Depth Estimation for Integral Imaging Microscopy Using a 3D–2D CNN with a Weighted Median Filter
Abstract
:1. Introduction
2. Background of IIMs and Depth Map
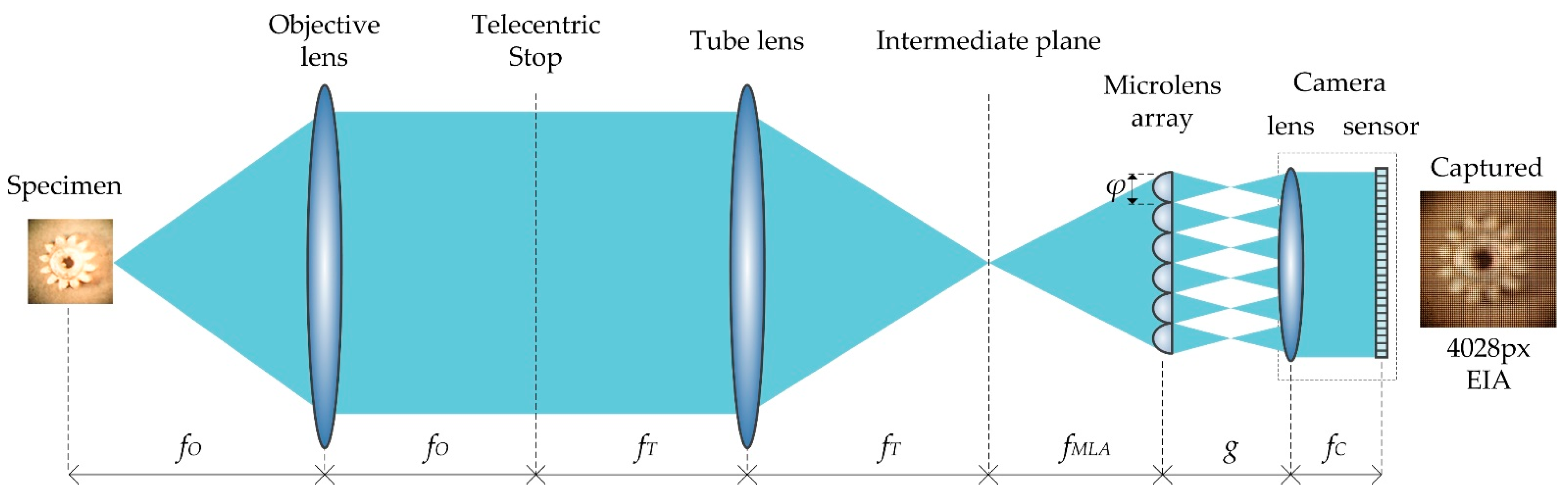
2.1. Integral Imaging Microscopy
2.2. Deep Learning-Based Depth Estimation Method
3. Depth Estimation Methodology
3.1. Data Generalization
3.2. Network Architecture
3.3. Loss Function
4. Experimental Setup and Quality Measurement Metrics
4.1. Root-Mean-Square Error
4.2. Discrete Entropy
4.3. Power Spectrum Density
5. Result and Discussion of the Proposed Depth Enhancement Method
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| CNN | Convolutional neural network |
| DOF | Depth of field |
| EI | Elemental image |
| EIA | Elemental image array |
| EL | Elemental lens |
| EPI | Epipolar plane image |
| FCN | Fully convolutional network |
| II | Integral imaging |
| IIMs | Integral imaging microscope system |
| LFM | Light field microscopy |
| MLA | Microlens array |
| OVI | Orthographic-view image |
| ReLU | Rectified linear unit |
| ROI | Region of interest |
| WMF | Weighted median filtering |
References
- Lippmann, G. Reversible prints giving the sensation of relief. J. Phys. Arch. 1908, 7, 821–825. [Google Scholar] [CrossRef]
- Okano, F.; Hoshino, H.; Arai, J.; Yuyama, I. Real-time pickup method for a three-dimensional image based on integral photography. Appl. Opt. 1997, 36, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Corral, M.; Javidi, B. Fundamentals of 3D imaging and displays: A tutorial on integral imaging, light-field, and plenoptic systems. Adv. Opt. Photonics 2018, 10, 512. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.; Kwon, K.-C.; Erdenebat, M.-U.; Lim, Y.-T.; Imtiaz, S.; Sufian, M.A.; Jeon, S.-H.; Kim, N. Resolution enhancement of an integral imaging microscopy using generative adversarial network. In Proceedings of the 14th Pacific Rim Conference on Lasers and Electro-Optics (CLEO PR 2020), Sydney, Australia, 3–5 August 2020. [Google Scholar]
- Javidi, B.; Moon, I.; Yeom, S. Three-dimensional identification of biological microorganism using integral imaging. Opt. Express 2006, 14, 12096. [Google Scholar] [CrossRef]
- Xiao, X.; Javidi, B.; Martinez-Corral, M.; Stern, A. Advances in three-dimensional integral imaging: Sensing, display, and applications. Appl. Opt. 2013, 52, 546–560. [Google Scholar] [CrossRef]
- Nepijko, S.A.; Schǒnhense, G. Electron holography for electric and magnetic field measurements and its application for nanophysics. In Advances in Imaging and Electron Physics; Elsevier: Amsterdam, The Netherlands, 2011; Volume 169, pp. 173–240. [Google Scholar]
- Jang, J.S.; Javidi, B. Three-dimensional integral imaging of micro-objects. Opt. Lett. 2004, 29, 1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Lu, Y.; Su, M. Light field based digital refocusing using a DSLR camera with a pinhole array mask. In Proceedings of the 2010 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Dallas, TX, USA, 15–19 March 2010. [Google Scholar]
- Lim, Y.-T.; Park, J.-H.; Kwon, K.-C.; Kim, N. Resolution-enhanced integral imaging microscopy that uses lens array shifting. Opt. Express 2009, 17, 19253. [Google Scholar] [CrossRef] [PubMed]
- Industrial Microscope OLYMPUS Stream. Available online: https://www.olympus-ims.com/en/microscope/stream2/ (accessed on 3 June 2022).
- Stereo Microscopes. Available online: https://www.olympus-lifescience.com/en/microscopes/stereo/ (accessed on 3 June 2022).
- Levoy, M.; Ng, R.; Adams, A.; Footer, M.; Horowitz, M. Light field microscopy. In Proceedings of the ACM SIGGRAPH 2006 papers (SIGGRAPH ’06), San Diego, CA, USA, 30 July–3 August 2006. [Google Scholar]
- Kwon, K.-C.; Erdenebat, M.-U.; Khuderchuluun, A.; Kwon, K.H.; Kim, M.Y.; Kim, N. High-quality 3d display system for an integral imaging microscope using a simplified direction-inversed computation based on user interaction. Opt. Lett. 2021, 46, 5079–5082. [Google Scholar] [CrossRef]
- Farhood, H.; Perry, S.; Cheng, E.; Kim, J. Enhanced 3D point cloud from a light field image. Remote Sens. 2020, 12, 1125. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xiao, X.; Hua, H.; Javidi, B. Augmented reality 3d displays with micro integral imaging. J. Disp. Technol. 2015, 11, 889–893. [Google Scholar] [CrossRef]
- Martínez-Corral, M.; Javidi, B.; Martínez-Cuenca, R.; Saavedra, G. Integral imaging with improved depth of field by use of amplitude-modulated microlens arrays. Appl. Opt. 2004, 43, 5806–5813. [Google Scholar] [CrossRef]
- Kwon, K.-C.; Erdenebat, M.-U.; Alam, M.A.; Lim, Y.-T.; Kim, K.G.; Kim, N. Integral imaging microscopy with enhanced depth-of-field using a spatial multiplexing. Opt. Express 2016, 24, 2072–2083. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Banks, M.S. Resolution of temporal-multiplexing and spatial-multiplexing stereoscopic televisions. Curr. Opt. Photonics 2017, 1, 34–44. [Google Scholar] [CrossRef] [Green Version]
- Kwon, K.-C.; Lim, Y.-T.; Shin, C.-W.; Erdenebat, M.-U.; Hwang, J.-M.; Kim, N. Enhanced depth-of-field of an integral imaging microscope using a bifocal holographic optical element-micro lens array. Opt. Lett. 2017, 42, 3209–3212. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.-C.; Erdenebat, M.-U.; Lim, Y.-T.; Joo, K.-I.; Park, M.-K.; Park, H.; Jeong, J.-R.; Kim, H.-R.; Kim, N. Enhancement of the depth-of-field of integral imaging microscope by using switchable bifocal liquid-crystalline polymer micro lens array. Opt. Express 2017, 25, 30503–30512. [Google Scholar] [CrossRef]
- Alam, M.S.; Kwon, K.-C.; Erdenebat, M.-U.; Abbass, M.Y.; Alam, M.A.; Kim, N. Super-resolution enhancement method based on generative adversarial network for integral imaging microscopy. Sensors 2021, 21, 2164. [Google Scholar] [CrossRef]
- Yang, Q.; Tan, K.-H.; Culbertson, B.; Apostolopoulos, J. Fusion of active and passive sensors for fast 3D capture. In Proceedings of the 2010 IEEE International Workshop on Multimedia Signal Processing (MMSP), Saint-Malo, France, 4–6 October 2010. [Google Scholar]
- Honauer, K.; Johannsen, O.; Kondermann, D.; Goldluecke, B. A dataset and evaluation methodology for depth estimation on 4d light fields. In Proceedings of the 13th Asian Conference on Computer Vision (ACCV), Taipei, Taiwan, 20–24 November 2016. [Google Scholar]
- Wang, Q.; Zhang, L.; Bertinetto, L.; Hu, W.; Torr, P.H.S. Fast online object tracking and segmentation: A unifying approach. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Kim, S.; Jun, D.; Kim, B.-G.; Lee, H.; Rhee, E. Single image super-resolution method using cnn-based lightweight neural networks. Appl. Sci. 2021, 11, 1092. [Google Scholar] [CrossRef]
- Shin, C.; Jeon, H.-G.; Yoon, Y.; Kweon, I.S.; Kim, S.J. EPINET: A fully-convolutional neural network using epipolar geometry for depth from light field images. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Heber, S.; Yu, W.; Pock, T. Neural epi-volume networks for shape from light field. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Rogge, S.; Schiopu, I.; Munteanu, A. Depth estimation for light-field images using stereo matching and convolutional neural networks. Sensors 2020, 20, 6188. [Google Scholar] [CrossRef]
- Han, L.; Huang, X.; Shi, Z.; Zheng, S. Depth estimation from light field geometry using convolutional neural networks. Sensors 2021, 21, 6061. [Google Scholar] [CrossRef]
- Wu, G.; Zhao, M.; Wang, L.; Dai, Q.; Chai, T.; Liu, Y. Light field reconstruction using deep convolutional network on EPI. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Li, K.; Zhang, J.; Sun, R.; Zhang, X.; Gao, J. EPI-based oriented relation networks for light field depth estimation. arXiv 2020, arXiv:2007.04538. [Google Scholar]
- Luo, Y.; Zhou, W.; Fang, J.; Liang, L.; Zhang, H.; Dai, G. EPI-patch based convolutional neural network for depth estimation on 4D light field. In Proceedings of the International Conference on Neural Information Processing, Guangzhou, China, 14–18 November 2017. [Google Scholar]
- Shi, J.; Jiang, X.; Guillemot, C. A framework for learning depth from a flexible subset of dense and sparse light field views. IEEE Trans. Image Process. 2019, 28, 5867–5880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilg, E.; Mayer, N.; Saikia, T.; Keuper, M.; Dosovitskiy, A.; Brox, T. FlowNet 2.0: Evolution of optical flow estimation with deep networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Feng, M.; Wang, Y.; Liu, J.; Zhang, L.; Zaki, H.F.M.; Mian, A. Benchmark data set and method for depth estimation from light field images. IEEE Trans. Image Process. 2018, 27, 3586–3598. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tao, C.; Wu, R.; Tao, X.; Sun, P.; Li, Y.; Zheng, Z. Light-field-depth-estimation network based on epipolar geometry and image segmentation. J. Opt. Soc. Am. A 2020, 37, 1236. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Wang, Q.; Lafruit, G. MANET: Multi-scale aggregated network for light field depth estimation. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020. [Google Scholar]
- Faluvegi, A.; Bolsee, Q.; Nedevschi, S.; Dadarlat, V.-T.; Munteanu, A. A 3D convolutional neural network for light field depth estimation. In Proceedings of the 2019 International Conference on 3D Immersion (IC3D), Brussels, Belgium, 11–12 December 2019. [Google Scholar]
- Leistner, T.; Schilling, H.; Mackowiak, R.; Gumhold, S.; Rother, C. Learning to think outside the box: Wide-baseline light field depth estimation with EPI-shift. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Québec City, QC, Canada, 16–19 September 2019. [Google Scholar]
- Imtiaz, S.I.; Kwon, K.-C.; Shahinur Alam, M.; Biddut Hossain, M.; Changsup, N.; Kim, N. Identification and correction of microlens-array error in an integral-imaging-microscopy system. Curr. Opt. Photonics 2021, 5, 524–531. [Google Scholar]
- Kwon, K.C.; Kwon, K.H.; Erdenebat, M.U.; Piao, Y.L.; Lim, Y.T.; Zhao, Y.; Kim, M.Y.; Kim, N. Advanced three-dimensional visualization system for an integral imaging microscope using a fully convolutional depth estimation network. IEEE Photonics J. 2020, 12, 1–14. [Google Scholar] [CrossRef]
- Kwon, K.-C.; Jeong, J.-S.; Erdenebat, M.-U.; Lim, Y.-T.; Yoo, K.-H.; Kim, N. Real-time interactive display for integral imaging microscopy. Appl. Opt. 2014, 53, 4450. [Google Scholar] [CrossRef]
- Ma, Z.; He, K.; Wei, Y.; Sun, J.; Wu, E. Constant time weighted median filtering for stereo matching and beyond. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Sydney, Australia, 1–8 December 2013. [Google Scholar]
- Kim, B.H.; Bohak, C.; Kwon, K.H.; Kim, M.Y. Cross fusion-based low dynamic and saturated image enhancement for infrared search and tracking systems. IEEE Access 2020, 8, 15347–15359. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [Green Version]









| Test Images | EPI-ORM [32] | EPINET [27] | MANET [38] | 3D-CNN-LF Depth [39] | Proposed |
|---|---|---|---|---|---|
| boxes | 0.9046 | 0.7707 | 1.0247 | 0.6942 | 0.6851 |
| dino | 0.8601 | 0.6789 | 0.9286 | 0.7035 | 0.6186 |
| dots | 0.8484 | 0.4624 | 0.5414 | 0.6552 | 0.6526 |
| pyramids | 0.7748 | 0.525 | 0.9285 | 0.6487 | 0.6789 |
| town | 0.8301 | 0.6991 | 1.2327 | 0.6608 | 0.6559 |
| Serial | Specimen | 2D Image | Center Directional View | Reconstructed Point Cloud | ||||
|---|---|---|---|---|---|---|---|---|
| Proposed | 3D-CNN-LF [39] | MANET [38] | EPINET [27] | EPI-ORM [32] | ||||
| 01 | Gear | 6.64 | 5.41 | 6.28 | 5.92 | 5.9 | 5.89 | 5.91 |
| 02 | Microchip | 6.81 | 5.66 | 6.03 | 5.67 | 5.66 | 5.69 | 5.69 |
| 03 | Seedpod | 6.88 | 5.04 | 5.95 | 5.42 | 5.43 | 5.44 | 5.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imtiaz, S.M.; Kwon, K.-C.; Hossain, M.B.; Alam, M.S.; Jeon, S.-H.; Kim, N. Depth Estimation for Integral Imaging Microscopy Using a 3D–2D CNN with a Weighted Median Filter. Sensors 2022, 22, 5288. https://doi.org/10.3390/s22145288
Imtiaz SM, Kwon K-C, Hossain MB, Alam MS, Jeon S-H, Kim N. Depth Estimation for Integral Imaging Microscopy Using a 3D–2D CNN with a Weighted Median Filter. Sensors. 2022; 22(14):5288. https://doi.org/10.3390/s22145288
Chicago/Turabian StyleImtiaz, Shariar Md, Ki-Chul Kwon, Md. Biddut Hossain, Md. Shahinur Alam, Seok-Hee Jeon, and Nam Kim. 2022. "Depth Estimation for Integral Imaging Microscopy Using a 3D–2D CNN with a Weighted Median Filter" Sensors 22, no. 14: 5288. https://doi.org/10.3390/s22145288









