An Automated System for Classification of Chronic Obstructive Pulmonary Disease and Pneumonia Patients Using Lung Sound Analysis
Abstract
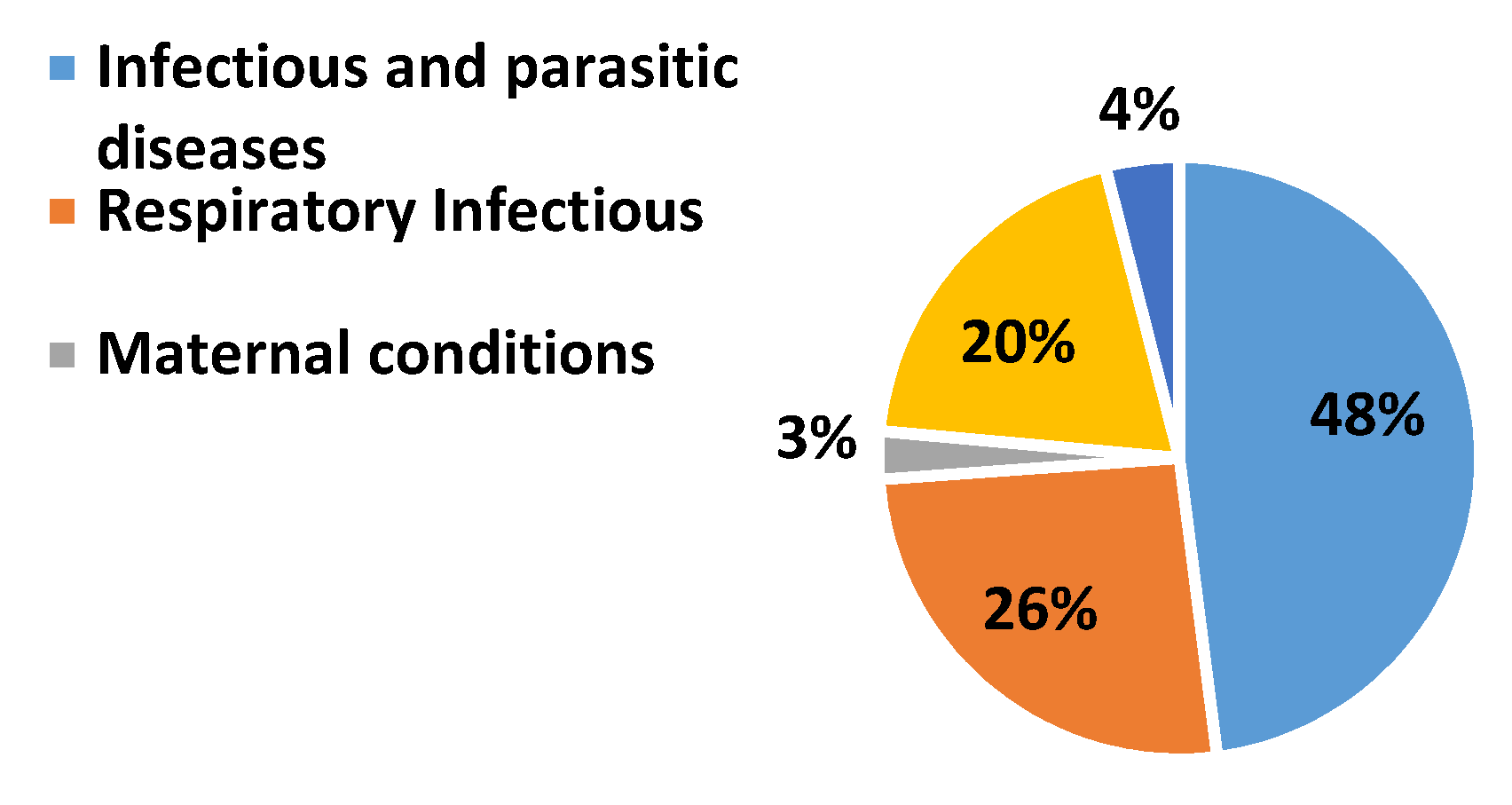
:1. Introduction
2. Literature Review
- (i)
- Data mining to extract the relevant and significant data of LS signals which should help to develop the diagnosis methods for COPD, pneumonia, and healthy LS.
- (ii)
- The design of the diagnosis method must be with simple statistical features that should not burden the system with computational cost and acquaint its performance with significant robustness.
- (iii)
- Investigation of minimum significant features required can be prolific to perform the classification of COPD, pneumonia, and healthy LS.
- (iv)
- Performance analysis of various classification methodologies would be required on selected features that are computationally smart.
3. Materials and Methods
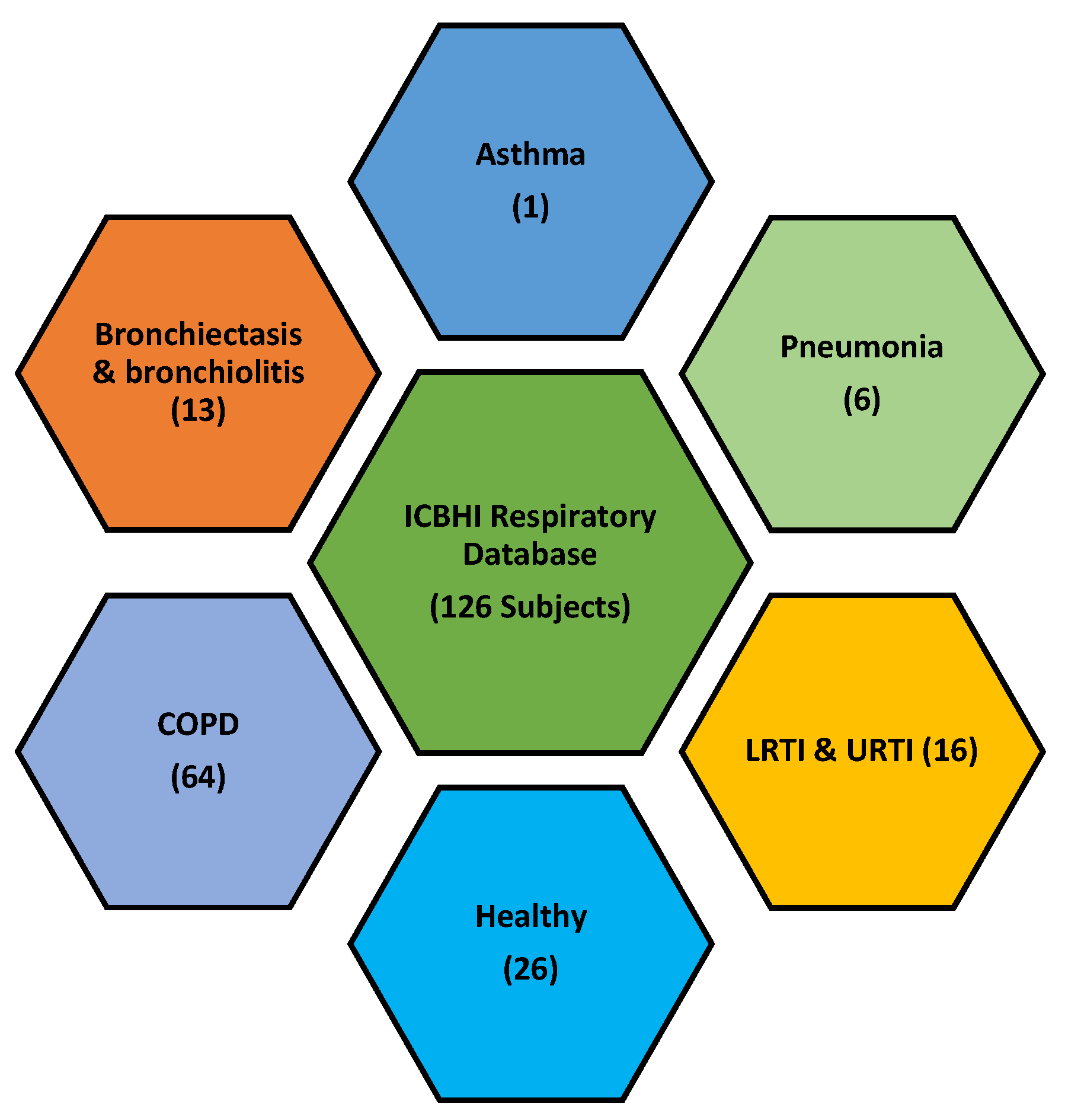
3.1. Database
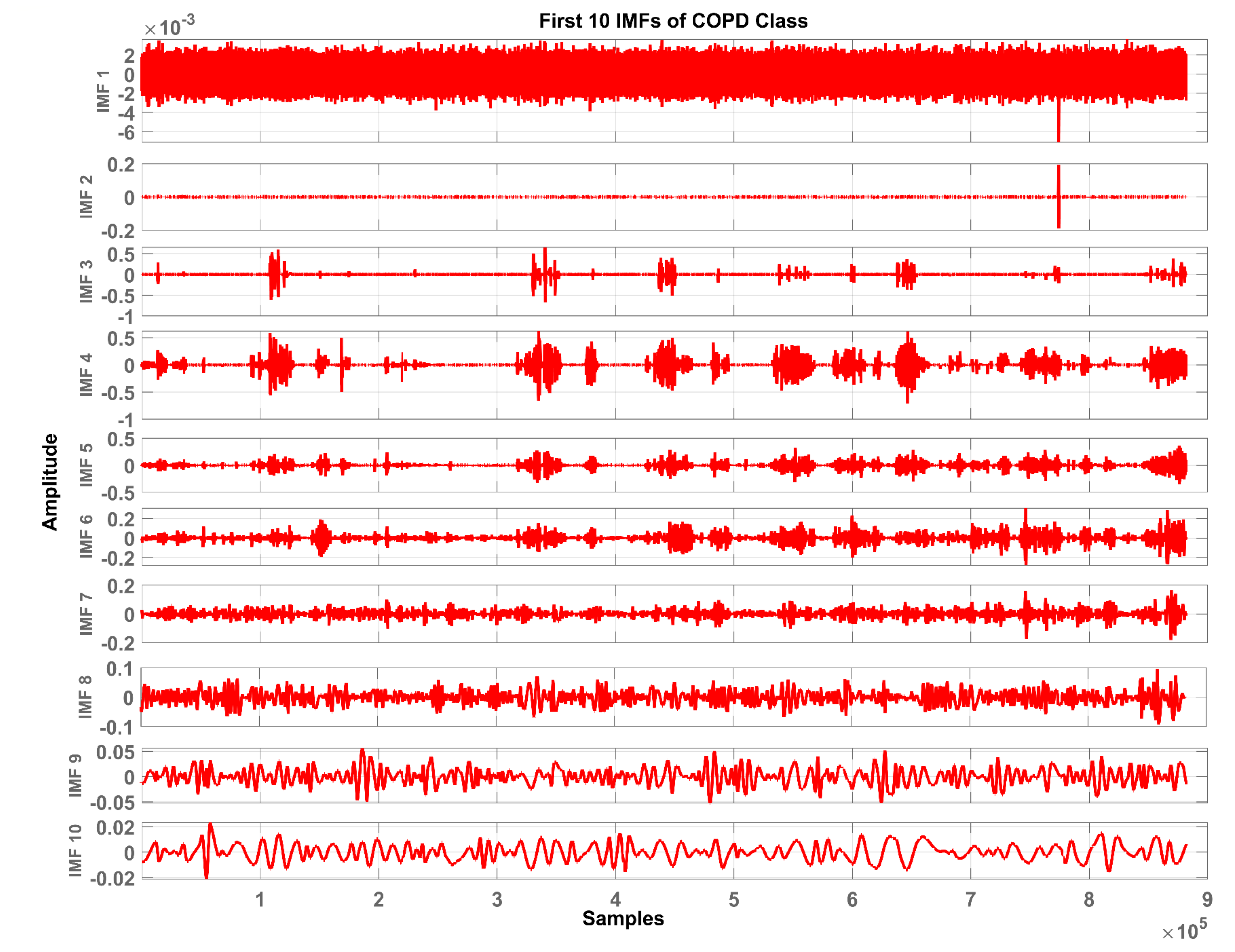
3.2. Pre-Processing and Segmentation
- (a)
- In the whole data set, the number of extreme and the number of zero-crossings must either be equal or differ at most by one.
- (b)
- At any point, the mean value of the envelope defined by the local maxima and the envelope defined by the local minima is zero.
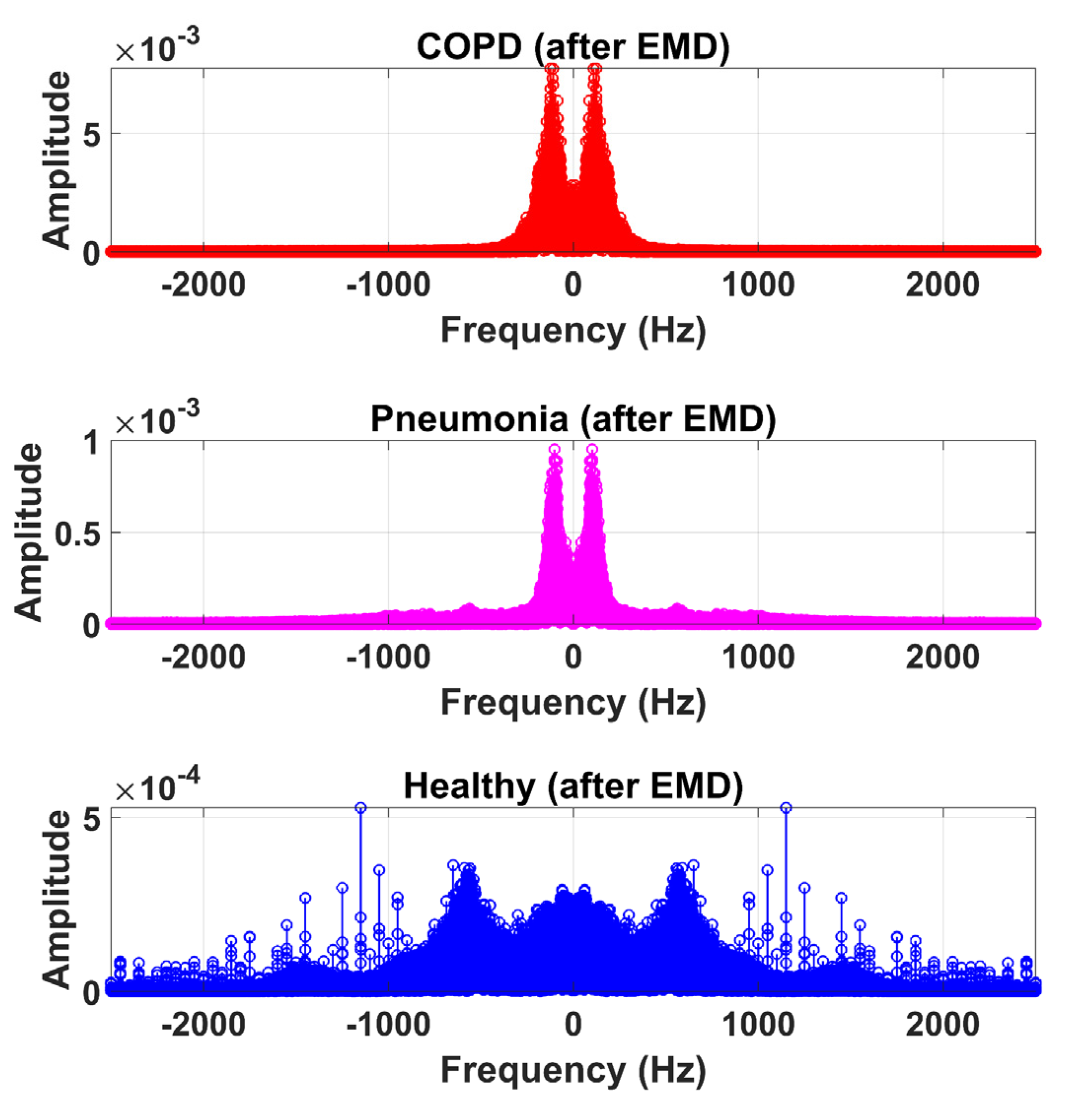
3.3. Feature Extraction
3.4. Classification
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Feature Statistics of All Classes
| Features | COPD | Pneumonia | Normal |
|---|---|---|---|
| Mean | |||
| Standard Deviation | |||
| Skewness | |||
| Kurtosis | |||
| Peak_to_Peak | |||
| Root_Mean_Square | |||
| Crest_Factor | |||
| Shape_Factor | |||
| Impulse_Factor | |||
| Margin_Factor | |||
| Energy | |||
| Peak_to_Root_Mean_Square | |||
| Root_Sum_of_Squares | |||
| Shannon_Energy | |||
| Log_Energy | |||
| Mean_Absolute_Deviation | |||
| Median_Absolute_Deviation | |||
| Average_Frequency | |||
| Jitter | |||
| Spectral Mean | |||
| Spectral Std_Deviation | |||
| Spectral Skewness | |||
| Spectral Kurtosis | |||
| Spectral Centriod | |||
| Spectral Flux | |||
| Spectral Rolloff | |||
| Spectral Flateness | |||
| Spectral Crest | |||
| Spectral Decrease | |||
| Spectral Slope | |||
| Spectral Spread | |||
| MFCC_1 | |||
| MFCC_2 | |||
| MFCC_3 | |||
| MFCC_4 | |||
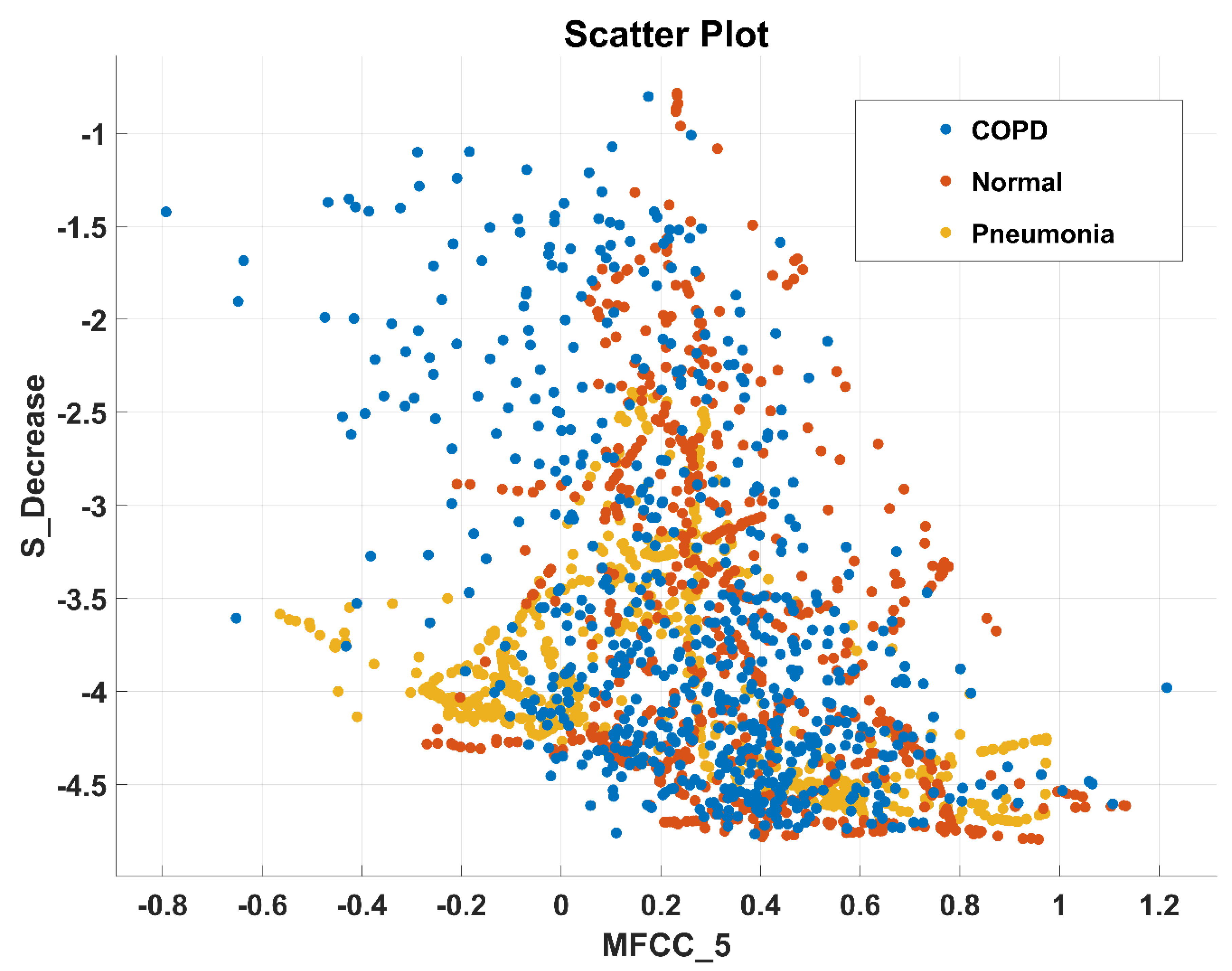
| MFCC_5 | |||
| MFCC_6 | |||
| MFCC_7 | |||
| MFCC_8 | |||
| MFCC_9 | |||
| MFCC_10 | |||
| MFCC_11 | |||
| MFCC_12 | |||
| MFCC_13 | |||
| GFCC_1 | |||
| GFCC_2 | |||
| GFCC_3 | |||
| GFCC_4 | |||
| GFCC_5 | |||
| GFCC_6 | |||
| GFCC_7 | |||
| GFCC_8 | |||
| GFCC_9 | |||
| GFCC_10 | |||
| GFCC_11 | |||
| GFCC_12 | |||
| GFCC_13 |
References
- Watkins, K. The State of the World’s Children 2016: A Fair Chance for Every Child; UNICEF: New York, NY, USA, 2016. [Google Scholar]
- Lieberman, D.; Gelfer, Y.; Varshavsky, R.; Dvoskin, B.; Leinonen, M.; Friedman, M.G. Pneumonic vs. nonpneumonic acute exacerbations of COPD. Chest 2002, 122, 1264–1270. [Google Scholar] [CrossRef]
- World Health Organization. Health Statistics and Information Systems; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Quach, A.; Giovannelli, J.; Cherot-Kornobis, N.; Ciuchete, A.; Clément, G.; Matran, R.; Amouyel, P.; Edme, J.-L.; Dauchet, L. Prevalence and underdiagnosis of airway obstruction among middle-aged adults in northern France: The ELISABET study 2011–2013. Respir. Med. 2015, 109, 1553–1561. [Google Scholar] [CrossRef]
- Serbes, G.; Ulukaya, S.; Kahya, Y.P. An Automated Lung Sound Preprocessing and Classification System Based Onspectral Analysis Methods. In Precision Medicine Powered by pHealth and Connected Health; Springer: Berlin/Heidelberg, Germany, 2018; pp. 45–49. [Google Scholar]
- Song, I. Diagnosis of Pneumonia from Sounds Collected using Low Cost Cell Phones. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–16 July 2015; pp. 1–8. [Google Scholar]
- Amrulloh, Y.; Abeyratne, U.; Swarnkar, V.; Triasih, R. Cough sound analysis for pneumonia and asthma classification in pediatric population. In Proceedings of the 6th International Conference on Intelligent Systems, Modelling and Simulation, Kuala Lumpur, Malaysia, 9–12 February 2015; pp. 127–131. [Google Scholar]
- Gupta, N.; Gupta, D.; Khanna, A.; Filho, P.P.R.; De Albuquerque, V.H.C. Evolutionary algorithms for automatic lung disease detection. Measurement 2019, 140, 590–608. [Google Scholar] [CrossRef]
- Rabe, F. Global Initiative for Chronic Obstructive Lung Disease. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. GOLD executive summary. Am. J. Respir. Crit. Care Med. 2007, 176, 532–555. [Google Scholar] [CrossRef]
- Reichert, S.; Gass, R.; Brandt, C.; Andrès, E. Analysis of respiratory sounds: State of the art. Clin. Med. Circ. Respir. Pulmon. Med. 2008, 2. [Google Scholar] [CrossRef]
- Bellos, C.C.; Papadopoulos, A.; Rosso, R.; Fotiadis, D.I. Identification of COPD patients’ health status using an intelligent system in the CHRONIOUS wearable platform. IEEE J. Biomed. Health Inform. 2013, 18, 731–738. [Google Scholar] [CrossRef]
- Kosasih, K.; Abeyratne, U.R.; Swarnkar, V.; Triasih, R. Wavelet Augmented Cough Analysis for Rapid Childhood Pneumonia Diagnosis. IEEE Trans. Biomed. Eng. 2014, 62, 1185–1194. [Google Scholar] [CrossRef]
- Abeyratne, U.R.; Swarnkar, V.; Setyati, A.; Triasih, R. Cough Sound Analysis Can Rapidly Diagnose Childhood Pneumonia. Ann. Biomed. Eng. 2013, 41, 2448–2462. [Google Scholar] [CrossRef]
- Shen, F.; Chao, J.; Zhao, J. Forecasting exchange rate using deep belief networks and conjugate gradient method. Neurocomputing 2015, 167, 243–253. [Google Scholar] [CrossRef]
- Morillo, D.S.; Fernandez-Granero, M.A.; Leon-Jimenez, A. Use of predictive algorithms in-home monitoring of chronic obstructive pulmonary disease and asthma. Chronic Respir. Dis. 2016, 13, 264–283. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.I.; Ahmed, V. Study of adventitious lung sounds of paediatric population using artificial neural network approach. Int. J. Cur. Res. Rev. 2017, 9, 37. [Google Scholar]
- Pingale, T.H.; Patil, H. Analysis of Cough Sound for Pneumonia Detection Using Wavelet Transform and Statistical Parameters. In Proceedings of the International Conference on Computing, Communication, Control. and Automation (ICCUBEA), Pune, India, 17–18 August 2017; pp. 1–6. [Google Scholar]
- Aziz, S.; Khan, M.U.; Shakeel, M.; Mushtaq, Z.; Khan, A.Z. An Automated System towards Diagnosis of Pneumonia using Pulmonary Auscultations. In Proceedings of the 2019 13th International Conference on Mathematics, Actuarial Science, Computer Science and Statistics (MACS), Karachi, Pakistan, 14–15 December 2019; pp. 1–7. [Google Scholar]
- Shi, Y.; Li, Y.; Cai, M.; Zhang, X.D. A Lung Sound Category Recognition Method Based on Wavelet Decomposition and BP Neural Network. Int. J. Biol. Sci. 2019, 15, 195. [Google Scholar] [CrossRef]
- Neili, Z.; Fezari, M.; Abdeghani, R. Analysis of Acoustic Parameters from Respiratory Signal in COPD and Pneumonia patients. In Proceedings of the International Conference on Signal, Image, Vision and their Applications (SIVA), Guelma, Algeria, 26–27 November 2018; pp. 1–4. [Google Scholar]
- Jindal, V.; Agarwal, V.; Kalaivani, S. Respiratory Sound Analysis for Detection of Pulmonary Diseases. In Proceedings of the IEEE Applied Signal Processing Conference (ASPCON), Kolkata, India, 7–9 December 2018; pp. 293–296. [Google Scholar]
- Islam, A.; Bandyopadhyaya, I.; Bhattacharyya, P.; Saha, G. Classification of Normal, Asthma and COPD Subjects Using Multichannel Lung Sound Signals. In Proceedings of the 2018 International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India, 3–5 April 2018; pp. 290–294. [Google Scholar]
- Nabi, F.G.; Sundaraj, K.; Lam, C.K. Identification of asthma severity levels through wheeze sound characterization and classification using integrated power features. Biomed. Signal Process. Control. 2019, 52, 302–311. [Google Scholar] [CrossRef]
- Naqvi, S.Z.H.; Choudhry, M.A.; Khan, A.Z.; Shakeel, M. Intelligent System for Classification of Pulmonary Diseases from Lung Sound. In Proceedings of the 2019 13th International Conference on Mathematics, Actuarial Science, Computer Science and Statistics (MACS), Karachi, Pakistan, 14–15 December 2019; pp. 1–6. [Google Scholar]
- Wang, Q.; Wang, H.; Wang, L.; Yu, F. Diagnosis of Chronic Obstructive Pulmonary Disease Based on Transfer Learning. IEEE Access 2020, 8, 47370–47383. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, H.; Wang, L.; Di, R.; Song, Y. Diagnosis of COPD Based on a Knowledge Graph and Integrated Model. IEEE Access 2019, 7, 46004–46013. [Google Scholar] [CrossRef]
- Sengupta, N.; Sahidullah, M.; Saha, G. Lung sound classification using cepstral-based statistical features. Comp. Biol. Med. 2016, 75, 118–129. [Google Scholar] [CrossRef]
- Demir, F.; Şengür, A.; Bajaj, V. Convolutional neural networks based efficient approach for classification of lung diseases. Heal. Inf. Sci. Syst. 2020, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- García-Ordás, M.T.; Benítez-Andrades, J.A.; García-Rodríguez, I.; Benavides, C.; Alaiz-Moretón, H. Detecting Respiratory Pathologies Using Convolutional Neural Networks and Variational Autoencoders for Unbalancing Data. Sensors 2020, 20, 1214. [Google Scholar] [CrossRef] [Green Version]
- Rocha, B.M.; Filos, D.; Mendes, L.; Serbes, G.; Ulukaya, S.; Kahya, Y.P.; Jakovljevic, N.; Turukalo, T.L.; Vogiatzis, I.M.; Perantoni, E.; et al. An open access database for the evaluation of respiratory sound classification algorithms. Physiol. Meas. 2019, 40, 035001. [Google Scholar] [CrossRef]
- Altuve, M.; Suárez, L.; Ardila, J. Fundamental heart sounds analysis using improved complete ensemble EMD with adaptive noise. Biocybern. Biomed. Eng. 2020, 40, 426–439. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B.K.; Behera, A.K. Comparative analysis of Lung sound denoising technique. In Proceedings of the First International Conference on Power, Control. and Computing Technologies (ICPC2T), Raipur, India, 3–5 January 2020; pp. 406–410. [Google Scholar]
- Gross, V.; Dittmar, A.; Penzel, T.; Schüttler, F.; Von Wichert, P. The Relationship between Normal Lung Sounds, Age, and Gender. Am. J. Respir. Crit. Care Med. 2000, 162, 905–909. [Google Scholar] [CrossRef]
- Sengupta, N.; Sahidullah, M.; Saha, G. Lung sound classification using local binary pattern. arXiv 2017, arXiv:1710.01703. [Google Scholar]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive Synthetic Sampling Approach for Imbalanced Learning. In Proceedings of the IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–6 June 2008; pp. 1322–1328. [Google Scholar]
- Khalid, S.; Khalil, T.; Nasreen, S. A Survey of Feature Selection and Feature Extraction Techniques in Machine Learning. In Proceedings of the Science and Information Conference, London, UK, 27–29 August 2014; pp. 372–378. [Google Scholar]
- Zhao, X.; Wang, D. Analyzing Noise Robustness of MFCC and GFCC Features in Speaker Identification. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, Vancuver, Canada, 26–31 May 2013; pp. 7204–7208. [Google Scholar]











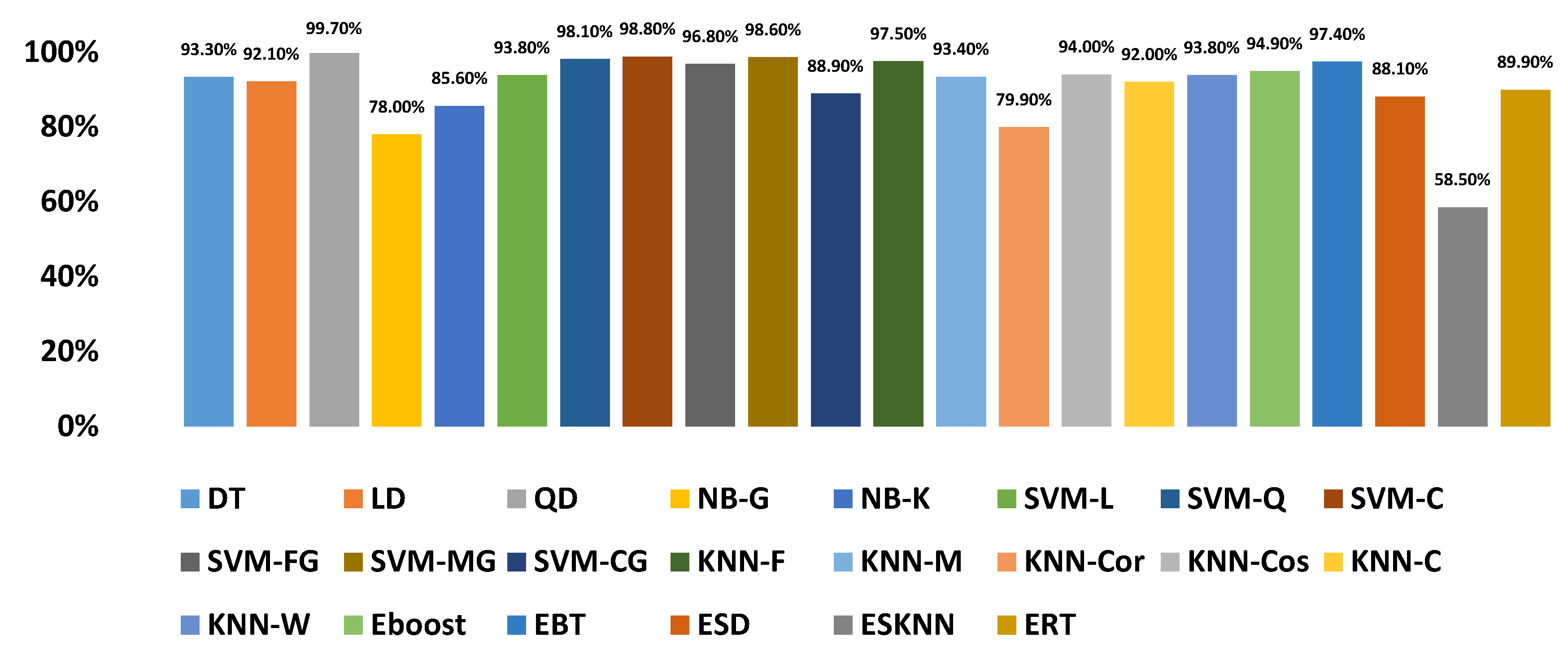


| Sr. No | Equipment |
|---|---|
| 1. | Welch Allyn Meditron Master Elite Plus Stethoscope Model 5079-400 |
| 2. | 3M Littmann 3200 |
| 3. | 3M Littmann Classic II SE |
| Description | Detail |
|---|---|
| Total number of LS signal sets (COPD, pneumonia, healthy) | 703 |
| The sampling frequency of recording equipment | 44.1 kHz |
| Bits/ sample | 16 |
| Average recording duration | 21.5s |
| Number of participants | 57 (50 adults & 7 children) |
| Gender | 40 males, 17 females |
| Age | Adults: 69.44 ± 8.24 Years, Children: 6.06 ± 5.98 Years |
| Time Domain Features (19) | Spectral (S) Domain Features (12) | Cepstral Features (26) | Texture Features (59) |
|---|---|---|---|
| Mean, St. Deviation, Skewness, Kurtosis, Peak to Peak, Root Mean Square, Crest Factor, Shape Factor, Impulse Factor, Margin Factor, Energy, Peak to RMS, Root Sum of Squares, Shannon Energy, Log Energy, Mean Abs Deviation, Median Abs Deviation, Average Frequency, Jitter | S.Mean, S.St. Deviation, S.Skewness, S.Kurtosis, S.Centriod, S.Flux, S.Rolloff, S.Flateness, S.Crest, S.Decrease, S.Slope, S.Spread | MFCC, GCC | LBP |
| FEATURE GROUPS | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLASSIFIERS | TD | FD | CD | Texture | TD+FD | TD+CD | FD+CD | CD+Texture | TD+Texture | TD+FD+CD | TD+FD+Texture | FD+CD+Texture | TD+FD+CD+Texture | |
| DT | 87.30% | 86.80% | 94.50% | 92.50% | 89.50% | 95.40% | 94.60% | 94.60% | 93.70% | 95.00% | 94.20% | 94.60% | 95.00% | |
| LD | 76.40% | 76.50% | 91.10% | 73.90% | 82.70% | 93.40% | 93.60% | 92.00% | - | 94.20% | - | 93.70% | - | |
| QD | - | 84.70% | 98.60% | - | - | - | - | - | - | - | - | - | - | |
| LR | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| NB-G | 59.30% | 68.00% | 82.40% | - | 69.20% | 73.40% | 79.80% | - | - | 75.10% | - | - | - | |
| NB-K | 66.80% | 71.10% | 88.30% | 41.70% | 73.00% | 85.50% | 88.90% | 47.10% | 43.50% | 85.90% | 44.70% | 48.00% | 48.50% | |
| SVM-L | 79.30% | 80.10% | 94.90% | 83.90% | 85.30% | 96.00% | 95.30% | 95.50% | 91.30% | 96.00% | 91.90% | 96.10% | 95.90% | |
| SVM-Q | 91.00% | 88.70% | 97.90% | 92.10% | 94.40% | 98.10% | 98.30% | 97.80% | 95.60% | 98.10% | 96.00% | 97.90% | 98.00% | |
| SVM-C | 93.60% | 93.50% | 98.50% | 58.00% | 97.00% | 98.70% | 98.50% | 96.00% | 88.00% | 98.60% | 94.20% | 98.60% | 98.60% | |
| SVM-FG | 94.60% | 91.30% | 96.20% | 91.40% | 97.30% | 97.20% | 95.80% | 99.70% | 97.20% | 97.20% | 98.10% | 99.40% | 99.20% | |
| SVM-MG | 85.60% | 82.90% | 98.40% | 79.60% | 91.40% | 98.60% | 98.80% | 97.10% | 90.40% | 98.70% | 92.20% | 97.80% | 97.80% | |
| SVM-CG | 70.00% | 75.40% | 90.80% | 61.40% | 79.70% | 94.00% | 93.10% | 85.70% | 73.50% | 93.20% | 78.70% | 89.90% | 90.70% | |
| KNN-F | 92.70% | 91.60% | 97.90% | 93.80% | 95.00% | 97.50% | 97.50% | 98.00% | 94.30% | 97.00% | 95.50% | 97.80% | 97.00% | |
| KNN-M | 87.50% | 84.90% | 94.10% | 86.70% | 89.40% | 94.00% | 92.60% | 94.30% | 90.00% | 92.90% | 89.80% | 93.20% | 93.00% | |
| KNN-Cor | 65.80% | 71.40% | 80.60% | 71.30% | 74.80% | 84.10% | 81.00% | 81.80% | 70.30% | 84.20% | 76.30% | 81.30% | 87.70% | |
| KNN-Cos | 88.10% | 86.00% | 94.90% | 87.50% | 90.20% | 94.60% | 94.60% | 95.20% | 90.90% | 94.40% | 91.00% | 94.30% | 94.80% | |
| KNN-C | 87.90% | 85.00% | 93.80% | 86.60% | 88.80% | 93.60% | 92.40% | 94.30% | 89.90% | 92.00% | 89.50% | 92.90% | 92.40% | |
| KNN-W | 89.60% | 88.40% | 9.60% | 90.10% | 91.10% | 94.20% | 93.30% | 94.50% | 91.40% | 93.30% | 91.00% | 93.70% | 93.20% | |
| Eboost | 84.80% | 83% | 96.40% | 89.30% | 90.50% | 96.60% | 96.60% | 96.60% | 94.70% | 96.30% | 94.50% | 96.70% | 96.70% | |
| EBT | 93.80% | 91.50% | 97.10% | 94.90% | 95.70% | 97.30% | 96.90% | 97.90% | 96.30% | 97.20% | 97.00% | 97.50% | 97.60% | |
| ESD | 71.70% | 72.30% | 88.70% | 71.40% | 79.50% | 92.30% | 91.60% | 90.00% | 81.40% | 93.10% | 85.30% | 93.30% | 94.00% | |
| ESKNN | 68.70% | 77.30% | 97.50% | 92.00% | 74.90% | 69.30% | 83.40% | 97.70% | 69.00% | 74.60% | 75.40% | 84.20% | 74.70% | |
| ERT | 77.00% | 78.80% | 91.20% | 84.40% | 84.20% | 92.40% | 92.90% | 92.40% | 88.40% | 92.80% | 88.30% | 92.80% | 93.00% | |
| Selected Features from Time, Frequency, and Cepstral Domain | Classifier | Performance Outcome |
|---|---|---|
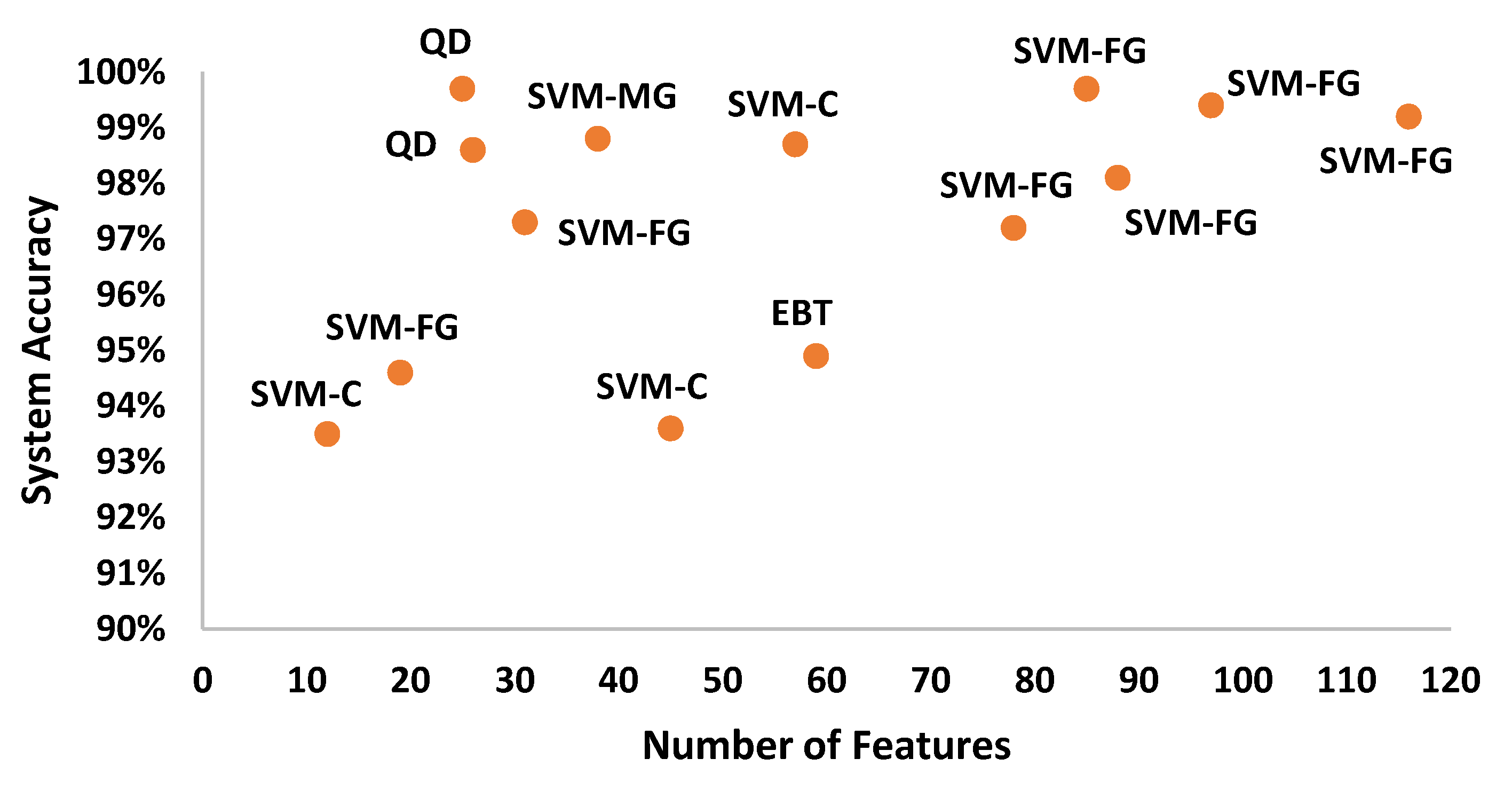
| Time (Standard Deviation, Peak to Peak, Log Energy), Spectral (Spectral Standard Deviation, Spectral Skewness, Spectral Kurtosis, Spectral Flux, Spectral Roll Off, Spectral Decrease), Cepstral (MFCC (3-10), GFCC (3-10)) | Quadratic discriminant | Overall Accuracy 99.70% |
| Feature | Mathematical Representation |
|---|---|
| Standard Deviation (SD) | |
| Peak to Peak (PP) | Where and is the minimum and maximum value in the time domain |
| Log Energy (LE) | |
| Spectral Standard Deviation (SSD) | |
| Spectral Skewness (SSkw) | |
| Spectral Kurtosis(SK) | |
| Spectral Flux (SF) | |
| Spectral Roll Off (SRO) | If DFT coefficient corresponds to the spectral roll-off of the frame, then C is the adapted percentage: 95% and |
| Spectral Decrease (SDec) | |
| Mel frequency cepstral coefficient (MFCC) | In MFCC, (i) Frame blocking or windowing to get 50 to 60ms. (ii) Performing a discrete Fourier transform (iii) computing logarithm of the signal. (iv) Deforming the frequencies on a Mel scale, followed by applying the discrete cosine transform (DCT). Mel scale is calculated as follows: ‘f’ refers to frequency ranges from 0 to fs. |
| Gammatone Frequency Cepstral Coefficient (GFCC) | In GCC, (i) Firstly, the signal is passed through gammatone filter bank which consists of 64 Channels. (ii) Take the absolute value at each channel and reduce it to 100 Hz as a way of time windowing. (iii) Take cubic root on the time-frequency representation. (iv) Deforming the frequencies on an equivalent rectangular bandwidth (ERB) scale Apply DCT to derive cepstral features. ERB scale is calculated as follows. where ‘hz’ refers to frequency ranges i.e. 0-fs. |
| Evaluation | Classes | ACC % | TPR % | FNR % | PPV % | FDR % |
|---|---|---|---|---|---|---|
| (5, 10, 15, and 20) Fold Cross-Validation | COPD | 99.6 | >99 | <1 | 99 | 1 |
| Normal | >99 | <1 | 100 | 0 | ||
| Pneumonia | >99 | <1 | >99 | <1 | ||
| 20% Hold Out Validation | COPD | 99.7 | 99 | 1 | 100 | 0 |
| Normal | 100 | 0 | 100 | 0 | ||
| Pneumonia | 100 | 0 | 99 | 1 | ||
| 25% Hold Out Validation | COPD | 99.8 | 100 | 0 | 99 | 1 |
| Normal | 99 | 1 | 100 | 0 | ||
| Pneumonia | 100 | 0 | 100 | 0 |
| Class | Number of Features | Accuracy (%) |
|---|---|---|
| Pneumonia [12] | 13 | 87.87 |
| Pneumonia [6] | 18 | 90.06 |
| Pneumonia [18] | 7 | 99.70 |
| COPD [25] | 25 | 85.10 |
| COPD [26] | 27 | 95.10 |
| COPD, pneumonia (This method) | 25 | 99.70 |
| Classes | Method | Results (%) |
|---|---|---|
| Crackles, Crackles+ Wheeze, Normal, Wheeze [5] | STFT, WT, SVM | ACC: 49.86 |
| Normal, Pneumonia [6] | SA | ACC: 91.98 SEN:92.06 SPE: 90.68 |
| Pneumonia and Asthma [7] | NN | SEN: 89, SPE:100 |
| Normal, Pneumonia [12] | WT, LR ANN | SEN: 94 SPE:63 |
| Normal, Pneumonia [18] | EMD, KNN | ACC: 99.7 |
| Normal, COPD and Pneumonia [20] | SA | - |
| Normal Asthma and COPD [22] | ANN | ACC:60.33 SEN: 65 SPE:54.2 |
| Normal, Asthma, Bronchitis [24] | EMD, KNN | ACC: 99.3 |
| COPD [25] | KT | ACC: 85.1 |
| COPD [26] | KG, ML | ACC:95.1 |
| COPD, Healthy, Pneumonia, Asthma, Bronchiectasis, Bronchiolitis [29] | CNN | SEN: 98.8 SPE:98.6 |
| Crackles, Crackles+ Wheeze, Normal, Wheeze [28] | CNN | ACC i: 65.5 ii: 63.09 |
| Normal, COPD, Pneumonia (This Method) | EMD, WT, QD | ACC: 99.8% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naqvi, S.Z.H.; Choudhry, M.A. An Automated System for Classification of Chronic Obstructive Pulmonary Disease and Pneumonia Patients Using Lung Sound Analysis. Sensors 2020, 20, 6512. https://doi.org/10.3390/s20226512
Naqvi SZH, Choudhry MA. An Automated System for Classification of Chronic Obstructive Pulmonary Disease and Pneumonia Patients Using Lung Sound Analysis. Sensors. 2020; 20(22):6512. https://doi.org/10.3390/s20226512
Chicago/Turabian StyleNaqvi, Syed Zohaib Hassan, and Mohammad Ahmad Choudhry. 2020. "An Automated System for Classification of Chronic Obstructive Pulmonary Disease and Pneumonia Patients Using Lung Sound Analysis" Sensors 20, no. 22: 6512. https://doi.org/10.3390/s20226512
APA StyleNaqvi, S. Z. H., & Choudhry, M. A. (2020). An Automated System for Classification of Chronic Obstructive Pulmonary Disease and Pneumonia Patients Using Lung Sound Analysis. Sensors, 20(22), 6512. https://doi.org/10.3390/s20226512






