-automatic FAZ area, the FAZ irregularity and the density of the parafoveal avascular region using fractal dimensions. Guo et al. [
15] segmented and quantified avascular ischemic zones with map probabilities that were extracted by a deep convolutional neural network. Wang et al. [
16] proposed a method to produce mosaics using different overlapping OCTA images, increasing the wide field of vision for the posterior clinical analysis. Wei et al. [
17] improved the quality of the OCTA scans in their work whereas Camino et al. [
18] identified existing shadow artifacts that typically appear in the OCTA images to improve the visualization and facilitate their inspection.
Given that there are several clinical studies that demonstrate the correlation of the VA with different parameters of the OCTA images, and the complexity of the VA manual calculation, a faster and more reliable alternative to automatically determinate this measurement is desired. Nowadays, no automatic approach is available that obtains the VA of the patient without the clinical intervention. For this reason, we propose a novel method to automatically estimate the VA of a patient using biomarkers that are directly extracted from the OCTA images. In fact, those biomarkers are barely used in existing state of the art works. The proposed method is mainly based on the extraction of two representative groups of biomarkers over the OCTA images and their posterior adaptation to be used as input in a Support Vector Machine (SVM) to estimate the target VA. The first biomarker is the FAZ area [
13], given the popularity of the FAZ characteristics in relation with the vision problems. The second group of biomarkers measure the VD. The VD represents an important metric of the OCTA images, given the quality and precision of the vascular visualization in this image modality. Also, its manual measurement is not feasible, given the necessity of labeling each pixel that could be considered as a vessel. In that line, the difficulty of obtaining a manual labeling imply the problem of performing the validation process by the comparison with an expert ground truth. As an alternative, we propose a method to extract the VD, comparing the results with the performance of a semi-automatic module that provides a capture device of reference. Finally, using these validated biomarkers, we perform the estimation of the VA using an adapted and tested SVM, demonstrating that both the automatic FAZ area and the VD extraction are reliable and that the VA can be predicted from the OCTA images.
In line with this, a preliminary version of this methodology has been included in a clinical study (Díez-Sotelo et al. [
19]) about how the visual acuity is affected in a particular cohort of ophthalmological patients. Once the automatic proposal has demonstrated its potential in a real case, the main objective of this work is to perform an exhaustive analysis of the performance of the methodology analyzing its main components as well as the most relevant computational characteristics in order to have an optimized and generalized methodology.
The paper is organized as follows:
Section 2 details the used dataset for validation purposes as well as the characteristics of the proposed methodology;
Section 3 exposes the performed experiments and their motivations; and, finally,
Section 4 discusses the obtained results and the contribution of our proposal to the state of the art.
2. Materials and Methods
The proposed methodology is generally structured as presented in
Figure 1, and is explained in the following sections. First of all, using the OCTA images as input, the method extract representative avascular and vascular biomarkers: the FAZ area is extracted identifying avascular regions followed by the FAZ region selection and the improvement of its contour [
13]. Moreover, the VD is extracted using the skeletonization of a previously extracted vascular mask using an adapted thresholding process. Next, we adapt and train an SVM regression model to finally estimate the target VA using the extracted biomarkers as input features.
2.1. OCTA Image Acquisition
The OCTA image acquisition process is based on the OCT extraction and the detection of the blood movement in successive OCT scans at the same depth of the eye fundus. In particular, it is generally based on the OCT acquisition process, which is performed following the process that we can graphically see in
Figure 2 and that is explained below. In particular, OCT imaging is based on the interference of light over the retinal layers. For this reason, the first step implies the direct interference of light (Step 1 in
Figure 2). The light is sent to the
optical coupler and split into different optical fibers (Step 2 in
Figure 2) that transmit the light to a mirror and to the retina, at the same time. The light that is reflected in the mirror returns in a constant way, and for this reason it is used as reference (Step 3 in
Figure 2). By contrast, the reflected light in the retina differs in each retinal layer, as we can see in Step 4 of
Figure 2. Then, the reflected lights from both the mirror and the retinal layers are compared. The difference between the light strength of each layer and the reference is measured (Step 5 in
Figure 2) to obtain the final signal that is represented as the OCT image before the processed step of the measured information.
Once we capture the OCT images, the required steps to obtain the final target OCTA image are graphically represented in
Figure 3. To do this, first, it is necessary to perform the explained OCT acquisition at the same positions and different progressive times (in the range of seconds). To obtain the spatial resolution of the OCTA images, it is also necessary the OCT acquisition at different positions from all the foveal zone. In Step 1 of
Figure 3, we can see the graphical representation of the required data to obtain the OCTA images and their distribution: different groups of OCT image sets in the same place and in the nearby areas, using the 3-dimensional characteristics of the OCT images. For each group of OCT images that are extracted from the same position, the average is measured to obtain images with high quality and the blood flow measurement is made over the obtained OCT image (Step 2 in
Figure 3) based on the differences over the OCT images at the same position. By using the blood flow in the nearest OCT scans, it is possible to construct the final target OCTA image (Step 3 in
Figure 3).
2.2. Used Image Dataset
The methodology was validated using an image dataset that contains 860 OCTA images that were extracted by the capture device DRI OCT Triton; Topcon Corp. These OCTA images were obtained in progressive consultations of different patients that suffered RVO, with 3 months between each consultation. The dataset also includes the manual measurement of the VA of the patient in the pathological eye, extracted with the previously mentioned VA test procedure [
9]. In particular, we organized the dataset into 215 samples, each one containing the information extracted from one patient consultation, including:
The 4 different OCTA images that we use as reference: 3 mm mm Superficial Capillary Plexus (SCP), 3 mm mm Deep Capillary Plexus (DCP), 6 mm mm SCP and 6 mm mm DCP.
The VA of the patient at the consultation, manually measured by a clinical specialist.
In summary, as indicated, the image dataset contains 860 images that we distributed into 215 samples, where each one represents the information of a patient in one ophthalmic consultation.
Moreover, in order to test the suitability and robustness of the automatically extracted vascular and avascular biomarkers, we further obtained the manual measurement of the FAZ area as well as the semi-automatic VD extraction that includes the used capture device of reference. Again, both tasks were performed by an expert clinician.
The used dataset was retrieved in clinical practice, where several images present some motion artifacts that are typical in this image modality. Despite that, these artifacts do not represent a significant proportion of the images to compromise the robustness of the method. In that line, OCTA images with severe deteriorations in the capturing process that are largely deformed were excluded from the study, as they represent images that are normally omitted from analysis by the expert clinicians given they constitute an unfeasible scenario to perform the manual measurement of the FAZ.
2.3. Automatic Estimation of the Avascular and Vascular Biomarkers
2.3.1. FAZ Area Measurement
The FAZ area [
13] is automatically extracted from each 3 mm
mm SCP OCTA image, resolution where this region is more clearly identifiable. Firstly, we apply a top-hat morphological operator as preprocessing step, obtaining an enhanced OCTA image. Then, we use an edge detector followed by morphological operators to obtain a binary image with different areas that represents the main existing avascular zones. Given the characteristics of the FAZ region, we coherently select the largest and centered region, posteriorly applying a region growing process to refine the extracted contour. Finally, we calculate the area of this region as the target FAZ area as we see in Equation (
1).
where
p represents the number of pixels of the extracted FAZ region,
mm represents the image size in millimeters, and
H and
W indicate the dimensions of the analyzed OCT-A image. To produce a global measurement independent from the resolution, this value represents the area in millimeters. In
Figure 4, we can see a representative progressive example of these mentioned steps to obtain the target FAZ segmentation and the corresponding FAZ area measurement.
2.3.2. VD Measurement
In order to extract the VD, we implemented a fully automatic method that precisely segments the vascular regions and measures their constituent density. In particular, this method is based on the analysis of the main structure of the retinal vascularity. Given the high level of noise that is typically present in the OCTA images and the irregularities in its background, we can not use directly the raw OCTA image to measure the VD. In such a case, it is desirable a previous improvement of the analyzed region, reducing the levels of noise and to only preserve the target vascularity. Moreover, it is also remarkable that the OCTA images typically present a high variability in terms of intensities: two OCTA scans taken at the same time and spatial zone of the retina can present variations in terms of intensities. For these reasons, we need a method that contemplates both characteristics: omitting the background noise and being robust to the variability of intensities.
Considering the previous requirements, we first performed a threshold step to segment the higher intensity pixels as vessels by using an adaptive thresholding process [
20]. This method provides the robustness that is necessary in this problem given that is based on the intra-class variation minimization. The method organizes the image in two classes where the procedure to obtain the desired threshold between both classes implies an exhaustive search that is based on minimizing the variation intensity in each class and maximizing the variation intensity between both classes. In particular, the equation that is necessary to minimize and consequently obtain this optimal threshold value is the following (Equation (
2)):
where
and
indicate the proportion of pixels in each of the classes generated with the threshold
t, and
,
are the variances of these classes.
Once we performed the thresholding process, we obtained a binary OCTA image that represents the zones that contains vascular information. Given that this vascular information still includes representative levels of noise, a further filtering process is necessary to analyze those real pixels of interest. Given the nature of the OCTA images, the main vessels are surrounded by a gradual loss of intensity. This means that the most interesting pixels of the region are the central ones. For this reason, the next step of this method is the skeleton extraction. The skeletonization process is based on a object thinning approach, and it is calculated by the Hit or Miss morphological operator, as we can see in Equation (
3), that is iteratively repeated over the binary image until the target skeleton is extracted:
where
I is the image,
e is the structural element necessary in the morphological operation and
Hit-or-Miss is the morphological operator used in the skeleton extraction process. In
Figure 5, a few representative examples are presented including the VD extraction steps that we explained before.
Finally, we measure the VD biomarker by the calculus of its proportion over the image, as follows:
where
p is the count of pixels that are inside of the extracted skeleton, and
and
are the dimensions of the image, measured in pixels.
2.4. VA Estimation
To fulfil the final stage of the methodology and perform the automatic VA estimation, we used a Support Vector Machine (SVM) [
21] regression model. To do so, we organized the described vascular and avascular extracted biomarkers to, then, be used to train and test the selected model.
2.4.1. Data Preparation
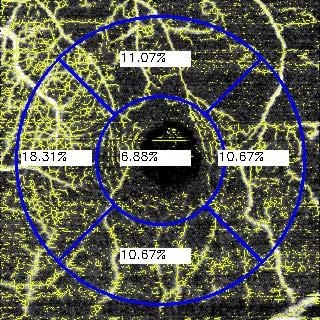
The data that we used to perform the VA estimation are both the automatically measured FAZ area and the VD. In particular, the previously defined VD represents a unique metric that measures the vascular information of an entire OCTA image. However, there are many diseases that present a local impact in particular regions of the eye fundus. For this reason, we decided to expand this metric and calculate the VD in different representative spatial regions of interest of the OCTA images, considering the central one including the fovea center. Also, this decision is reinforced by the mentioned fact that there are different diseases that are represented by the vascular loss in a particular zone. For example, the RVO can be presented more significantly in the superior or inferior regions. Performing the VD as a global metric, the high or low vascularity in a particular zone may loose impact and be diluted in the global metric. On the other hand, if we divide the OCTA image in different representative spatial regions, it is possible to analyze more precisely each one of them. For this reason, we decided to use the traditional circular quadrant grid that is commonly used in the ophthalmic field, analyzing the OCTA images and their corresponding VDs in each of the five quadrants of the grid: central, temporal, nasal, superior and inferior. We show representative examples of the used grid in
Figure 6.
In particular, each OCTA image is decomposed into 6 values: FAZ area, central VD, temporal VD, nasal VD, superior VD and inferior VD. As we said, each sample is formed by the information of an unique clinical consultation. This means that each sample takes the data extracted from 4 different OCTA images at 3 mm mm and 6 mm mm both in SCP and DCP. It is also remarkable that the VD in temporal and nasal zones are right or left sections depending on which is the pathological eye, therefore we also considered this fact. Given that the FAZ area for SCP and DCP are similar, we only used the extracted FAZ area in superficial mode given its higher precision. In summary, we used 22 features per patient to perform the estimation of the corresponding VA.
However, the experiments were restricted by the limitation of the expert to manually segment some FAZ cases that presented an advanced disease severity. In particular, this means that the expert can not segment the FAZ area given the difficult of the manual segmentation process in these mentioned cases. Given that, in the comparative step, we used both the manual segmentation and our automatic approach, we used the same samples in both methods to perform a reliable and fair comparative. For this reason, in this comparative study, we reduced the used samples from 215 to 127.
2.4.2. SVM Regression Model to Estimate the VA
To perform the VA estimation, we decided to use a SVM regression model. We used the described extracted biomarkers as the input feature vector to extract the predicted VA. In particular, to select the hyper-parameters, we used an automatic exhaustive search to find the best model parameter configuration. This exhaustive search is based on a Cross-Validation (CV) [
22] model that minimizes the error function, selecting the parameters that satisfy this minimization. This means that each model is performed with the hyper-parameters that allow the best results with the input data. This is an important characteristic, given that in the experiments we compare the performance of the different methods with the best created model for each input data. For the CV that is performed during the hyper-parameters search, the data was distributed as follows:
80% of the dataset was used to perform the CV in the training step with a 5-fold. In this CV, the hyper-parameters were selected, based on the best score over the CV process.
20% of the data was utilized to test the model. With the selected hyper-parameters, the model with the best score was selected
4. Conclusions
Many systemic and eye diseases affect the retinal micro-circulation and the characteristics of its vascularity. Hence, RD is one of the main diseases that may cause ocular micro-circulation problems. It is characteristic of regional vascular deterioration in the advanced stages of the pathology, as well as the consequent gradual vision loss. Also, ocular diseases such as RVO, for reference, may deteriorate the eye micro-circulation, with the corresponding visual penalization. In that context, early pathological identification is crucial but challenging. The disease is commonly detected in advanced stages when the clear symptoms are experienced. For these reasons, it is necessary to detect these vascular diseases in their initial stages by identifying slight changes in the retinal vascular circulation in order to avoid the drastic consequences of the advanced pathological stages and revert the condition.
The principal pathological changes in the vascular structure are mainly represented by the loss of vascular zones in the retina. In this context, the recent appearance of the OCTA image modality and its significant increase in popularity is motivated by its characteristic detailed visualization of the vascular structure of the eye fundus, representing an accurate source of information for the analysis of the retinal vascular diseases. However, the manual analysis of this vascular structure is tedious and requires a time-consuming process.
Commonly, the visual loss is measured by the VA of the patient, which is typically extracted by different manual tests with the patient and the expert clinician. The VA extraction is a slow process and it is conditioned by the non deterministic reply of the patient and the assessment of the clinician. The fast, deterministic and objectivity factors that generally provide an automatic approach are desirable in this problem.
In the present work, we propose a novel fully automatic methodology to estimate the VA of the patient, which is supported by the extraction of representative vascular and avascular biomarkers captured from the OCTA images. In particular, we measure representative biomarkers as the FAZ area and the VD. We use them as genuine biomarkers to estimate the VA of the patient, dividing the analysis of the VD into 5 circular quadrants to improve the precision of the information provided by the input features of the VA estimation model. Given that the VD extraction process removes all the possible regions that are most certainly not vessels, our proposal is robust to the presence of noise or motion artifacts. Also, the fact of using 5 different circular quadrants to create the feature vector that is used to estimate the VA provides a positive tolerance to particular local imperfections in the capture process. In the case that any significant noise is included in any region of the circular quadrant, this region only represents 1 of the 21 total used features to estimate the VA, given that we are also using four complementary OCTA images to perform this estimation. In summary, this means that the method is highly robust and tolerant to the presence of noise and artifacts in the OCTA images, which is still a common situation in this novel image modality.
Then, to perform the validation process, we compared our approach with the state of the art and the created capture device model, based on the existing Topcon semi-automatic tools. With these comparisons, we demonstrated that our VD extraction provides robust results to perform the VA estimation that can be accurately combined with the analysis of the FAZ area. The target VA is estimated with an error of 0.1713, providing an accurate system to evaluate the visual loss of the patient. This way, the proposed method offers automatically the estimation of the VA of the patient extracting representative computational biomarkers related with the FAZ region and the VD.
As future work, we propose the improvement of this VA estimation by adding new complementary biomarkers from the OCTA images. Additionally, it would be interesting to extend the analysis of the VA to patients with other relevant diseases and measure the relation of the vision loss and the vascular and avascular characteristics of the eye fundus. Indeed, it would also be interesting to use this estimation complementarily to other extracted biomarkers to obtain a model that aids in the clinical diagnostic process in the clinical field using a novel image modality as is the case of OCTA.