A Review on Bio- and Chemosensors for the Detection of Biogenic Amines in Food Safety Applications: The Status in 2022
Abstract
:1. Introduction
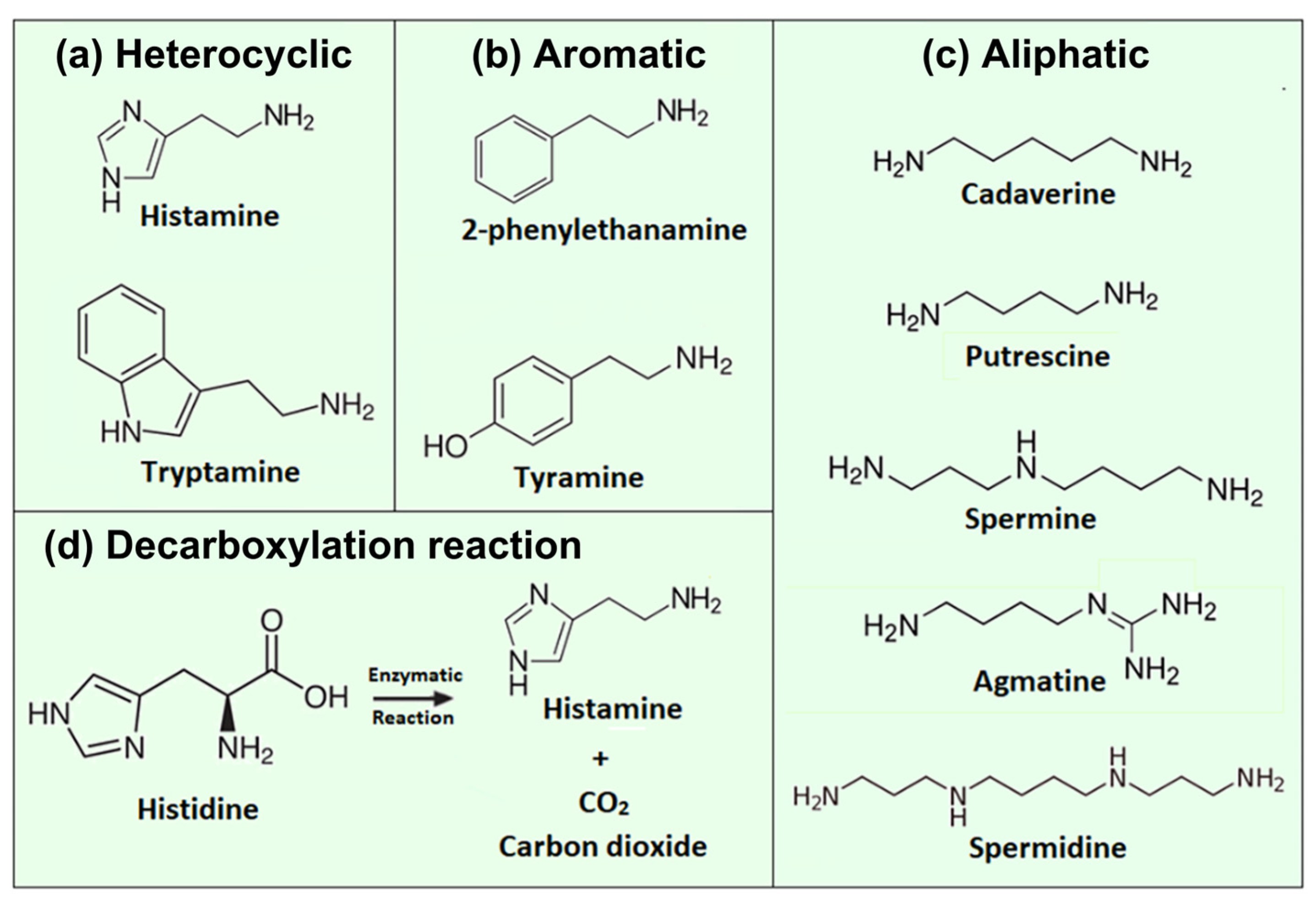
1.1. Biogenic Amines from a Biochemical Perspective
1.2. Occurrence of BAs in Food and Beverages
1.3. Adverse Health Effects Due to Intake of BAs
1.4. Legal Regulations on the BA Contents in Food Products
2. Sensor-Based Techniques for BA Detection
2.1. Analytical Reference Methods for BA Quantification and Identification
2.2. Receptor-Free Chemosensors for Biogenic Amines
2.3. Towards BA Sensors with Bio-Recognition Elements
3. Biosensors for Biogenic Amines Using Aptamer-Type Receptors
3.1. Selection Techniques for Aptamers
3.2. Aptamers for Selective Biogenic Amines Recognition
3.3. Aptasensors for BA Detection with Optical Transducers
| Biogenic Amines | Sample | Sensor Architecture | Transducer | LoD | Analytical Range | Reference |
|---|---|---|---|---|---|---|
| Histamine | Tuna samples | Fluorescently labeled RNA apta-mer, quencher displacement | Fluorescence of Cyanine 5 | 1 µM | 1–1000 µM | [142] |
| Tuna and sardines | Aptamer adsorbed at gold NPs, salt-induced aggregation | Colorimetric (UV–Vis) | 8 nM | 8–2000 nM | [143] | |
| Buffer, synth. urine | Competitive assay with histamine and histamine-coated magn. beads | Colorimetric (UV–Vis) | 18 pM in buffer, | N/A | [132] | |
| Spiked PBS solution | Biotinylated aptamers are attached to streptavidin-modified surfaces | Impedimetric (faradaic) | 4.83 mM | 4.83–60 mM | [131] | |
| Spermine | Urine | Aptamers adsorbed on AuNP, aggregation in presence of spermine | UV–Vis absorption | 473 nM | 0–18 µM | [144] |
| Dopamine | Swine feed, chicken liver | AuNPs solution in combination with the fluorophore Rhodamine B | FRET-based fluorescence | 2 nM | 26 nM–2.9 μM | [146] |
| Human body fluids | Thiolated aptamer and ferrocene immobilized on gold electrode | Cyclic voltammetry, Impedance | 60 pM, 20 pM | 0.1–10 nM | [148] | |
| Human serum | Amplification based on Ce-MOF and methylene blue | Square wave voltammetry | 6 pM | 6 pM–100 nM | [149] | |
| Filtered serum | Amplification by AgNP solution with nanocarbon modification | Diff. pulse voltammetry | 700 pM | 3–110 nM | [150] | |
| Human blood | Thiolated aptamer immobilized on gold electrode | Square wave voltammetry | 1 nM | 5–150 nM | [151] | |
| Ethanol-amine | Tap water, serum | G-quadruplex films immobilized on gold electrode | Impedimetric (faradaic) | 0.08 nM | 0.16–16 nM | [152] |
3.4. Aptamer-Based BA Sensors Based on Electrochemical Transducers
4. Biomimetic Sensors for Biogenic Amines Using Molecularly Imprinted Polymers
4.1. Molecular-Imprinting Technology
4.2. MIP-Type Receptors for Biogenic Amines
4.3. MIP-Based BA Sensors with Mass-Sensitive Transducers
4.4. MIP-Based BA Sensors with Electrochemical Transducers
4.5. MIP-Based BA Sensors with Optical Transducers
4.6. MIP-Based BA Sensors Based on the Heat Transfer Method HTM
5. Biosensors for Biogenic Amines Using Antibodies and Enzymes
5.1. Synthesis of Antibodies and Immobilization Strategies
5.2. Immunosensors for Biogenic-Amine Detection
| Biogenic Amine | Sample | Sensor Architecture | Transducer | LoD | Analytical Range | Reference |
|---|---|---|---|---|---|---|
| Histamine | Buffer | Carbon nanoparticles-based colorimetric competitive immunoassays on a microarray format | Colorimetric | 8 µg/mL | 15–101 µg/mL | [243] |
| Spiked fish samples | Competitive immunoassay using hydroquinone as electron mediator to measure the catalytic reaction between HRP and H2O2. | Amperometric | 1.25 pg/mL | 0.01–100 µg/mL | [247] | |
| Cod fish, red wine, yogurt | Nanozyme-mediated ratiometric fluorescence immunoassay using histamine-specific antibody labeled with Prussian blue nanoparticles | Fluorescence based on carbon dots | 1.2 ng/mL | 1.6 ng/mL– 125 µg/mL | [248] | |
| Spiked PBS buffer | Impedimetric and SPR-based immunosensors using reduced graphene oxide to measure binding of histamine-BSA conjugate | Impedimetric, SPR | 0.1 µM | 0.1–1 µM | [241] | |
| Spiked PBS, spiked fish samples | Screen-printed competitive immunosensor based on a silver electrode coated with single-walled carbon nanotubes | Chronoampero-metic | 2.48 pg/mL | 0.005–50 ng/mL | [242] | |
| Red wine | Lateral flow colorimetric and magnetic immunoassay using superparamagnetic particles (gold and iron oxide) labels | Magnetic, colorimetric | 1.2 mg/L, 1.5 mg/L | 1–100 mg/L | [245] | |
| Saury fish | Nanoporous alumina membrane-based biosensor using biofunctionalized magnetic nanoparticles conjugated with antihistamine antibody | Impedimetric | 3 nM | 1 µM–40 mM | [246] |
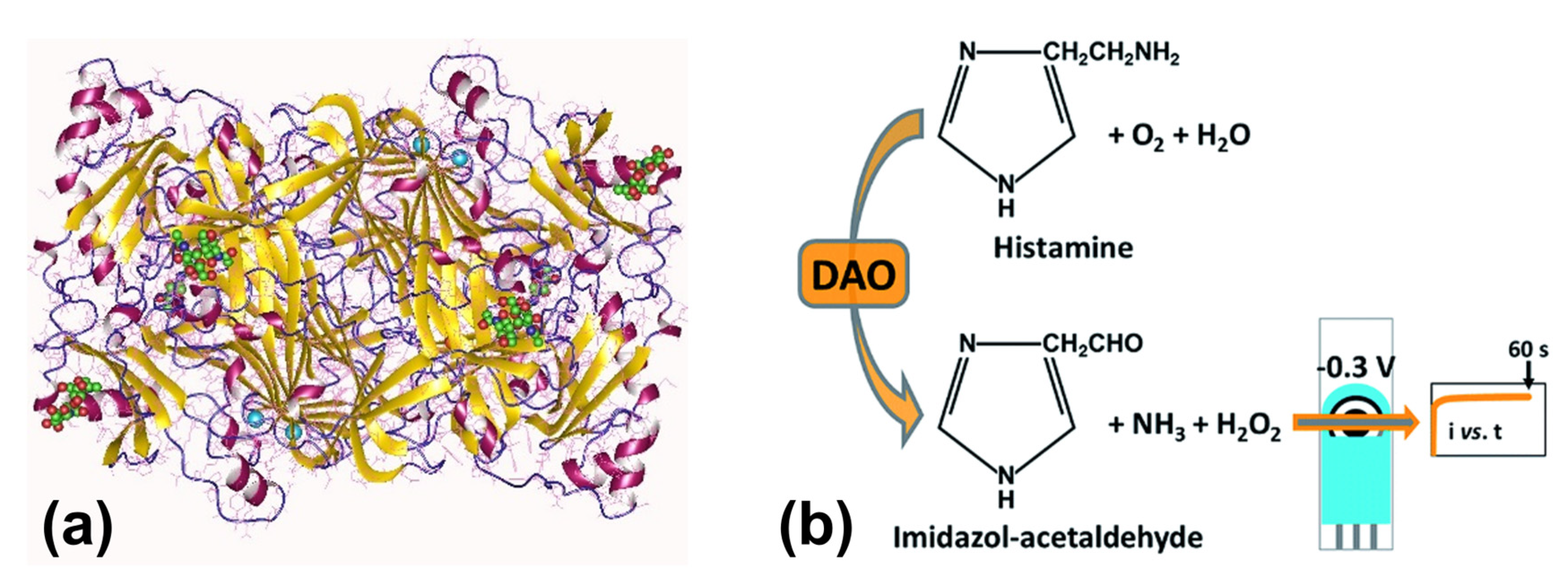
5.3. Enzymatic Detection of Biogenic Amines
5.4. Commercially Available Fast Tests for Histamine Detection
6. Summary, Outlook and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rhee, J.E.; Rhee, J.H.; Ryu, P.Y.; Choi, H.S. Identification of the cadBA Operon From Vibrio vulnificus and its Influence on Survival to Acid Stress. FEMS Microbiol. Lett. 2002, 208, 245–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkachenko, A.; Nesterova, L.; Pshenichnov, M. The Role of the Natural Polyamine Putrescine in Defense Against Oxidative Stress in Escherichia coli. Arch. Microbiol. 2001, 176, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, D.; Bosscher, J.S.; ten Brink, B.; Driessen, A.J.; Konings, W.N. Generation of a Proton Motive Force by Histidine Decarboxylation and Electrogenic Histidine/Histamine Antiport in Lactobacillus buchneri. J. Bacteriol. 1993, 175, 2864–2870. [Google Scholar] [CrossRef] [Green Version]
- Borriello, F.; Iannone, R.; Marone, G. Histamine Release from Mast Cells and Basophils. Handb. Exp. Pharmacol. 2017, 241, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Prinz, C.; Zanner, R.; Gratz, M. Physiology of Gastric Enterochromaffin-Like Cells. Annu. Rev. Physiol. 2003, 65, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Akdis, C.A.; Simons, F.E.R. Histamine Receptors are Hot in Immunopharmacology. Eur. J. Pharmacol. 2006, 533, 69–76. [Google Scholar] [CrossRef]
- Passani, M.B.; Panula, P.; Lin, J.-S. Histamine in the Brain. Front. Syst. Neurosci. 2014, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Bjornsdottir, K.; Bolton, G.E.; McClellan-Green, P.D.; Jaykus, L.-A.; Green, D.P. Detection of Gram-Negative Histamine-Producing Bacteria in Fish: A Comparative Study. J. Food Prot. 2009, 72, 1987–1991. [Google Scholar] [CrossRef]
- Kim, S.-H.; Field, K.G.; Morrissey, M.T.; Price, R.J.; Wei, C.-I.; An, H. Source and Identification of Histamine-Producing Bacteria from Fresh and Temperature-Abused Albacore. J. Food Prot. 2001, 64, 1035–1044. [Google Scholar] [CrossRef]
- López-Sabater, E.I.; Rodríguez-Jerez, J.J.; Hernádez-Herrero, M.; Roig-Sagués, A.X.; Mora-Ventura, M.T. Sensory Quality and Histamine Formation During Controlled Decomposition of Tuna (Thunnus thynnus). J. Food Prot. 1996, 59, 167–174. [Google Scholar] [CrossRef]
- Okuzumi, M.; Hiraishi, A.; Kobayashi, T.; Fujii, T. Photobacterium histaminum sp. nov., a Histamine-Producing Marine Bacterium. Int. J. Syst. Evol. 1994, 44, 631–636. [Google Scholar] [CrossRef] [Green Version]
- Ababouch, L.; Afilal, M.E.; Rhafiri, S.; Busta, F.F. Identification of Histamine-Producing Bacteria Isolated from Sardine (Sardina pilchardus) Stored in Ice and at Ambient Temperature (25 °C). Food Microbiol. 1991, 8, 127–136. [Google Scholar] [CrossRef]
- Stratton, J.E.; Hutkins, R.W.; Sumner, S.S.; Taylor, S.L. Histamine and Histamine-Producing Bacteria in Retail Swiss and Low-Salt Cheeses. J. Food Prot. 1992, 55, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Ferrer, S.; Pardo, I. Which Lactic Acid Bacteria are Responsible for Histamine Production in Wine? J. Appl. Microbiol. 2005, 99, 580–586. [Google Scholar] [CrossRef]
- Lonvaud-Funel, A.; Joyeux, A. Histamine Production by Wine Lactic Acid Bacteria: Isolation of a Histamine-Producing Strain of Leuconostoc oenos. J. Appl. Bacteriol. 1994, 77, 401–407. [Google Scholar] [CrossRef]
- Oktariani, A.F.; Ramona, Y.; Sudaryatma, P.E.; Dewi, I.A.M.M.; Shetty, K. Role of Marine Bacterial Contaminants in Histamine Formation in Seafood Products: A Review. Microorganisms 2022, 10, 1197. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic Acid Bacteria as Functional Starter Cultures for the Food Fermentation Industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Capozzi, V.; Ladero, V.; Beneduce, L.; Fernández, M.; Alvarez, M.A.; Benoit, B.; Laurent, B.; Grieco, F.; Spano, G. Isolation and Characterization of Tyramine-Producing Enterococcus faecium Strains from Red Wine. Food Microbiol. 2011, 28, 434–439. [Google Scholar] [CrossRef]
- Moreno-Arribas, V.; Torlois, S.; Joyeux, A.; Bertrand, A.; Lonvaud-Funel, A. Isolation, Properties and Behaviour of Tyramine-Producing Lactic Acid Bacteria from Wine. J. Appl. Microbiol. 2000, 88, 584–593. [Google Scholar] [CrossRef]
- González de Llano, D.; Cuesta, P.; Rodríguez, A. Biogenic Amine Production by Wild Lactococcal and Leuconostoc Strains. Lett. Appl. Microbiol. 1998, 26, 270–274. [Google Scholar] [CrossRef]
- Diaz, M.; del Rio, B.; Sanchez-Llana, E.; Ladero, V.; Redruello, B.; Fernández, M.; Martin, M.C.; Alvarez, M.A. Histamine-Producing Lactobacillus parabuchneri Strains Isolated From Grated Cheese Can Form Biofilms on Stainless Steel. Food Microbiol. 2016, 59, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, M.; Ladero, V.; del Rio, B.; Redruello, B.; Fernández, M.; Martin, M.C.; Alvarez, M.A. Biofilm-Forming Capacity in Biogenic Amine-Producing Bacteria Isolated from Dairy Products. Front. Microbiol. 2016, 7, 591. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lu, S. The Importance of Amine-degrading Enzymes on the Biogenic Amine Degradation in Fermented Foods: A Review. Process Biochem. 2020, 99, 331–339. [Google Scholar] [CrossRef]
- Jairath, G.; Singh, P.K.; Dabur, R.S.; Rani, M.; Chaudhari, M. Biogenic Amines in Meat and Meat Products and its Public Health Significance: A Review. J. Food Sci. Technol. 2015, 52, 6835–6846. [Google Scholar] [CrossRef]
- Linares, D.M.; del Río, B.; Ladero, V.; Martínez, N.; Fernández, M.; Martín, M.C.; Alvarez, M.A. Factors Influencing Biogenic Amines Accumulation in Dairy Products. Front. Microbiol. 2012, 3, 180. [Google Scholar] [CrossRef] [Green Version]
- Visciano, P.; Schirone, M.; Tofalo, R.; Suzzi, G. Biogenic Amines in Raw and Processed Seafood. Front. Microbiol. 2012, 3, 188. [Google Scholar] [CrossRef] [Green Version]
- Latorre-Moratalla, M.; Bover-Cid, S.; Veciana-Nogués, M.T. Vidal-Carou, Control of Biogenic Amines in Fermented Sausages: Role of Starter Cultures. Front. Microbiol. 2012, 3, 169. [Google Scholar] [CrossRef] [Green Version]
- Ladero, V.; Calles-Enriquez, M.; Fernandez, M.; Alvarez, M.A. Toxicological Effects of Dietary Biogenic Amines. Curr. Nutr. Food Sci. 2010, 6, 145–156. [Google Scholar] [CrossRef]
- Ruiz-Capillas, C.; Jiménez-Colmenero, F. Biogenic Amines in Meat and Meat Products. Crit. Rev. Food Sci. Nutr. 2004, 44, 489–499. [Google Scholar] [CrossRef] [Green Version]
- Suzzi, G.; Gardini, F. Biogenic Amines in Dry Fermented Sausages: A Review. Int. J. Food Microbiol. 2003, 88, 41–54. [Google Scholar] [CrossRef]
- Feng, C.; Teuber, S.; Gershwin, M.E. Histamine (Scombroid) Fish Poisoning: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 50, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Vinci, G.; Maddaloni, L. Biogenic Amines in Alcohol-Free Beverages. Beverages 2020, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Moniente, M.; Botello-Morte, L.; García-Gonzalo, D.; Pagán, R.; Ontañón, I. Analytical strategies for the determination of biogenic amines in dairy products. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3612–3646. [Google Scholar] [CrossRef] [PubMed]
- Benkerroum, N. Biogenic Amines in Dairy Products: Origin, Incidence, and Control Means. Compr. Rev. Food Sci. Food Saf. 2016, 15, 801–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Oceanic and Atmospheric Administration NOAA, US Department of Commerce. Available online: https://www.fisheries.noaa.gov/find-species (accessed on 20 December 2022).
- Morrow, J.D.; Margolies, G.R.; Rowland, J.; Roberts, L.J. Evidence That Histamine is the Causative Toxin of Scombroid-Fish Poisoning. N. Engl. J. Med. 1991, 324, 716–720. [Google Scholar] [CrossRef]
- D’Aloia, A.; Vizzardi, E.; Della Pina, P.; Bugatti, S.; Del Magro, F.; Raddino, R.; Curnis, A.; Dei Cas, L. A Scombroid Poisoning Causing a Life-Threatening Acute Pulmonary Edema and Coronary Syndrome in a Young Healthy Patient. Cardiovasc. Toxicol. 2011, 11, 280–283. [Google Scholar] [CrossRef]
- Borade, P.S.; Ballary, C.C.; Lee, D.K.C. A Fishy Cause of Sudden Near Fatal Hypotension. Resuscitation 2007, 72, 158–160. [Google Scholar] [CrossRef]
- Lehane, L.; Olley, J. Histamine Fish Poisoning Revisited. Int. J. Food Microbiol. 2000, 58, 1–37. [Google Scholar] [CrossRef]
- Rauscher-Gabernig, E.; Grossgut, R.; Bauer, F.; Paulsen, P. Assessment of Alimentary Histamine Exposure of Consumers in Austria and Development of Tolerable Levels in Typical Foods. Food Control 2009, 20, 423–429. [Google Scholar] [CrossRef]
- Stratta, P.; Badino, G. Scombroid Poisoning. Can. Med. Assoc. J. 2012, 184, 674. [Google Scholar] [CrossRef]
- Guizani, N.; Al-Busaidy, M.A.; Al-Belushi, I.M.; Mothershaw, A.; Rahman, M.S. The Effect of Storage Temperature on Histamine Production and the Freshness of Yellowfin Tuna (Thunnus albacares). Food Res. Int. 2005, 38, 215–222. [Google Scholar] [CrossRef]
- Kim, S.H.; Price, R.J.; Morrissey, M.T.; Field, K.G.; Wei, C.I.; An, H. Histamine Production by Morganella morganii in Mackerel, Albacore, Mahi-mahi, and Salmon at Various Storage Temperatures. J. Food Sci. 2002, 67, 1522–1528. [Google Scholar] [CrossRef]
- Vosikis, V.; Papageorgopoulou, A.; Economou, V.; Frillingos, S.; Papadopoulou, C. Survey of the Histamine Content in Fish Samples Randomly Selected From the Greek Retail Market. Food Addit. Contam. B Surveill. 2008, 1, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Wöhrl, S.; Hemmer, W.; Focke, M.; Rappersberger, K.; Jarisch, R. Histamine Intolerance-Like Symptoms in Healthy Volunteers After Oral Provocation with Liquid Histamine. Allergy Asthma Proc. 2004, 25, 305–311. [Google Scholar] [PubMed]
- Maintz, L.; Novak, N. Histamine and histamine intolerance. Am. J. Clin. Nutr. 2007, 85, 1185–1196. [Google Scholar] [CrossRef] [Green Version]
- Schwelberger, H.G. Histamine Intolerance: Overestimated or underestimated? Inflamm. Res. 2009, 58, 51–52. [Google Scholar] [CrossRef]
- Kovacova-Hanuskova, E.; Buday, T.; Gavliakova, S.; Plevkova, J. Histamine, Histamine Intoxication and Intolerance. Allergol. Immunopathol. 2015, 43, 498–506. [Google Scholar] [CrossRef]
- Hui, J.Y.; Taylor, S.L. Inhibition of in vivo Histamine Metabolism in Rats by Foodborne and Pharmacologic Inhibitors of Diamine Oxidase, Histamine N-Methyltransferase, and Monoamine Oxidase. Toxicol. Appl. Pharmacol. 1985, 81, 241–249. [Google Scholar] [CrossRef]
- Taylor, S.L.; Lieber, E.R. In vitro Inhibition of Rat Intestinal Histamine-Metabolizing Enzymes. Food Cosmet. Toxicol. 1979, 17, 237–240. [Google Scholar] [CrossRef]
- Bjeldanes, L.F.; Schutz, D.E.; Morris, M.M. On the Aetiology of Scombroid Poisoning: Cadaverine Potentiation of Histamine Toxicity in the Guinea-Pig. Food Cosmet. Toxicol. 1978, 16, 157–159. [Google Scholar] [CrossRef]
- Lyons, D.E.; Beery, J.T.; Lyons, S.A.; Taylor, S.L. Cadaverine and Aminoguanidine Potentiate the Uptake of Histamine in vitro in Perfused Intestinal Segments of Rats. Toxicol. Appl. Pharmacol. 1983, 70, 445–458. [Google Scholar] [CrossRef]
- Bulushi, I.A.; Poole, S.; Deeth, H.C.; Dykes, G.A. Biogenic Amines in Fish: Roles in Intoxication, Spoilage, and Nitrosamine Formation—A Review. Crit. Rev. Food Sci. Nutr. 2009, 49, 369–377. [Google Scholar] [CrossRef]
- Bills, D.D.; Hildrum, K.I.; Scanlan, R.A.; Libbey, L.M. Potential Precursors of N-Nitrosopyrrolidine in Bacon and Other Fried Foods. J. Agric. Food Chem. 1973, 21, 876–877. [Google Scholar] [CrossRef]
- Pegg, A.E. The Function of Spermine. IUBMB Life 2014, 66, 8–18. [Google Scholar] [CrossRef]
- Pegg, A.E. Toxicity of Polyamines and Their Metabolic Products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef]
- Til, H.P.; Falke, H.E.; Prinsen, M.K.; Willems, M.I. Acute and Subacute Toxicity of Tyramine, Spermidine, Spermine, Putrescine and Cadaverine in Rats. Food Chem. Toxicol. 1997, 35, 337–348. [Google Scholar] [CrossRef]
- Finberg, J.P.M.; Rabey, J.M. Inhibitors of MAO-A and MAO-B in Psychiatry and Neurology. Front. Pharmacol. 2016, 7, 340. [Google Scholar] [CrossRef] [Green Version]
- Novella-Rodríguez, S.; Veciana-Nogués, M.T.; Izquierdo-Pulido, M.; Vidal-Carou, M.C. Distribution of Biogenic Amines and Polyamines in Cheese. J. Food Sci. 2003, 68, 750–756. [Google Scholar] [CrossRef]
- Önal, A. A Review: Current Analytical Methods for the Determination of Biogenic Amines in Foods. Food Chem. 2007, 103, 1475–1486. [Google Scholar] [CrossRef]
- Blackwell, B.; Mabbitt, L.A. Tyramine in Cheese Related to Hypertensive Crises after Monoamine-Oxidase Inhibition. Lancet 1965, 285, 938–940. [Google Scholar] [CrossRef]
- Sathyanarayana Rao, T.S.; Yeragani, V.K. Hypertensive Crisis and Cheese. Indian J. Psychiatry 2009, 51, 65–66. [Google Scholar] [CrossRef]
- EU. EU Commission Regulation (EC) no. 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, 338, 1–25. [Google Scholar]
- FDA (CFSAN). Scombrotoxin (histamine) formation. In Fish and Fishery Products Hazards and Controls Guide, 3rd ed.; Department of Health and Human Services, Public Health Service, Food and Drug Administration, Center for Food Safety and Applied Nutr., Office of Seafood: Washington, DC, USA, June 2001; p. 73. [Google Scholar]
- European Food Safety Authority, EFSA Panel on Biological Hazards (BIOHAZ). Scientific Opinion on Risk Based Control of Biogenic Amine Formation in Fermented Foods. EFSA J. 2011, 9, 2393. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, P.; Grossgut, R.; Bauer, F.; Rauscher-Gabernig, E. Estimates of Maximum Tolerable Levels of Tyramine Content in Foods in Austria. J. Food Nutr. Res. 2012, 51, 52–59. [Google Scholar]
- EU Commission Regulation, no. 1019/2013 of 23 October 2013, amending Annex I to Regulation (EC) No 2073/2005 as regards histamine in fishery products. Available online: http://data.europa.eu/eli/reg/2013/1019/oj (accessed on 28 November 2022).
- Pennotti, R.; Scallan, E.; Backer, L.; Thomas, J.; Angulo, F.J. Ciguatera and Scombroid Fish Poisoning in the United States. Foodborne Pathog. Dis. 2013, 10, 1059–1066. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC); Davis, J.; Henry, S.A.; Rowland, J.; Ripley, D.; Jacobson, G.; Brunkard, J.; Carpenter, L.R. Scombroid fish poisoning associated with tuna steaks—Louisiana and Tennessee, 2006. MMWR Morb. Mortal. Wkly. Rep. 2007, 56, 817–819. [Google Scholar] [CrossRef] [Green Version]
- Feldman, K.A.; Werner, S.B.; Cronan, S.; Hernandez, M.; Horvath, A.R.; Lea, C.S.; Au, A.M.; Vugia, D.J. A Large Outbreak of Scombroid Fish Poisoning Associated with Eating Escolar Fish (Lepidocybium flavobrunneum). Epidemiol. Infect. 2005, 133, 29–33. [Google Scholar] [CrossRef]
- Gould, D.; Kraa, E.; Dalton, C.B.; Givney, R.; Gregory, J.; Stafford, R.J.; Kirk, M.D. Foodborne Disease Outbreaks in Australia, 1995 to 2000. Commun. Dis. Intell. Q. Rep. 2004, 28, 211–224. [Google Scholar]
- Lavon, O.; Lurie, Y.; Bentur, Y. Scombroid fish poisoning in Israel, 2005–2007. Isr. Med. Assoc. J. 2008, 10, 789–792. [Google Scholar] [PubMed]
- Demoncheaux, J.-P.; Michel, R.; Mazenot, C.; Duflos, G.; Iacini, C.; Delaval, F.; Saware, E.M.; Renard, J.-C. A Large Outbreak of Scombroid Fish Poisoning Associated with Eating Yellowfin Tuna (Thunnus albacares) at a Military Mass Catering in Dakar, Senegal. Epidemiol. Infect. 2012, 140, 1008–1012. [Google Scholar] [CrossRef]
- Gonzaga, V.E.; Lescano, A.G.; Huamán, A.A.; Salmón-Mulanovich, G.; Blazes, D.L. Histamine Levels in Fish from Markets in Lima, Perú. J. Food Prot. 2009, 72, 1112–1115. [Google Scholar] [CrossRef]
- Wu, S.-F.; Chen, W. An Outbreak of Scombroid Fish Poisoning in a Kindergarten. Acta Paediatr. Taiwan 2003, 44, 297–299. [Google Scholar] [PubMed]
- Tsai, Y.-H.; Lin, C.-Y.; Chang, S.-C.; Chen, H.-C.; Kung, H.-F.; Wei, C.-I.; Hwang, D.-F. Occurrence of Histamine and Histamine-Forming Bacteria in Salted Mackerel in Taiwan. Food Microbiol. 2005, 22, 461–467. [Google Scholar] [CrossRef]
- Tao, Z.; Sato, M.; Zhang, H.; Yamaguchi, T.; Nakano, T. A Survey of Histamine Content in Seafood Sold in Markets of Nine Countries. Food Control 2011, 22, 430–432. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Zhang, Y.; Zhou, Y.; Li, G.-H.; Yang, W.-Z.; Feng, X.-S. A Review of Pretreatment and Analytical Methods of Biogenic Amines in Food and Biological Samples Since 2010. J. Chromatogr. A 2019, 1605, 360361. [Google Scholar] [CrossRef]
- Gagic, M.; Jamroz, E.; Krizkova, S.; Milosavljevic, V.; Kopel, P.; Adam, V. Current Trends in Detection of Histamine in Food and Beverages. J. Agric. Food Chem. 2019, 67, 773–783. [Google Scholar] [CrossRef]
- Önal, A.; Tekkeli, S.E.K.; Önal, C. A Review of the Liquid Chromatographic Methods for the Determination of Biogenic Amines in Foods. Food Chem. 2013, 138, 509–515. [Google Scholar] [CrossRef]
- Bedia Erim, F. Recent Analytical Approaches to the Analysis of Biogenic Amines in Food Samples. TrAC-Trends Anal. Chem. 2013, 52, 239–247. [Google Scholar] [CrossRef]
- Lázaro de la Torre, C.A.; Conte-Júnior, C.A. Chromatographic Methods for Biogenic Amines Determination in Foods of Animal Origin. Braz. J. Vet. Res. Anim. Sci. 2013, 50, 430–446. [Google Scholar] [CrossRef] [Green Version]
- Ordóñez, J.L.; Troncoso, A.M.; García-Parrilla, M.D.C.; Callejón, R.M. Recent Trends in the Determination of Biogenic Amines in Fermented Beverages—A Review. Anal. Chim. Acta 2016, 939, 10–25. [Google Scholar] [CrossRef]
- Kivirand, K.; Rinken, T. Biosensors for Biogenic Amines: The Present State of Art Mini-Review. Anal. Lett. 2011, 44, 2821–2833. [Google Scholar] [CrossRef]
- Verma, N.; Hooda, V.; Gahlaut, A.; Gothwal, A.; Hooda, V. Enzymatic Biosensors for the Quantification of Biogenic Amines: A Literature Update. Crit. Rev. Biotechnol. 2020, 40, 1–14. [Google Scholar] [CrossRef]
- Vasconcelos, H.; Coelho, L.C.C.; Matias, A.; Saraiva, C.; Jorge, P.A.S.; de Almeida, J.M.M.M. Biosensors for Biogenic Amines: A Review. Biosensors 2021, 11, 82. [Google Scholar] [CrossRef]
- Gomes Müller, D.; Quadro Oreste, E.; Heinemann, M.G.; Dias, D.; Kessler, F. Biogenic Amine Sensors and its Building Materials: A Review. Eur. Polym. J. 2022, 175, 111221. [Google Scholar] [CrossRef]
- Danchuk, A.I.; Komova, N.S.; Mobarez, S.N.; Doronin, S.Y.; Burmistrova, N.A.; Markin, A.V.; Duerkop, A. Optical Sensors for Determination of Biogenic Amines in Food. Anal. Bioanal. Chem. 2020, 412, 4023–4036. [Google Scholar] [CrossRef]
- Stojanović, Z.S.; Mehmeti, E.; Kalcher, K.; Guzsvány, V.; Stanković, D.M. SWCNT-Modified Carbon Paste Electrode as an Electrochemical Sensor for Histamine Determination in Alcoholic Beverages. Food Anal. Methods 2016, 9, 2701–2710. [Google Scholar] [CrossRef]
- Young, J.A.; Jiang, X.; Kirchhoff, J.R. Amperometric Detection of Histamine with a Pyrroloquinoline-Quinone Modified Electrode. Electroanalysis 2013, 25, 1589–1593. [Google Scholar] [CrossRef]
- Emahi, I.; Mitchell, M.P.; Baum, D.A. Electrochemistry of Pyrroloquinoline Quinone (PQQ) on Multi-Walled Carbon Nanotube-Modified Glassy Carbon Electrodes in Biological Buffers. J. Electrochem. Soc. 2017, 164, H3097–H3102. [Google Scholar] [CrossRef] [Green Version]
- Katz, E.; Schlereth, D.D.; Schmidt, H.-L. Electrochemical Study of Pyrroloquinoline Quinone Covalently Immobilized as a Monolayer onto a Cystamine-Modified Gold Electrode. J. Electroanal. Chem. 1994, 367, 59–70. [Google Scholar] [CrossRef]
- Oubrie, A. Structure and Mechanism of Soluble Glucose Dehydrogenase and Other PQQ-Dependent Enzymes. Biochim. Biophys. Acta-Proteins Proteom. 2003, 1647, 143–151. [Google Scholar] [CrossRef]
- Uchida, W.; Wakabayashi, M.; Ikemoto, K.; Nakano, M.; Ohtani, H.; Nakamura, S. Mechanism of Glycine Oxidation Catalyzed by Pyrroloquinoline Quinone in Aqueous Solution. Chem. Phys. Lett. 2015, 620, 13–18. [Google Scholar] [CrossRef]
- Han, H.; Tachikawa, H. Electrochemical Determination of Thiols at Single-Wall Carbon Nanotubes and PQQ Modified Electrodes. Front. Biosci. 2005, 10, 931–939. [Google Scholar] [CrossRef] [Green Version]
- Houen, G.; Larsen, C.; Larsson, L.-I. Oxidation of Polyamines by Pyrroloquinoline Quinone (PQQ). Tetrahedron 1989, 45, 4235–4242. [Google Scholar] [CrossRef]
- Yilmaz, U.T.; Inan, D. Quantification of Histamine in Various Fish Samples Using Square Wave Stripping Voltammetric Method. J. Food Sci. Technol. 2015, 52, 6671–6678. [Google Scholar] [CrossRef] [Green Version]
- Torreggiani, A.; Tamba, M.; Bonora, S.; Fini, G. Raman and IR Study on Copper Binding of Histamine. Biopolymers 2003, 72, 290–298. [Google Scholar] [CrossRef]
- Geto, A.; Tessema, M.; Admassie, S. Determination of Histamine in Fish Muscle at Multi-Walled Carbon Nanotubes Coated Conducting Polymer Modified Glassy Carbon Electrode. Synth. Met. 2014, 191, 135–140. [Google Scholar] [CrossRef]
- Degefu, H.; Amare, M.; Tessema, M.; Admassie, S. Lignin Modified Glassy Carbon Electrode for the Electrochemical Determination of Histamine in Human Urine and Wine Samples. Electrochim. Acta 2014, 121, 307–314. [Google Scholar] [CrossRef]
- Akbari-adergani, B.; Norouzi, P.; Ganjali, M.R.; Dinarvand, R. Ultrasensitive Flow-Injection Electrochemical Method for Determination of Histamine in Tuna Fish Samples. Food Res. Int. 2010, 43, 1116–1122. [Google Scholar] [CrossRef]
- Sarada, B.V.; Rao, T.N.; Tryk, D.A.; Fujishima, A. Electrochemical Oxidation of Histamine and Serotonin at Highly Boron-Doped Diamond Electrodes. Anal. Chem. 2000, 72, 1632–1638. [Google Scholar] [CrossRef]
- Švarc-Gajić, J.; Stojanović, Z. Electrocatalytic Determination of Histamine on a Nickel-Film Glassy Carbon Electrode. Electroanalysis 2010, 22, 2931–2939. [Google Scholar] [CrossRef]
- Stojanović, Z.S.; Švarc-Gajić, J.V. A Simple and Rapid Method for Histamine Determination in Fermented Sausages by Mediated Chronopotentiometry. Food Control 2011, 22, 2013–2019. [Google Scholar] [CrossRef]
- Carralero, V.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Pulsed Amperometric Detection of Histamine at Glassy Carbon Electrodes Modified with Gold Nanoparticles. Electroanalysis 2005, 17, 289–297. [Google Scholar] [CrossRef]
- Švarc-Gajić, J.; Stojanović, Z. Determination of Histamine in Cheese by Chronopotentiometry on a Thin Film Mercury Electrode. Food Chem. 2011, 124, 1172–1176. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, G.; Li, H.; Du, D.; Lin, Y. Electrochemical Sensors and Biosensors Based on Nanomaterials and Nanostructures. Anal. Chem. 2015, 87, 230–249. [Google Scholar] [CrossRef]
- Amorim, C.G.; Souza, R.C.; Araújo, A.N.; Montenegro, M.C.B.S.M.; Silva, V.L. SI Lab-on-Valve Analysis of Histamine Using Potentiometric Detection for Food Quality Control. Food Chem. 2010, 122, 871–876. [Google Scholar] [CrossRef]
- Veseli, A.; Vasjari, M.; Arbneshi, T.; Hajrizi, A.; Švorc, L.; Samphao, A.; Kalcher, K. Electrochemical Determination of Histamine in Fish Sauce Using Heterogeneous Carbon Electrodes Modified with Rhenium(IV) Oxide. Sens. Actuators B Chem. 2016, 228, 774–781. [Google Scholar] [CrossRef]
- Butwong, N.; Khajonklin, J.; Thongbor, A.; Luong, J.H.T. Electrochemical Sensing of Histamine Using Glassy Carbon Electrode Modified with Multiwalled Carbon Nanotubes Decorated with Ag-Ag2O Nanoparticles. Microchim. Acta 2019, 186, 714. [Google Scholar] [CrossRef]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent Advances in Bacteriophage Based Biosensors for Food-Borne Pathogen Detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [Green Version]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The Application of Whole Cell-Based Biosensors for Use in Environmental Analysis and in Medical Diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef] [Green Version]
- Acha, V.; Andrews, T.; Huang, Q.; Sardar, D.K.; Hornsby, P.J. Tissue-Based Biosensors. In Recognition Receptors in Biosensors; Zourob, M., Ed.; Springer: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- Cieplak, M.; Kutner, W. Artificial Biosensors: How Can Molecular Imprinting Mimic Biorecognition? Trends Biotechnol. 2016, 34, 922–941. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro Selection of RNA Molecules that Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.-i.; Yokoyama, S.; Hirao, I. Generation of High-Affinity DNA Aptamers Using an Expanded Genetic Alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef]
- Cox, J.C.; Hayhurst, A.; Hesselberth, J.; Bayer, T.S.; Georgiou, G.; Ellington, A.D. Automated Selection of Aptamers Against Protein Targets Translated in vitro: From Gene to Aptamer. Nucleic Acids Res. 2002, 30, e108. [Google Scholar] [CrossRef]
- Tran, D.T.; Knez, K.; Janssen, K.P.; Pollet, J.; Spasic, D.; Lammertyn, J. Selection of Aptamers against Ara h 1 Protein for FO-SPR Biosensing of Peanut Allergens in Food Matrices. Biosens. Bioelectron. 2013, 43, 245–251. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Schubert, T.; Strehlitz, B. In vitro Selection and Interaction Studies of a DNA Aptamer Targeting Protein A. PLoS ONE 2015, 10, e0134403. [Google Scholar] [CrossRef] [Green Version]
- Duan, N.; Li, C.; Zhu, X.; Qi, S.; Wang, Z.; Wu, S. Simultaneous coupled with Separate SELEX for heterocyclic biogenic amine-specific aptamers screening and their application in establishment of an effective aptasensor. Sens. Actuators B Chem. 2022, 352, 130985. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. Adaptive Recognition by Nucleic Acid Aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [Green Version]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y.; et al. Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications. ACS Appl. Mater. Interfaces 2021, 13, 9500–9519. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Dreymann, N.; Wuensche, J.; Sabrowski, W.; Moeller, A.; Czepluch, D.; Vu Van, D.; Fuessel, S.; Menger, M.M. Inhibition of Human Urokinase-Type Plasminogen Activator (uPA) Enzyme Activity and Receptor Binding by DNA Aptamers as Potential Therapeutics through Binding to the Different Forms of uPA. Int. J. Mol. Sci. 2022, 23, 4890. [Google Scholar] [CrossRef] [PubMed]
- Valenzano, S.; De Girolamo, A.; DeRosa, M.C.; McKeague, M.; Schena, R.; Catucci, L.; Pascale, M. Screening and Identification of DNA Aptamers to Tyramine Using in Vitro Selection and High-Throughput Sequencing. ACS Comb. Sci. 2016, 18, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.; Reinemann, C.; Stoltenburg, R.; Strehlitz, B. In vitro Selection of DNA Aptamers Binding Ethanolamine. Biochem. Biophys. Res. Commun. 2005, 338, 1928–1934. [Google Scholar] [CrossRef]
- Reinemann, C.; Stoltenburg, R.; Strehlitz, B. Investigations on the Specificity of DNA Aptamers Binding to Ethanolamine. Anal. Chem. 2009, 81, 3973–3978. [Google Scholar] [CrossRef]
- Oguro, A.; Yanagida, A.; Fujieda, Y.; Amano, R.; Otsu, M.; Sakamoto, T.; Kawai, G.; Matsufuji, S. Two Stems with Different Characteristics and an Internal Loop in an RNA Aptamer Contribute to Spermine-Binding. J. Biochem. 2017, 161, 197–206. [Google Scholar] [CrossRef]
- Ho, L.S.J.; Fogel, R.; Limson, J.L. Generation and Screening of Histamine-Specific Aptamers for Application in a Novel Impedimetric Aptamer-Based Sensor. Talanta 2020, 208, 120474. [Google Scholar] [CrossRef]
- Mairal Lerga, T.; Jauset-Rubio, M.; Skouridou, V.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. High Affinity Aptamer for the Detection of the Biogenic Amine Histamine. Anal. Chem. 2019, 91, 7104–7111. [Google Scholar] [CrossRef]
- Tian, H.; Duan, N.; Wu, S.; Wang, Z. Selection and Application of ssDNA Aptamers Against Spermine Based on Capture-SELEX. Anal. Chim. Acta 2019, 1081, 168–175. [Google Scholar] [CrossRef]
- Mannironi, C.; Di Nardo, A.; Fruscoloni, P.; Tocchini-Valentini, G.P. In vitro Selection of Dopamine RNA Ligands. Biochemistry 1997, 36, 9726–9734. [Google Scholar] [CrossRef]
- Walsh, R.; DeRosa, M.C. Retention of Function in the DNA Homolog of the RNA Dopamine Aptamer. Biochem. Biophys. Res. Commun. 2009, 388, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Martos, I.; Ferapontova, E.E. A DNA Sequence Obtained by Replacement of the Dopamine RNA Aptamer Bases is not an Aptamer. Biochem. Biophys. Res. Commun. 2017, 489, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Andreola, M.-L.; Calmels, C.; Michel, J.; Toulme, J.-J.; Litvak, S. Towards the Selection of Phosphorothioate Aptamers—Optimizing in vitro Selection Steps with Phosphorothioate Nucleotides. Eur. J. Biochem. 2000, 267, 5032–5040. [Google Scholar] [CrossRef]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, C.D.; et al. Chemically Modified DNA Aptamers Bind Interleukin-6 with High Affinity and Inhibit Signaling by Blocking Its Interaction with Interleukin-6 Receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef] [Green Version]
- Briones, C.; Moreno, M. Applications of Peptide Nucleic Acids (PNAs) and Locked Nucleic Acids (LNAs) in Biosensor Development. Anal. Bioanal. Chem. 2012, 402, 3071–3089. [Google Scholar] [CrossRef]
- Liu, M.; Khan, A.; Wang, Z.; Liu, Y.; Yang, G.; Deng, Y.; He, N. Aptasensors for Pesticide Detection. Biosens. Bioelectron. 2019, 130, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-Based Biosensors for Antibiotic Detection: A Review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Dwidar, M.; Yokobayashi, Y. Development of a Histamine Aptasensor for Food Safety Monitoring. Sci. Rep. 2019, 9, 16659. [Google Scholar] [CrossRef] [Green Version]
- Mairal Lerga, T.; Skouridou, V.; Bermudo, M.C.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. Gold Nanoparticle Aptamer Assay for the Determination of Histamine in Foodstuffs. Microchim. Acta 2020, 187, 452. [Google Scholar] [CrossRef]
- Tsoi, T.H.; Gu, Y.J.; Lo, W.S.; Wong, W.T.; Wong, W.T.; Ng, C.F.; Lee, C.S.; Wong, K.L. Study of the Aggregation of DNA-Capped Gold Nanoparticles: A Smart and Flexible Aptasensor for Spermine Sensing. ChemPlusChem 2017, 82, 802–809. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Yang, X. Aptamer-Based Colorimetric Biosensing of Dopamine Using Unmodified Gold Nanoparticles. Sens. Actuators B Chem. 2011, 156, 95–99. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, L.; Huang, Y.; Liu, D.; Sun, R.; Luo, J.; Sun, C.A. Facile Aptamer-Based Sensing Strategy for Dopamine Through the Fluorescence Resonance Energy Transfer Between Rhodamine B and Gold Nanoparticles. Dye. Pigm. 2015, 123, 55–63. [Google Scholar] [CrossRef]
- Huang, H.; Shi, S.; Gao, X.; Gao, R.; Zhu, Y.; Wu, X.; Zang, R.; Yao, T. A Universal Label-Free Fluorescent Aptasensor Based on Ru Complex and Quantum Dots for Adenosine, Dopamine and 17β-Estradiol Detection. Biosens. Bioelectron. 2016, 79, 198–204. [Google Scholar] [CrossRef]
- Abu-Ali, H.; Ozkaya, C.; Davis, F.; Walch, N.; Nabok, A. Electrochemical Aptasensor for Detection of Dopamine. Chemosensors 2020, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; You, X.; Li, Y.; Zuo, Y.; Wang, W.; Li, D.; Huang, S.; Hu, H.; Yuan, F.; Shao, F.; et al. A Novel Electrochemical Aptasensor for Serum Dopamine Detection Based on Methylene Blue-Integrated m-PdNFs Signal Material. Sens. Actuators B Chem. 2022, 354, 131233. [Google Scholar] [CrossRef]
- Bahrami, S.; Abbasi, A.R.; Roushani, M.; Derikvand, Z.; Azadbakht, A. An Electrochemical Dopamine Aptasensor Incorporating Silver Nanoparticle, Functionalized Carbon Nanotubes and Graphene Oxide for Signal Amplification. Talanta 2016, 159, 307–316. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, W.; Yu, P.; Xiong, E.; Zhang, X.; Chen, J. A Simple Label-Free Electrochemical Aptasensor for Dopamine Detection. RSC Adv. 2014, 4, 52250–52255. [Google Scholar] [CrossRef]
- Liang, G.; Man, Y.; Jin, X.; Pan, L.; Liu, X. Aptamer-Based Biosensor for Label-Free Detection of Ethanolamine by Electrochemical Impedance Spectroscopy. Anal. Chim. Acta 2016, 936, 222–228. [Google Scholar] [CrossRef]
- Talemi, R.P.; Mousavi, S.M.; Afruzi, H. Using Gold Nanostars Modified Pencil Graphite Electrode as a Novel Substrate for Design a Sensitive and Selective Dopamine Aptasensor. Mater. Sci. Eng. C 2017, 73, 700–708. [Google Scholar] [CrossRef]
- Azadbakht, A.; Roushani, M.; Abbasi, A.R.; Derikvand, Z. Design and Characterization of Electrochemical Dopamine–Aptamer as Convenient and Integrated Sensing Platform. Anal. Biochem. 2016, 507, 47–57. [Google Scholar] [CrossRef]
- Yang, W.; Ozsoz, M.; Hibbert, D.B.; Gooding, J.J. Evidence for the Direct Interaction Between Methylene Blue and Guanine Bases Using DNA-Modified Carbon Paste Electrodes. Electroanalysis 2002, 14, 1299–1302. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.; Davis, J.J.; Luo, X. Ultrasensitive and Selective Voltammetric Aptasensor for Dopamine Based on a Conducting Polymer Nanocomposite Doped with Graphene Oxide. Microchim. Acta 2015, 182, 1123–1129. [Google Scholar] [CrossRef]
- Liu, L.; Xia, N.; Meng, J.-J.; Zhou, B.-B.; Li, S.-J. An Electrochemical Aptasensor for Sensitive and Selective Detection of Dopamine Based on Signal Amplification of Electrochemical-Chemical Redox Cycling. J. Electroanal. Chem. 2016, 775, 58–63. [Google Scholar] [CrossRef]
- Haupt, K.; Mosbach, K. Plastic Antibodies: Developments and Applications. Trends Biotechnol. 1998, 16, 468–475. [Google Scholar] [CrossRef]
- Haupt, K. (Ed.) Molecular imprinting, Topics in Current Chemistry 325; Spinger: Heidelberg, Germany; Dordrecht, The Netherlands; London, UK; New York, NY, USA, 2012; ISBN 978-3-642-28420-5. [Google Scholar]
- Nguy, T.P.; Phi, T.V.; Tram, D.T.N.; Eersels, K.; Wagner, P.; Lien, T.T.N. Development of an Impedimetric Sensor for the Label-Free Detection of the Amino Acid Sarcosine with Molecularly Imprinted Polymer Receptors. Sens. Actuators B Chem. 2017, 246, 461–470. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, G.; Fu, C.; Liu, X. Tyrosine Imprinted Polymer Beads with Different Functional Monomers via Seed Swelling and Suspension Polymerization. Polym. Eng. Sci. 2003, 43, 965–974. [Google Scholar] [CrossRef]
- Scorrano, S.; Mergola, L.; Del Sole, R.; Vasapollo, G. Synthesis of Molecularly Imprinted Polymers for Amino Acid Derivates by Using Different Functional Monomers. Int. J. Mol. Sci. 2011, 12, 1735–1743. [Google Scholar] [CrossRef] [Green Version]
- Kempe, M.; Mosbach, K. Separation of Amino Acids, Peptides and Proteins on Molecularly Imprinted Stationary Phases. J. Chromatogr. A 1995, 691, 317–323. [Google Scholar] [CrossRef]
- Burow, M.; Minoura, N. Molecular Imprinting: Synthesis of Polymer Particles with Antibody-like Binding Characteristics for Glucose Oxidase. Biochem. Biophys. Res. Commun. 1996, 227, 419–422. [Google Scholar] [CrossRef]
- Guo, T.Y.; Xia, Y.Q.; Hao, G.J.; Song, M.D.; Zhang, B.H. Adsorptive Separation of Hemoglobin by Molecularly Imprinted Chitosan Beads. Biomaterials 2004, 25, 5905–5912. [Google Scholar] [CrossRef]
- Moro, G.; Bottari, F.; Sleegers, N.; Florea, A.; Cowen, T.; Moretto, L.M.; Piletsky, S.; De Wael, K. Conductive Imprinted Polymers for the Direct Electrochemical Detection of beta-Lactam Antibiotics: The Case of Cefquinome. Sens. Actuators B Chem. 2019, 297, 126786. [Google Scholar] [CrossRef]
- Pichon, V.; Chapuis-Hugon, F. Role of Molecularly Imprinted Polymers for Selective Determination of Environmental Pollutants—A Review. Anal. Chim. Acta 2008, 622, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ping, H.; Xiang, L.; Han, P.; Wang, J.; Pan, L. Application of Molecular Imprinting Technique in Organophosphorus Pesticides Detection. In Computer and Computing Technologies in Agriculture IV; Springer: Berlin/Heidelberg, Germany, 2010; pp. 290–295. [Google Scholar] [CrossRef] [Green Version]
- Eersels, K.; Lieberzeit, P.; Wagner, P. A Review on Synthetic Receptors for Bioparticle Detection Created by Surface-Imprinting Techniques: From Principles to Applications. ACS Sens. 2016, 1, 1171–1187. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Molecular Imprinting Techniques Used for the Preparation of Biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef] [Green Version]
- Wackerlig, J.; Lieberzeit, P.A. Molecularly Imprinted Polymer Nanoparticles in Chemical Sensing—Synthesis, Characterisation and Application. Sens. Actuators B Chem. 2015, 207, 144–157. [Google Scholar] [CrossRef]
- Turner, N.W.; Jeans, C.W.; Brain, K.R.; Allender, C.J.; Hlady, V.; Britt, D.W. From 3D to 2D: A Review of the Molecular Imprinting of Proteins. Biotechnol. Prog. 2006, 22, 1474–1489. [Google Scholar] [CrossRef]
- Byrne, M.E.; Park, K.; Peppas, N.A. Molecular Imprinting within Hydrogels. Adv. Drug Deliv. Rev. 2002, 54, 149–161. [Google Scholar] [CrossRef]
- Verheyen, E.; Schillemans, J.P.; van Wijk, M.; Demeniex, M.-A.; Hennink, W.E.; van Nostrum, C.F. Challenges for the Effective Molecular Imprinting of Proteins. Biomaterials 2011, 32, 3008–3020. [Google Scholar] [CrossRef] [Green Version]
- Saylan, Y.; Akgönüllü, S.; Yavuz, H.; Ünal, S.; Denizli, A. Molecularly Imprinted Polymer Based Sensors for Medical Applications. Sensors 2019, 19, 1279. [Google Scholar] [CrossRef] [Green Version]
- Spivak, D.A. Optimization, Evaluation, and Characterization of Molecularly Imprinted Polymers. Adv. Drug Deliv. Rev. 2005, 57, 1779–1794. [Google Scholar] [CrossRef]
- García-Calzón, J.A.; Díaz-García, M.E. Characterization of Binding Sites in Molecularly Imprinted Polymers. Sens. Actuators B Chem. 2007, 123, 1180–1194. [Google Scholar] [CrossRef]
- Ansell, R.J. Characterization of the Binding Properties of Molecularly Imprinted Polymers. In Molecularly Imprinted Polymers in Biotechnology; Mattiasson, B., Ye, L., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Cham, Switzerland, 2015; Volume 150. [Google Scholar] [CrossRef]
- Umpleby, R.J., II; Baxter, S.C.; Rampey, A.M.; Rushton, G.T.; Chen, Y.; Shimizu, K.D. Characterization of the Heterogeneous Binding Site Affinity Distributions in Molecularly Imprinted Polymers. J. Chromatogr. B 2004, 804, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Peeters, M.; Troost, F.J.; van Grinsven, B.; Horemans, F.; Alenus, J.; Murib, M.S.; Keszthelyi, D.; Ethirajan, A.; Thoelen, R.; Cleij, T.J.; et al. MIP-Based Biomimetic Sensor for the Electronic Detection of Serotonin in Human Blood Plasma. Sens. Actuators B Chem. 2012, 171–172, 602–610. [Google Scholar] [CrossRef]
- Sehit, E.; Drzazgowska, J.; Buchenau, D.; Yesildag, C.; Lensen, M.; Altintas, Z. Ultrasensitive Nonenzymatic Electrochemical Glucose Sensor Based on Gold Nanoparticles and Molecularly Imprinted Polymers. Biosens. Bioelectron. 2020, 165, 112432. [Google Scholar] [CrossRef]
- Feng, X.; Ashley, J.; Zhou, T.; Halder, A.; Sun, Y. A Facile Molecularly Imprinted Polymer-Based Fluorometric Assay for Detection of Histamine. RSC Adv. 2018, 8, 2365–2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horemans, F.; Alenus, J.; Bongaers, E.; Weustenraed, A.; Thoelen, R.; Duchateau, J.; Lutsen, L.; Vanderzande, D.; Wagner, P.; Cleij, T.J. MIP-Based Sensor Platforms for the Detection of Histamine in the Nano- and Micromolar Range in Aqueous Media. Sens. Actuators B Chem. 2010, 148, 392–398. [Google Scholar] [CrossRef]
- Huang, J.; Xing, X.; Zhang, X.; He, X.; Lin, Q.; Lian, W.; Zhu, H. A Molecularly Imprinted Electrochemical Sensor Based on Multiwalled Carbon Nanotube-Gold Nanoparticle Composites and Chitosan for the Detection of Tyramine. Food Res. Int. 2011, 44, 276–281. [Google Scholar] [CrossRef]
- Meng, X.; Guo, W.; Qin, X.; Liu, Y.; Zhu, X.; Pei, M.; Wang, L. A Molecularly Imprinted Electrochemical Sensor Based on Gold Nanoparticles and Multiwalled Carbon Nanotube–Chitosan for the Detection of Tryptamine. RSC Adv. 2014, 4, 38649–38654. [Google Scholar] [CrossRef]
- Ying, X.; Yoshioka, H.-T.; Liu, C.; Sassa, F.; Hayashi, K. Molecular Imprinting Technique in Putrescine Visualized Detection. Sens. Actuators B Chem. 2018, 258, 870–880. [Google Scholar] [CrossRef]
- Ying, X.; Huang, M.; Li, X. Synthesis of Putrescine-Imprinted Double-Layer Nanofiber Membrane by Electrospinning for the Selective Recognition of Putrescine. J. Appl. Polym. Sci. 2020, 137, 48932. [Google Scholar] [CrossRef]
- Song, W.; Chen, Y.; Xu, J.; Yang, X.-R.; Tian, D.-B. Dopamine Sensor Based on Molecularly Imprinted Electrosynthesized Polymers. J. Solid State Electrochem. 2010, 14, 1909–1914. [Google Scholar] [CrossRef]
- Jiang, S.; Peng, Y.; Ning, B.; Bai, J.; Liu, Y.; Zhang, N.; Gao, Z. Surface Plasmon Resonance Sensor Based on Molecularly Imprinted Polymer Film for Detection of Histamine. Sens. Actuators B Chem. 2015, 221, 15–21. [Google Scholar] [CrossRef]
- Allender, C.J.; Richardson, C.; Woodhouse, B.; Heard, C.M.; Brain, K.R. Pharmaceutical Applications for Molecularly Imprinted Polymers. Int. J. Pharm. 2000, 195, 39–43. [Google Scholar] [CrossRef]
- Trikka, F.A.; Yoshimatsu, K.; Ye, L.; Kyriakidis, D.A. Molecularly Imprinted Polymers for Histamine Recognition in Aqueous Environment. Amino Acids 2012, 43, 2113–2124. [Google Scholar] [CrossRef] [PubMed]
- Akhoundian, M.; Rüter, A.; Shinde, S. Ultratrace Detection of Histamine Using a Molecularly-Imprinted Polymer-Based Voltammetric Sensor. Sensors 2017, 17, 645. [Google Scholar] [CrossRef] [Green Version]
- Bongaers, E.; Alenus, J.; Horemans, F.; Weustenraed, A.; Lutsen, L.; Vanderzande, D.; Cleij, T.J.; Troost, F.J.; Brummer, R.-J.; Wagner, P. A MIP-Based Biomimetic Sensor for the Impedimetric Detection of Histamine in Different pH Environments. Phys. Stat. Sol. A 2010, 207, 837–843. [Google Scholar] [CrossRef]
- Peeters, M.; Csipai, P.; Geerets, B.; Weustenraed, A.; van Grinsven, B.; Thoelen, R.; Gruber, J.; De Ceuninck, W.; Cleij, T.J.; Troost, F.J.; et al. Heat-Transfer-Based Detection of L-Nicotine, Histamine, and Serotonin Using Molecularly Imprinted Polymers as Biomimetic Receptors. Anal. Bioanal. Chem. 2013, 405, 6453–6460. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Liu, S.; Yu, J.; Lian, W.; Huang, J. Electrochemical Sensor Based on Molecularly Imprinted Film at Polypyrrole-Sulfonated Graphene/Hyaluronic Acid-Multiwalled Carbon Nanotubes Modified Electrode for Determination of Tryptamine. Biosens. Bioelectron. 2012, 31, 277–283. [Google Scholar] [CrossRef]
- Peeters, M.; Troost, F.J.; Mingels, R.H.G.; Welsch, T.; van Grinsven, B.; Vranken, T.; Ingebrandt, S.; Thoelen, R.; Cleij, T.J.; Wagner, P. Impedimetric Detection of Histamine in Bowel Fluids Using Synthetic Receptors with pH-Optimized Binding Characteristics. Anal. Chem. 2013, 85, 1475–1483. [Google Scholar] [CrossRef]
- Wackers, G.; Cornelis, P.; Putzeys, T.; Peeters, M.; Tack, J.; Troost, F.; Doll, T.; Verhaert, N.; Wagner, P. Electropolymerized Receptor Coatings for the Quantitative Detection of Histamine with a Catheter-Based, Diagnostic Sensor. ACS Sens. 2021, 6, 100–110. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; Banks, C.E.; Peeters, M. Recent Advances in Electrosynthesized Molecularly Imprinted Polymer Sensing Platforms for Bioanalyte Detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakers, G.; Wackers, G.; Trouillet, V.; Welle, A.; Wagner, P.; Junkers, T. Laser-Grafted Molecularly Imprinted Polymers for the Detection of Histamine from Organocatalyzed Atom Transfer Radical Polymerization. Macromolecules 2019, 52, 2304–2313. [Google Scholar] [CrossRef]
- Diltemiz, S.E.; Keçili, R.; Ersöz, A.; Say, R. Molecular Imprinting Technology in Quartz Crystal Microbalance (QCM) Sensors. Sensors 2017, 17, 454. [Google Scholar] [CrossRef] [PubMed]
- Pietrzyk, A.; Suriyanarayanan, S.; Kutner, W.; Chitta, R.; D’Souza, F. Selective Histamine Piezoelectric Chemosensor Using a Recognition Film of the Molecularly Imprinted Polymer of Bis(Bithiophene) Derivatives. Anal. Chem. 2009, 81, 2633–2643. [Google Scholar] [CrossRef]
- Freundlich, H. Über die Adsorption in Lösungen. Z. Phys. Chem. 1906, 57, 385–470. [Google Scholar] [CrossRef]
- Dai, J.; Zhang, Y.; Pan, M.; Kong, L.; Wang, S. Development and Application of Quartz Crystal Microbalance Sensor Based on Novel Molecularly Imprinted Sol-Gel Polymer for Rapid Detection of Histamine in Foods. J. Agric. Food Chem. 2014, 62, 5269–5274. [Google Scholar] [CrossRef]
- Ratautaite, V.; Nesládek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A. Evaluation of Histamine Imprinted Polypyrrole Deposited on Boron Doped Nanocrystalline Diamond. Electroanalysis 2014, 26, 2458–2464. [Google Scholar] [CrossRef]
- Croux, D.; Vangerven, T.; Broeders, J.; Boutsen, J.; Peeters, M.; Duchateau, S.; Deferme, W.; Cleij, T.; Wagner, P.; Thoelen, R.; et al. Molecular Imprinted Polymer Films on RFID Tags—A First Step Toward Disposable Packaging Sensors. Phys. Stat. Sol. A 2013, 210, 938–944. [Google Scholar] [CrossRef]
- Gao, F.; Grant, E.; Lu, X. Determination of Histamine in Canned Tuna by Molecularly Imprinted Polymers-Surface Enhanced Raman Spectroscopy. Anal. Chim. Acta 2015, 901, 68–75. [Google Scholar] [CrossRef]
- Basozabal, I.; Guerreiro, A.; Gomez-Caballero, A.; Aranzazu, M.G.; Barrio, R.J. Direct Potentiometric Quantification of Histamine Using Solid-Phase Imprinted Nanoparticles as Recognition Elements. Biosens. Bioelectron. 2014, 58, 138–144. [Google Scholar] [CrossRef]
- Mattsson, L.; Xu, J.; Preininger, C.; Tse Sum Bui, B.; Haupt, K. Competitive Fluorescent Pseudo-Immunoassay Exploiting Molecularly Imprinted Polymers for the Detection of Biogenic Amines in Fish Matrix. Talanta 2018, 181, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Y.; Geng, W.; Liu, H. Rapid Detection of Tryptamine by Optosensor with Molecularly Imprinted Polymers Based on Carbon Dots-Embedded Covalent-Organic Frameworks. Sens. Actuators B Chem. 2019, 285, 546–552. [Google Scholar] [CrossRef]
- Li, Y.; Hsieh, C.-H.; Lai, C.-W.; Chang, Y.-F.; Chan, H.-Y.; Tsai, C.-F.; Ho, J.-A.A.; Wu, L.-C. Tyramine Detection Using PEDOT:PSS/AuNPs/1-Methyl-4-Mercaptopyridine Modified Screen-Printed Carbon Electrode with Molecularly Imprinted Polymer Solid Phase Extraction. Biosens. Bioelectron. 2017, 87, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-J.; Chang, R.; Zhu, Q.-J. Synthesis and Characterization of a Molecularly Imprinted Polymer of Spermidine and the Exploration of its Molecular Recognition Properties. Polymers 2018, 10, 1389. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Jiang, P.; Dong, X. Molecularly Imprinted Fluorescent Chemosensor Synthesized Using quinoline-Modified-β-Cyclodextrin as Monomer for Spermidine Recognition. RSC Adv. 2015, 5, 5566–5574. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Z.; Xiong, C.; Zheng, L.; Ding, Y.; Lu, H.; Zhang, G.; Qiu, L. Selective Recognition of Histidine Enantiomers Using Novel Molecularly Imprinted Organic Transistor Sensor. Org. Electron. 2018, 61, 254–260. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, B.; Wei, S.; Xu, J.; Wang, J.; Li, T. Histidine Detection and Signal Amplification Based on Magnetic Molecularly Imprinted Particle and Cu2+ Coordination. Sens. Actuators B Chem. 2022, 358, 131487. [Google Scholar] [CrossRef]
- Wackers, G.; Putzeys, T.; Peeters, M.; Van de Cauter, L.; Cornelis, P.; Wübbenhorst, M.; Tack, J.; Troost, F.; Verhaert, N.; Doll, T.; et al. Towards a Catheter-Based Impedimetric Sensor for the Assessment of Intestinal Histamine Levels in IBS Patients. Biosens. Bioelectron. 2020, 158, 112152. [Google Scholar] [CrossRef]
- Stilman, W.; Wackers, G.; Bakhshi Sichani, S.; Khorshid, M.; Theßeling, F.; Vereman, J.; Andruck, L.; Elian, D.; Van Cornelis, P.; Impe, J.; et al. A Table-Top Sensor for the Detection of Hydrophobins and Yeasts in Brewery Applications. Sens. Actuators B Chem. 2022, 373, 132690. [Google Scholar] [CrossRef]
- Phan, D.T.; Nguyen, C.H.; Nguyen, T.D.P.; Tran, L.H.; Park, S.; Choi, J.; Lee, B.; Oh, J. A Flexible, Wearable, and Wireless Biosensor Patch with Internet of Medical Things Applications. Biosensors 2022, 12, 139. [Google Scholar] [CrossRef]
- Steiner, M.-S.; Meier, R.J.; Duerkop, A.; Wolfbeis, O.S. Chromogenic Sensing of Biogenic Amines Using a Chameleon Probe and the Red−Green−Blue Readout of Digital Camera Images. Anal. Chem. 2010, 82, 8402–8405. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Reichert, C.L.; Schmid, M. Biogenic Amine Detection Systems for Intelligent Packaging Concepts: Meat and Meat Products. Food Rev. Int. 2021, 1–25. [Google Scholar] [CrossRef]
- Alexi, N.; Thamsborg, K.; Hvam, J.; Lund, B.W.; Nsubuga, L.; de Oliveira Hansen, R.M.; Byrne, D.V.; Leisner, J.J. Novel cadaverine non-invasive biosensor technology for the prediction of shelf life of modified atmosphere packed pork cutlets. Meat Sci. 2022, 192, 108876. [Google Scholar] [CrossRef] [PubMed]
- Atta, N.F.; Abdel-Mageed, A.M. Smart Electrochemical Sensor for Some Neurotransmitters Using Imprinted Sol–Gel Films. Talanta 2009, 80, 511–518. [Google Scholar] [CrossRef]
- Tong, A.; Dong, H.; Li, L. Molecular Imprinting-Based Fluorescent Chemosensor for Histamine Using Zinc(II)–Protoporphyrin as a Functional Monomer. Anal. Chim. Acta 2002, 466, 31–37. [Google Scholar] [CrossRef]
- Wu, G.; Dou, X.; Li, D.; Xu, S.; Zhang, J.; Ding, Z.; Xie, J. Recent Progress of Fluorescence Sensors for Histamine in Foods. Biosensors 2022, 12, 161. [Google Scholar] [CrossRef]
- Haran, Y.; Li, C.; Zhou, X.; Yan, S. Preparation of Quantum Dots and Fluorescent Molecularly Imprinted Composite Microspheres for Determination of Tyramine. J. Mater. Sci. Nanotechnol. 2017, 4, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Khorshid, M.; Bakhshi Sichani, S.; Cornelis, P.; Wackers, G.; Wagner, P. The Hot-Wire Concept: Towards a One-Element Thermal Biosensor Platform. Biosens. Bioelectron. 2021, 179, 113043. [Google Scholar] [CrossRef]
- van Grinsven, B.; Eersels, K.; Peeters, M.; Losada-Pérez, P.; Vandenryt, T.; Cleij, T.J.; Wagner, P. The Heat-Transfer Method: A Versatile Low-Cost, Label-Free, Fast, and User-Friendly Readout Platform for Biosensor Applications. ACS Appl. Mater. Interfaces 2014, 6, 13309–13318. [Google Scholar] [CrossRef] [Green Version]
- Wackers, G.; Vandenryt, T.; Cornelis, P.; Kellens, E.; Thoelen, R.; De Ceuninck, W.; Losada-Pérez, P.; van Grinsven, B.; Peeters, M.; Wagner, P. Array Formatting of the Heat-Transfer Method (HTM) for the Detection of Small Organic Molecules by Molecularly Imprinted Polymers. Sensors 2014, 14, 11016–11030. [Google Scholar] [CrossRef]
- Peeters, M.; Kobben, S.; Jiménez-Monroy, K.; Modesto, L.; Kraus, M.; Vandenryt, T.; Gaulke, A.; van Grinsven, B.; Ingebrandt, S.; Junkers, T.; et al. Thermal Detection of Histamine with a Graphene Oxide Based Molecularly Imprinted Polymer Platform Prepared by Reversible Addition–Fragmentation Chain Transfer Polymerization. Sens. Actuators B Chem. 2014, 203, 527–535. [Google Scholar] [CrossRef]
- Ricci, F.; Volpe, G.; Micheli, L.; Palleschi, G. A Review on Novel Developments and Applications of Immunosensors in Food Analysis. Anal. Chim. Acta 2007, 605, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.-J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef] [PubMed]
- Lipman, N.S.; Jackson, L.R.; Trudel, L.J.; Weis-Garcia, F. Monoclonal Versus Polyclonal Antibodies: Distinguishing Characteristics, Applications, and Information Resources. ILAR J. 2005, 46, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaleh, M.A.; Alsaab, H.O.; Mahmoud, A.B.; Alkayyal, A.A.; Jones, M.L.; Mahler, S.M.; Hashem, A.M. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front. Immunol. 2020, 11, 1986. [Google Scholar] [CrossRef]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, Antibody scFv, and Antibody Fab’ Fragments: An Overview and Comparison of Three of the Most Versatile Biosensor Biorecognition Elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef]
- Guesdon, J.-L.; Chevrier, D.; Mazié, J.-C.; David, B.; Avrameas, S. Monoclonal Anti-Histamine Antibody: Preparation, Characterization and Application to Enzyme Immunoassay of Histamine. J. Immunol. Methods 1986, 87, 69–78. [Google Scholar] [CrossRef]
- Cooreman, P.; Thoelen, R.; Manca, J.; vandeVen, M.; Vermeeren, V.; Michiels, L.; Ameloot, M.; Wagner, P. Impedimetric Immunosensors Based on the Conjugated Polymer PPV. Biosens. Bioelectron. 2005, 20, 2151–2156. [Google Scholar] [CrossRef]
- Wiseman, M.E.; Frank, C.W. Antibody Adsorption and Orientation on Hydrophobic Surfaces. Langmuir 2012, 28, 1765–1774. [Google Scholar] [CrossRef]
- Adesina, A.; Mashazi, P. Oriented Antibody Covalent Immobilization for Label-Free Impedimetric Detection of C-Reactive Protein via Direct and Sandwich Immunoassays. Front. Chem. 2021, 9, 587142. [Google Scholar] [CrossRef]
- Trilling, A.K.; Beekwilder, J.; Zuilhof, H. Antibody Orientation on Biosensor Surfaces: A Minireview. Analyst 2013, 138, 1619–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welch, N.G.; Scoble, J.A.; Muir, B.W.; Pigram, P.J. Orientation and Characterization of Immobilized Antibodies for Improved Immunoassays (Review). Biointerphases 2017, 12, 02D301. [Google Scholar] [CrossRef] [Green Version]
- Mutlu, S.; Çökeliler, D.; Shard, A.; Goktas, H.; Ozansoy, B.; Mutlu, M. Preparation and Characterization of Ethylenediamine and Cysteamine Plasma Polymerized Films on Piezoelectric Quartz Crystal Surfaces for a Biosensor. Thin Solid Film. 2008, 516, 1249–1255. [Google Scholar] [CrossRef]
- Delle, L.E.; Huck, C.; Bäcker, M.; Müller, F.; Grandthyll, S.; Jacobs, K.; Lilischkis, R.; Vu, X.T.; Schöning, M.J.; Wagner, P.; et al. Impedimetric Immunosensor for the Detection of Histamine Based on Reduced Graphene Oxide. Phys. Stat. Sol. A 2015, 212, 1327–1334. [Google Scholar] [CrossRef]
- Shkodra, B.; Abera, B.D.; Cantarella, G.; Douaki, A.; Avancini, E.; Petti, L.; Lugli, P. Flexible and Printed Electrochemical Immunosensor Coated with Oxygen Plasma Treated SWCNTs for Histamine Detection. Biosensors 2020, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattsson, L.; Jungmann, C.; Lieberzeit, P.A.; Preininger, C. Modified Carbon Black as Label in a Colorometric On-Chip Immunoassay for Histamine. Sens. Actuators B Chem. 2017, 246, 1092–1099. [Google Scholar] [CrossRef]
- Mattsson, L.; Doppler, S.; Preininger, C. Challenges in Developing a Biochip for Intact Histamine Using Commercial Antibodies. Chemosensors 2017, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Moyano, A.; Salvador, M.; Martínez-García, J.C.; Socoliuc, V.; Vékás, L.; Peddis, D.; Alvarez, M.A.; Fernández, M.; Rivas, M.; Blanco-López, C. Magnetic Immunochromatographic Test for Histamine Detection in Wine. Anal. Bioanal. Chem. 2019, 411, 6615–6624. [Google Scholar] [CrossRef]
- Ye, W.; Xu, Y.; Zheng, L.; Zhang, Y.; Yang, M.; Sun, P. A Nanoporous Alumina Membrane Based Electrochemical Biosensor for Histamine Determination with Biofunctionalized Magnetic Nanoparticles Concentration and Signal Amplification. Sensors 2016, 16, 1767. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.-X.; Yang, J.-Y.; Luo, L.; Zhang, Y.-F.; Mao, C.; Sun, Y.-M.; Lei, H.-T.; Shen, Y.-D.; Beier, R.C.; Xu, Z.-L. Portable Amperometric Immunosensor for Histamine Detection Using Prussian Blue-Chitosan-Gold Nanoparticle Nanocomposite Films. Biosens. Bioelectron. 2017, 98, 305–309. [Google Scholar] [CrossRef]
- Liao, C.-X.; Jia, B.-Z.; Wang, H.; Sun, Y.-M.; Xu, X.-Y.; Wei, X.-Q.; Shen, Y.-D.; Lei, H.-T.; Xu, Z.-L.; Luo, L. Prussian Blue Nanoparticles-Enabled Sensitive and Accurate Ratiometric Fluorescence Immunoassay for Histamine. Food Chem. 2022, 376, 131907. [Google Scholar] [CrossRef] [PubMed]
- Kenten, R.H.; Mann, P.J.G. The Oxidation of Amines by Extracts of Pea Seedlings. Biochem. J. 1952, 50, 360–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGowan, R.E.; Muir, R.M. Purification and Properties of Amine Oxidase from Epicotyls of Pisum sativum. Plant Physiol. 1971, 47, 644–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wimmerová, M.; Macholán, L. Sensitive Amperometric Biosensor for the Determination of Biogenic and Synthetic Amines Using Pea Seedlings Amine Oxidase: A Novel Approach for Enzyme Immobilisation. Biosens. Bioelectron. 1999, 14, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Telsnig, D.; Kalcher, K.; Leitner, A.; Ortner, A. Design of an Amperometric Biosensor for the Determination of Biogenic Amines Using Screen Printed Carbon Working Electrodes. Electroanalysis 2013, 25, 47–50. [Google Scholar] [CrossRef]
- Ahangari, H.; Kurbanoglu, S.; Ehsani, A.; Uslu, B. Latest Trends for Biogenic Amines Detection in Foods: Enzymatic Biosensors and Nanozymes Applications. Trends Food Sci. Technol. 2021, 112, 75–87. [Google Scholar] [CrossRef]
- Torre, R.; Costa-Rama, E.; Nouws, H.P.A.; Delerue-Matos, C. Screen-Printed Electrode-Based Sensors for Food Spoilage Control: Bacteria and Biogenic Amines Detection. Biosensors 2020, 10, 139. [Google Scholar] [CrossRef]
- McGrath, A.P.; Hilmer, K.M.; Collyer, C.A.; Shepard, E.M.; Elmore, B.O.; Brown, D.E.; Dooley, D.M.; Guss, J.M. Structure and Inhibition of Human Diamine Oxidase. Biochemistry 2009, 48, 9810–9822. [Google Scholar] [CrossRef] [Green Version]
- Torre, R.; Costa-Rama, E.; Lopes, P.; Nouws, H.P.A.; Delerue-Matos, C. Amperometric Enzyme Sensor for the Rapid Determination of Histamine. Anal. Methods 2019, 11, 1264. [Google Scholar] [CrossRef]
- Koçoğlu, İ.O.; Erden, P.E.; Kılıç, E. Disposable Biogenic Amine Biosensors for Histamine Determination in Fish. Anal. Methods 2020, 12, 3802–3812. [Google Scholar] [CrossRef]
- Kaçar, C.; Erden, P.E.; Dalkiran, B.; İnal, E.K.; Kiliç, E. Amperometric Biogenic Amine Biosensors Based on Prussian Blue, Indium Tin Oxide Nanoparticles and Diamine Oxidase– Or Monoamine Oxidase–Modified Electrodes. Anal. Bioanal. Chem. 2020, 412, 1933–1946. [Google Scholar] [CrossRef] [PubMed]
- Webpage. Available online: https://upload.wikimedia.org/wikipedia/commons/f/f3/3k5t.jpg (accessed on 21 November 2022).
- Keow, C.M.; Abu Bakar, F.; Salleh, A.B.; Heng, L.Y.; Wagiran, R.; Bean, L.S. An Amperometric Biosensor for the Rapid Assessment of Histamine Level in Tiger Prawn (Penaeus monodon) Spoilage. Food Chem. 2007, 105, 1636–1641. [Google Scholar] [CrossRef]
- Pérez, S.; Bartrolí, J.; Fàbregas, E. Amperometric Biosensor for the Determination of Histamine in Fish Samples. Food Chem. 2013, 141, 4066–4072. [Google Scholar] [CrossRef]
- Leonardo, S.; Campàs, M. Electrochemical Enzyme Sensor Arrays for the Detection of the Biogenic Amines Histamine, Putrescine and Cadaverine Using Magnetic Beads as Immobilisation Supports. Microchim. Acta 2016, 183, 1881–1890. [Google Scholar] [CrossRef]
- Alonso-Lomillo, M.A.; Domínguez-Renedo, O.; Matos, P.; Arcos-Martínez, M.J. Disposable Biosensors for Determination of Biogenic Amines. Anal. Chim. Acta 2010, 665, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Henao-Escobar, W.; del Torno-de Román, L.; Domínguez-Renedo, O.; Alonso-Lomillo, M.A.; Arcos-Martínez, M.J. Dual Enzymatic Biosensor for Simultaneous Amperometric Determination of Histamine and Putrescine. Food Chem. 2016, 190, 818–823. [Google Scholar] [CrossRef]
- Bóka, B.; Adányi, N.; Virág, D.; Sebela, M.; Kiss, A. Spoilage Detection with Biogenic Amine Biosensors, Comparison of Different Enzyme Electrodes. Electroanalysis 2012, 24, 181–186. [Google Scholar] [CrossRef]
- Bóka, B.; Adányi, N.; Szamos, J.; Virág, D.; Kiss, A. Putrescine Biosensor Based on Putrescine Oxidase From Kocuria rosea. Enzym. Microb. Technol. 2012, 51, 258–262. [Google Scholar] [CrossRef]
- Henao-Escobar, W.; Domínguez-Renedo, O.; Alonso-Lomillo, M.A.; Arcos-Martínez, M.J. Simultaneous Determination of Cadaverine and Putrescine Using a Disposable Monoamine Oxidase Based Biosensor. Talanta 2013, 117, 405–411. [Google Scholar] [CrossRef]
- Boffi, A.; Favero, G.; Federico, R.; Macone, A.; Antiochia, R.; Tortolini, C.; Sanzó, G.; Mazzei, F. Amine Oxidase-Based Biosensors for Spermine and Spermidine Determination. Anal. Bioanal. Chem. 2015, 407, 1131–1137. [Google Scholar] [CrossRef]
- Tortolini, C.; Favero, G.; Mazzei, F. Development of Amine-Oxidase-Based Biosensors for Spermine and Spermidine Analysis. Methods Mol. Biol. 2018, 1694, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Tian, L.; Wu, J.; Wu, Y.; Liu, Y.; Du, J.; Lu, X. A Highly Sensitive Electrochemiluminescence Spermine Biosensor Based on Au-Ag Bimetallic Nanoclusters. Electroanalysis 2021, 33, 2016–2024. [Google Scholar] [CrossRef]
- Erden, P.E.; Selvi, C.K.; Kılıç, E. A Novel Tyramine Biosensor Based on Carbon Nanofibers, 1-Butyl-3-Methylimidazolium Tetrafluoroborate and Gold Nanoparticles. Microchem. J. 2021, 170, 106729. [Google Scholar] [CrossRef]
- Erden, P.E.; Erdoğan, Z.Ö.; Öztürk, F.; Koçoğlu, İ.O.; Kılıç, E. Amperometric Biosensors for Tyramine Determination Based on Graphene Oxide and Polyvinylferrocene Modified Screen-printed Electrodes. Electroanalysis 2019, 31, 2368–2378. [Google Scholar] [CrossRef]
- Calvo-Pérez, A.; Domínguez-Renedo, O.; Alonso-Lomillo, M.A.; Arcos-Martínez, M.J. Disposable Amperometric Biosensor for the Determination of Tyramine Using Plasma Amino Oxidase. Microchim. Acta 2013, 180, 253–259. [Google Scholar] [CrossRef]
- Dalkıran, B.; Erden, P.E.; Kaçar, C.; Kılıç, E. Disposable Amperometric Biosensor Based on Poly-L-lysine and Fe3O4 NPs-chitosan Composite for the Detection of Tyramine in Cheese. Electroanalysis 2019, 31, 1324–1333. [Google Scholar] [CrossRef]
- Apetrei, I.M.; Apetrei, C. Amperometric Biosensor Based on Polypyrrole and Tyrosinase for the Detection of Tyramine in Food Samples. Sens. Actuators B Chem. 2013, 178, 40–46. [Google Scholar] [CrossRef]
- Apetrei, I.M.; Apetrei, C. The Biocomposite Screen-Printed Biosensor Based on Immobilization of Tyrosinase onto the Carboxyl Functionalised Carbon Nanotube for Assaying Tyramine in Fish Products. J. Food Eng. 2015, 149, 1–8. [Google Scholar] [CrossRef]
- da Silva, W.; Ghica, M.E.; Ajayi, R.F.; Iwuoha, E.I.; Brett, C.M.A. Tyrosinase Based Amperometric Biosensor for Determination of Tyramine in Fermented Food and Beverages with Gold Nanoparticle Doped Poly(8-Anilino-1-Naphthalene Sulphonic Acid) Modified Electrode. Food Chem. 2019, 282, 18–26. [Google Scholar] [CrossRef]
- Gonçalves da Silva, A.; Franco, D.L.; Santos, L.D. A Simple, Fast, and Direct Electrochemical Determination of Tyramine in Brazilian Wines Using Low-Cost Electrodes. Food Control 2021, 130, 108369. [Google Scholar] [CrossRef]
- Sato, T.; Horiuchi, T.; Nishimura, I. Simple and Rapid Determination of Histamine in Food Using a New Histamine Dehydrogenase from Rhizobium sp. Anal. Biochem. 2005, 346, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Pais, G.L.; Meloni, D.; Mudadu, A.G.; Crobu, L.; Pulina, A.; Chessa, G. Colorimetric Analysis and Determination of Histamine in Samples of Yellowfin Tuna (Thunnus albacares) Marketed in Sardinia (Italy) by a Combination of Rapid Screening Methods and LC-MS/MS. Foods 2022, 11, 639. [Google Scholar] [CrossRef] [PubMed]
- Köse, S.; Kaklıkkaya, N.; Koral, S.; Tufan, B.; Buruk, K.C.; Aydın, F. Commercial Test Kits and the Determination of Histamine in Traditional (Ethnic) Fish Products—Evaluation Against an EU Accepted HPLC Method. Food Chem. 2011, 125, 1490–1497. [Google Scholar] [CrossRef]













| Biogenic Amine | Selection Route | Nucleic Acid | KD (mol/L) | Reference |
|---|---|---|---|---|
| Histamine | SELEX | DNA, 49 bases | 72.8 ± 13.9 × 10−9 | [131] |
| Histamine | SELEX | DNA, 99 bases | 3.08 ± 1.13 × 10−9 | [132] |
| Histamine | Separate-SELEX | DNA, 80 bases | 11.62 ± 3.24 × 10−9 | [122] |
| Tryptamine | 6.30 ± 1.76 × 10−9 | |||
| Tyramine | SELEX | DNA, 38 bases | 0.2 ± 0.4 × 10−6 | [127] |
| Spermine | Capture-SELEX | DNA, 40 bases | 9.65 ± 0.90 × 10−9 | [133] |
| Ethanolamine | SELEX | DNA, 96 bases | 6 ± 3 × 10−9 | [128] |
| Dopamine | SELEX | RNA, 57 bases | 1.6 × 10−6 | [134] |
| A | Sample | Sensor Architecture | Transducer | LOD | Analytical Range | Reference |
|---|---|---|---|---|---|---|
| Spiked fish products | Hydrolytic cross-linking of silanol groups using a sol–gel process | QCM | 7.49 × 10−4 mg/kg | 0.11 × 10−2– 4.45 × 10−2 mg/L | [203] | |
| Canned tuna | Bulk polymerized MAA particles, OC1C10-PVV and PVC adhesive | Impedimetric QCM | Below 1 nM 1 µM | 0–12 nM 1–100 µM | [183] | |
| PBS buffer solution | Bulk polymerized MAA particles, OC1C10-PVV adhesive | Impedimetric | 2 nM | 3–12 nM | [193] | |
| PBS buffer solution | Bulk polymerized MAA particles, OC1C10-PVV adhesive | Impedimetric | 15 nM | 500–1000 nM | [197] | |
| PBS + KCl solution | Electro-polymerization of Pyrrole on B:NCD diamond film | Impedimetric | Not mentioned | Not mentioned | [204] | |
| Intestinal fluids | Electro-polymerization of Pyrrole on Titanium wires | Impedimetric | 1 nM | 1 nM–10 µM | [197] | |
| Histamine | Spiked buffer | Bulk polymerized MAA deposited on a PDMS stamp. | Admittance spectroscopy | 50 nM | 50 nM–1000 nM | [205] |
| Histamine buffer solution | Bulk polymerized MAA particles, OC1C10-PVV adhesive | Heat transfer resistance HTM | 30 nM | 0.2 µM–1 µM | [194] | |
| PBS solutions with K3[Fe(CN)6] | Bulk polymerization of MAA on CP electrodes | Voltammetry | 7.4 × 10−5 µM | 10−4–10−3 µM, 7 × 10−3–4 × 10−1 µM | [192] | |
| Canned tuna | Bulk polymerized MAA, MIPs-PVC film combined with AuNPs | SERS | Not mentioned | Not mentioned | [206] | |
| Carp fish | Bulk polymerized MAA on a thin-film gold SPR sensor chip | SPR | 24.9 µg/L | 25–1000 μg/L | [189] | |
| Wine and fish matrices | Solid-phase imprinted MAA, MINs in PVC membrane | SPR, potentiometric | 1.1 µM | 1 µM–10 mM | [207] | |
| Purified fish extract | Precipitation polymerization of MAA | Competitive fluoro-immunoassays, BODIPY C9H7BN2F2 | 1 µM | 1–430 µM | [208] | |
| B | Sample | Sensor architecture | Transducer | LOD | Analyticalrange | Reference |
| Tryptamine | Cheese, lacto-bacillus beverage | MIP electropolymerized on conductive, MWCNT modified GCE | Amperometric | 42 nM | 60 nM – 30 μM | [185] |
| Meat samples | MIP made of MAA and AM polymerization, based on CD-embedded COFs | Fluorimetric, luminescent carbon dots | 7 µg/kg | 0.025–0.4 mg/kg | [209] | |
| Tyramine | Milk | MIP-based Tyr extraction before detection with AuNP modified SPCE | Voltammetric | 0.00231 µM | 5–100 nM | [210] |
| Yoghurt | MIP by precipitation polymerization on MWCNT/AuNP modified GCE | Voltammetric | 0.057 µM | 0.108 µM–10 µM | [184] | |
| Spermidine | CCl4–spermidine solution | Noncovalent polymerization of MAA | Absorption spectroscopy | 0.3 mmol/L | Not mentioned | [211] |
| Spiked serum samples | MIP membrane by bulk polymerization of quinolyl-b-CD | Fluorimetric, quinoline C9H7N | 5 × 10−7 mol/L | 0.5–200 µM | [212] | |
| Putrescine | Putrescine aqueous solution | Bulk polymerization of PVA500 on glass slide and Petry dish | Colorimetric | 4 mg/g | 4–24 mg/g | [186] |
| Putrescine in ethanol | Bulk polymerization of PVA1799, 1,4-butanediol. MIP deposited on NFM | Colorimetric | Not mentioned | Not mentioned | [187] | |
| Histidine | Human urine | AM films on Au electrode by electro-polymerization (cyclic voltammetry) | Amperometric | D-His: 0.01 µM L-His: 0.1 µM | D-His: 100 nM–10 µM L-His: 100 nM–10 µM | [213] |
| Spiked water and serum samples | Bulk polymerization of TEOS bound to Fe3O4 particles | Potentiometric | 0.01 µM | 0.01 µM–1 µM 30 µM–70 µM | [214] |
| Recommend Sample Type | Name of Test | Concentration Range | Recognition Element | Time to Result | Manufacturer |
|---|---|---|---|---|---|
| Seafood, wine, juice, milk, dairy | QuantiQuik™ | 20 ppm–200 ppm | Enzyme: Histamine dehydrogenase | 15 min | BioAssay Systems, Hayward, CA, USA |
| Fish products (fresh, frozen, canned marine products) | HistaSure™ Fish Rapid Test | 10 ppm–200 ppm | Immunogold labeled histamine antibody | 5 min | BioSystems S.A., Barcelona, Spain |
| Seafood, fish sauce, fish meal, wine, milk | HistaStrip™ | 15 ppm–75 ppm | Histamine-specific enzyme | 4 min | PerkinElmer Inc., Waltham, MA, USA |
| Fresh, canned and salted fish, fish in oil, fish meals | AgraQuant® | 0.5 ppb–50 ppb | Antibody, competitive ELISA | not given | Romer Labs, Tulln an der Donau, Austria |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Givanoudi, S.; Heyndrickx, M.; Depuydt, T.; Khorshid, M.; Robbens, J.; Wagner, P. A Review on Bio- and Chemosensors for the Detection of Biogenic Amines in Food Safety Applications: The Status in 2022. Sensors 2023, 23, 613. https://doi.org/10.3390/s23020613
Givanoudi S, Heyndrickx M, Depuydt T, Khorshid M, Robbens J, Wagner P. A Review on Bio- and Chemosensors for the Detection of Biogenic Amines in Food Safety Applications: The Status in 2022. Sensors. 2023; 23(2):613. https://doi.org/10.3390/s23020613
Chicago/Turabian StyleGivanoudi, Stella, Marc Heyndrickx, Tom Depuydt, Mehran Khorshid, Johan Robbens, and Patrick Wagner. 2023. "A Review on Bio- and Chemosensors for the Detection of Biogenic Amines in Food Safety Applications: The Status in 2022" Sensors 23, no. 2: 613. https://doi.org/10.3390/s23020613
APA StyleGivanoudi, S., Heyndrickx, M., Depuydt, T., Khorshid, M., Robbens, J., & Wagner, P. (2023). A Review on Bio- and Chemosensors for the Detection of Biogenic Amines in Food Safety Applications: The Status in 2022. Sensors, 23(2), 613. https://doi.org/10.3390/s23020613









