Building Flexible, Scalable, and Machine Learning-Ready Multimodal Oncology Datasets
Abstract
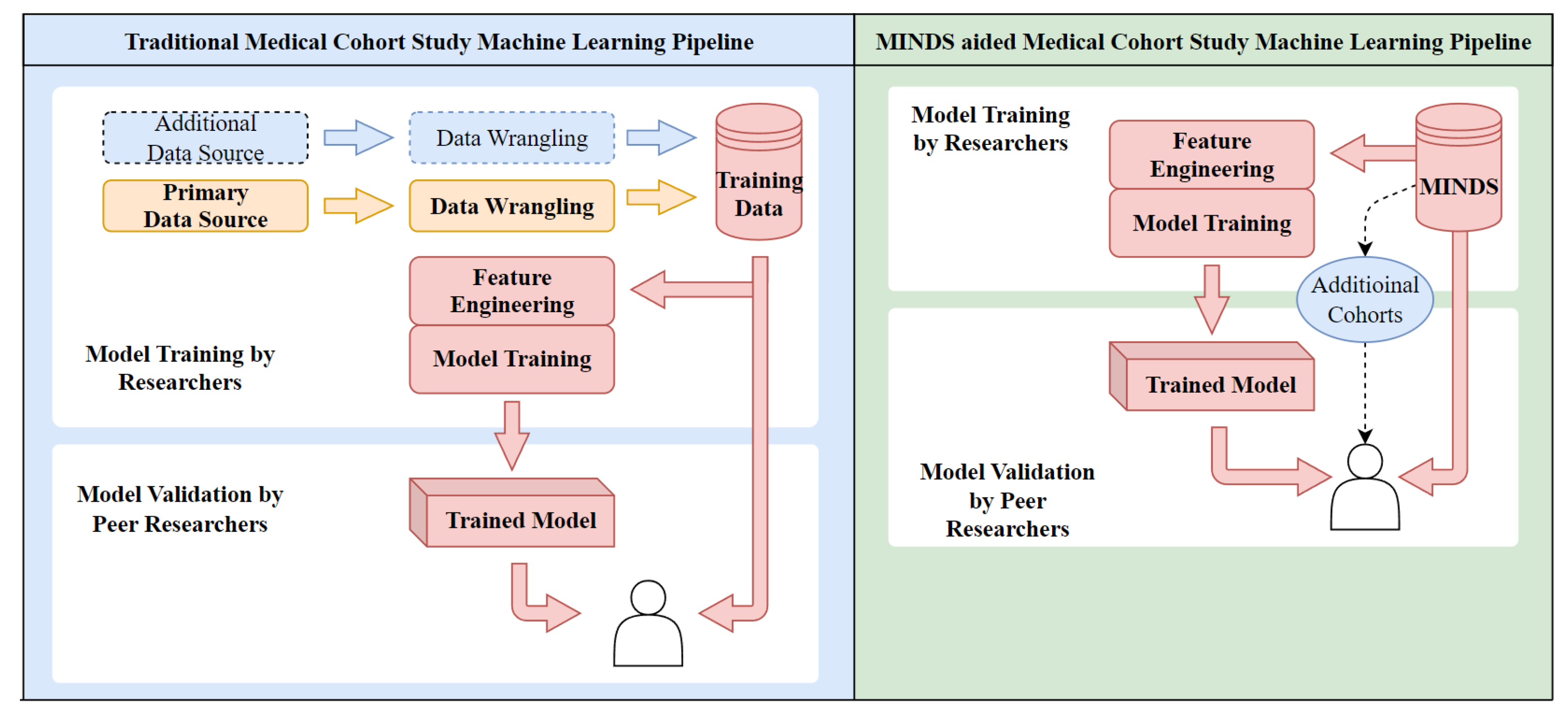
:1. Introduction
Contributions of MINDS
- 1.
- Integrating siloed multimodal data into a unified access point: By consolidating dispersed datasets across repositories and modalities, MINDS delivers a single unified interface for accessing integrated data. This overcomes fragmentation across disconnected silos.
- 2.
- Implementing robust data security and access control while supporting reproducibility: Strict access policies and controls safeguard sensitive data while enabling reproducibility via dataset versioning tied to cohort definitions.
- 3.
- An automated system to accommodate new data continually: Automated pipelines ingest updates and additions, ensuring analysts always have access to the latest data.
- 4.
- Enabling efficient, scalable multimodal machine learning: Cloud-based storage and compute scale elastically to handle growing data volumes while optimized warehousing delivers high-performance model training.
- The unprecedented scale of heterogeneous data consolidation enables new analysis paradigms. The cohort diversity in MINDS also surpasses existing systems.
- Tight integration between cohort definition and on-demand multimodal data assembly, not offered in current platforms.
- An industrial-strength cloud-native architecture delivers advanced translational informatics over a browser.
- Support for reproducibility via dataset versioning based on user cohort queries. This allows regenerating the same data even with newer updates.
- Option to build vector databases capturing data embeddings instead of actual data. This eliminates storage needs while ensuring patient privacy.
2. Background and Literature Review
2.1. Data Characterization Pipeline
- Tissue Collection and Processing: Tissue source sites, which include clinical trials and community oncology groups, collect tumor tissue samples and normal tissue from participating patients. These samples are either formalin-fixed paraffin-embedded (FFPE) tissues or frozen tissue. In CCG, Biospecimen Core Resource (BCR) is responsible for collecting and processing these samples and collecting, harmonizing, and curating clinical data [21].
- Genome Characterization: This stage involves generating data from the collected samples. At CCG, the Genome Characterization Centers (GCCs) generate data from the samples received from the BCR. Each GCC supports distinct genomic or epigenomic pipelines, including whole genome sequencing, total RNA and microRNA sequencing, methylation arrays, and single-cell sequencing [21].
- Genomic Data Analysis: The raw data from the previous stage is then transformed into meaningful biological information at this stage. In CCG, the Genomic Data Analysis Network (GDAN) transforms the raw data output from the GCCs into biological insights. The GDAN has a wide range of expertise, from identifying genomic abnormalities to integrating and visualizing multi-omics data [21].
- Data Sharing and Discovery: At this stage, the insightful genomic data is processed, shared, and unified at a central location. The NCI’s Genomic Data Commons (GDC) harmonizes genomic data by applying a standardized set of data processing protocols and bioinformatic pipelines. The data generated by the Genome Characterization Pipeline are made available to the public via the GDC [21,22].
2.2. Traditional Data Management—BioBanks
- Fragmented Data: One of the main issues is that data from different sources are often stored in separate biobanks, leading to fragmentation of information [34]. This makes integrating and analyzing data across different modalities difficult, limiting the potential for comprehensive, multi-dimensional analysis of patient data [33].
- Incoherent Data Management: How data is stored, formatted, and organized often varies significantly across biobanks, even for the same patient. For example, clinical data may be encoded differently, imaging data may use proprietary formats, and terminology can differ across systems. This heterogeneity and lack of unified standards make aggregating and analyzing data across multiple biobanks challenging [33].
- Data Synchronization: Over time, patient data stored in separate biobanks tends to go out of sync as patients undergo new tests and treatments, adding new data to different silos uncoordinatedly [33]. Piecing together a patient’s history timeline requires extensive manual effort to sync disparate records across systems [33].
- Data Governance: The increasing prevalence of bio-banking has sparked an extensive discussion regarding the ethical, legal, and social implications (ELSI) of utilizing vast quantities of human biological samples and their associated personal data [35]. Ensuring and safeguarding the fundamental ethical and legal principles concerning research involving human data in Biobanks becomes significantly more intricate and challenging than conducting ethical reviews for specific research projects [35].
2.3. Data Commons
- CDA’s mapping of the CRDC data is not real-time. For example, as of September of 2023, when querying patients with the primary diagnosis site being lung, only 4870 cases are present, despite there being 12,267 cases present in the GDC data portal. MINDS pulls source data directly from repositories like GDC to ensure real-time, up-to-date mapping of all cases.
- MINDS is designed as an end-to-end platform for users to build integrated multimodal datasets themselves rather than a fixed service. The open methodology enables full replication of huge multi-source datasets. To this end, anyone can replicate our method to generate the exact copy of over 40,000 public case data on their infrastructure.
- MINDS is flexible and incorporates diverse repositories and data sources, not just CRDC resources. Our proposed architecture can integrate new repositories as needed, unlike CDA, which is constrained to CRDC-managed data. For example, the cBioPortal for Cancer Genomics, a widely used platform for exploring, visualizing, and analyzing cancer genomics data, has its own data management and storage system separate from the CDA [37,38]. The data stored in cBioPortal cannot be directly queried or accessed through CDA, limiting the potential for integrated data analysis across platforms.
2.4. The “Big Data” Approach
2.5. Summary of Gaps in Existing Methods
- Prior consolidation is limited to structured data: Most prior efforts, like CDA, focused on consolidating structured clinical records. Support for aggregating unstructured imaging, -omics, and pathology data is lacking.
- Query interfaces have limited standardization: Different repositories have proprietary APIs and schemas. Unified interfaces for federated querying are needed.
- Scalability is constrained for large data: On-premises systems restrict scaling storage and compute for exponentially growing heterogeneous data.
- Minimal reproducibility without versioning: Dynamic dataset extracts make precise tracking of model data versions difficult, hampering reproducibility.
- Coarse-grained access controls: Most systems have limited options for fine-grained data access policies tailored to users.
3. Methodology
3.1. Requirements of a Flexible and Scalable Data Management System
- Requirement 1: Minimize large-scale unstructured data storage whenever possible. This requirement ensures the efficient use of storage resources and allows the user to access the data directly from the data provider.
- Requirement 2: The system should be horizontally and vertically scalable. Satisfying this requirement is crucial to handle the increasing volume of oncology data and ensure the system can accommodate data size and complexity growth.
- Requirement 3: The system should be interoperable, allowing for the easy integration of new data sources. This is important in oncology, where data is often distributed across various databases and systems.
- Requirement 4: The system should track data from the point of ingestion to the point of training. This ensures reproducibility, a key requirement in scientific research and machine learning.
- Requirement 5: Incorporate audit checkpoints in the data collection, pre-processing, storage, processing, and analysis stages of the data pipeline. This ensures data integrity, the prime consideration in delivering reliable machine learning outcomes.
3.2. MINDS Architecture
| Amazon S3 Ingest Bucket | Object storage bucket for staging raw data before loading into a data lake. |
| Amazon Web Services (AWS) | A cloud platform that provides scalable computing, storage, analytics, and machine learning services. |
| AWS Athena | Serverless interactive query service to analyze data in Amazon S3 using standard SQL. |
| AWS Big Data Analytics | Suite of services for processing and analyzing big data across storage, compute, and databases. |
| AWS Data Lake Formation | Service to set up and manage data lakes with indexing, security, and data governance. |
| AWS Data Warehouse | Fully-managed data warehousing service for analytics using standard SQL. |
| AWS Glue Crawler | Discovers data via classifiers and populates the AWS Glue Data Catalog. |
| AWS Glue Data Catalog | Central metadata store on AWS for datasets, schemas, and mappings. |
| AWS Lambda | Serverless compute to run code without managing infrastructure. |
| AWS QuickSight | Business intelligence service for easy visualizations and dashboards. |
| AWS RDS | Amazon Relational Database Service is a managed relational database service that handles database administration tasks like backup, patching, failure detection, and recovery. Including RDS MySQL, a managed relational database optimized for online transaction processing. |
| AWS Redshift | Petabyte-scale data warehouse for analytics and business intelligence. |
| JDBC | JDBC (Java Database Connectivity) is a standard API for connecting to traditional relational databases from Java. The JDBC was released as part of the Java Development Kit (JDK) in version 1.1 in 1997 and has since been part of every Java edition. |
3.2.1. Stage-1: Data Acquisition
3.2.2. Stage 2: Data Processing
- Fast Healthcare Interoperability Resources (FHIR): Specifies RESTful APIs, schemas, profiles, and formats for exchanging clinical, genomic, imaging, and other healthcare data. Offers platform-agnostic interconnection [43].
- Clinical Data Interchange Standards Consortium (CDISC): Develops data models, terminologies, and protocols focused specifically on clinical research and FDA submissions, including the Study Data Tabulation Model (SDTM) and the Clinical Data Acquisition Standards Harmonization (CDASH) [44].
- Systematized Nomenclature of MEDicine Clinical Terms (SNOMED CT): Provides consistent clinical terminology and codes for electronic health records. Enables semantic interoperability [46].
- National Cancer Institute (NCI) Thesaurus: Models cancer research domain semantics with 33 distinct hierarchies and 54,000 classes/properties. Binds related concepts for knowledge discovery [47].
3.2.3. Stage 3: Data Serving
3.3. Cloud Deployment
3.3.1. MINDS Infrastructure on AWS
- 1.
- Establish access-controlled database connections: The crawler first establishes secure connections to the databases from which data needs to be extracted. This ensures the safety and integrity of the data in transit.
- 2.
- Use custom classifiers: If any custom classifiers are defined, they catalog the data lake and generate the necessary metadata. These classifiers help in identifying the type and structure of the data.
- 3.
- Use built-in classifiers for ETL: AWS’s built-in classifiers perform ETL tasks for the rest of the data. This process involves extracting data from the source, transforming it into a more suitable format, and loading it into the data warehouse.
- 4.
- Merge catalog tables into a database: The catalog tables created from the previous steps are merged into a single database. During this process, any conflicts in the data are resolved to ensure consistency and deduplication.
- 5.
- Upload catalog to a data store: Finally, the catalog is uploaded to a data store to be accessed and utilized for analytics. This data store is a central repository where all the processed and cataloged data is stored.
3.3.2. Benefits of Cloud as a PaaS Platform
3.3.3. Scalability across Different Platforms
- Employing Cloud Data Fusion for data integration in place of AWS Glue
- Leveraging BigQuery for data warehousing rather than Redshift
- Using Cloud SQL over RDS for relational data
- Adopting Cloud Functions and Cloud Run for serverless compute instead of Lambda.
- Visualizing with Looker as an alternative to QuickSight
- Applying Cloud Data Loss Prevention for security rather than AWS options
3.4. On-Premise Deployment
3.5. User Application
4. Results and Discussion
4.1. Multimodal Data Consolidation
4.2. Cohort Building
- MINDS allows researchers to build cohorts tailored to the problem. This prevents sampling biases linked to the availability of pre-defined cohorts.
- SQL combines and consolidates disparate clinical, molecular, and outcomes data from the entire period of medical treatment. This provides a complete view of each patient.
- Version IDs uniquely label dataset variants to enable precise tracking of changes during iterative model development. Researchers can pinpoint the exact dataset used to generate each model version.
- JSON manifests comprehensively log the dataset composition, including the originating queries, data sources, and extraction workflows. This provides full documentation of the data provenance.
4.3. Data Standards
- Fast Healthcare Interoperability Resources (FHIR): Specifies RESTful APIs, schemas, profiles, and formats for exchanging clinical, genomic, imaging, and other healthcare data. Offers platform-agnostic interconnection.
- Clinical Data Interchange Standards Consortium (CDISC): Develops data models, terminologies, and protocols focused specifically on clinical research and FDA submissions, including the Study Data Tabulation Model (SDTM) and the Clinical Data Acquisition Standards Harmonization (CDASH).
- Health Level 7 (HL7): Defines structures and semantics for messaging healthcare data between computer systems, including Clinical Document Architecture (CDA) and Fast Healthcare Interoperable Resources (FHIR) specifications.
- Systematized Nomenclature of MEDicine Clinical Terms (SNOMED CT): Provides consistent clinical terminology and codes for EHR. Enables semantic interoperability.
- National Cancer Institute (NCI) Thesaurus: Models cancer research domain semantics with 33 distinct hierarchies and 54,000 classes/properties. Binds related concepts for knowledge discovery.
4.4. Data Tracking and Reproducibility
4.5. Integrated Analytics
- Rapid hypothesis testing during exploratory analysis to refine cohorts and features.
- Understanding model performance across cohorts reveals generalization capabilities.
- Uncovering correlations between clinical factors, assays, and predictions guides feature engineering.
- Visualizations build trust by providing direct views into model behaviors.
4.6. Limitations and Future Improvements
- Incorporating regulated data through privacy-preserving methods
- Migrating imagery via compact embeddings
- Absorbing unstructured notes through advanced NLP
- Expanding across diseases by reusing consolidation components
- Scaling across cloud platforms to prevent vendor lock-in
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application Programming Interface |
| AWS | Amazon Web Services |
| CDA | Cancer Data Aggregator |
| CRDC | Cancer Research Data Commons |
| CSV | Comma-Separated Values |
| ETL | Extract, Transform, Load |
| GDC | Genomic Data Commons |
| IAM | Identity and Access Management |
| IDC | Imaging Data Commons |
| JSON | JavaScript Object Notation |
| KMS | Key Management Service |
| MINDS | Multimodal Integration of Oncology Data System |
| PDC | Proteomics Data Commons |
| RDS | Relational Database Service |
| S3 | Amazon Simple Storage Service |
| SQL | Structured Query Language |
| TSV | Tab-Separated Values |
References
- Boehm, K.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer 2021, 22, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Waqas, A.; Dera, D.; Rasool, G.; Bouaynaya, N.C.; Fathallah-Shaykh, H.M. Brain Tumor Segmentation and Surveillance with Deep Artificial Neural Networks. In Deep Learning for Biomedical Data Analysis; Springer: Cham, Switzerland, 2021; pp. 311–350. [Google Scholar]
- Ektefaie, Y.; Dasoulas, G.; Noori, A.; Farhat, M.; Zitnik, M. Multimodal learning with graphs. Nat. Mach. Intell. 2023, 5, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Lipkova, J.; Chen, R.J.; Chen, B.; Lu, M.Y.; Barbieri, M.; Shao, D.; Vaidya, A.J.; Chen, C.; Zhuang, L.; Williamson, D.F.; et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 2022, 40, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Waqas, A.; Tripathi, A.; Ramachandran, R.P.; Stewart, P.; Rasool, G. Multimodal Data Integration for Oncology in the Era of Deep Neural Networks: A Review. arXiv 2023, arXiv:2303.06471. Available online: https://arxiv.org/abs/2303.06471 (accessed on 30 January 2024).
- El Naqa, I.; Rollison, D. Moffitt Cancer Center: Why we are building the first machine learning department in oncology. Cancer Lett. 2021, 47, 5–10. [Google Scholar]
- Andreu-Perez, J.; Poon, C.C.Y.; Merrifield, R.D.; Wong, S.T.C.; Yang, G.Z. Big Data for Health. IEEE J. Biomed. Health Inform. 2015, 19, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Fessele, K.L. The Rise of Big Data in Oncology. Semin. Oncol. Nurs. 2018, 34, 168–176. [Google Scholar] [CrossRef]
- Xu, P.; Zhu, X.; Clifton, D.A. Multimodal Learning with Transformers: A Survey. arXiv 2023, arXiv:2206.06488. [Google Scholar] [CrossRef]
- Waqas, A.; Bui, M.M.; Glassy, E.F.; El Naqa, I.; Borkowski, P.; Borkowski, A.A.; Rasool, G. Revolutionizing Digital Pathology with the Power of Generative Artificial Intelligence and Foundation Models. Lab. Investig. 2023, 103, 100255. [Google Scholar] [CrossRef]
- Common Crawl. 2023. Available online: https://commoncrawl.org/ (accessed on 18 September 2023).
- Bote-Curiel, L.; Muñoz-Romero, S.; Gerrero-Curieses, A.; Rojo-Álvarez, J.L. Deep Learning and Big Data in Healthcare: A Double Review for Critical Beginners. Appl. Sci. 2019, 9, 2331. [Google Scholar] [CrossRef]
- Khan, M.A.; Karim, M.R.; Kim, Y. A Two-Stage Big Data Analytics Framework with Real World Applications Using Spark Machine Learning and Long Short-Term Memory Network. Symmetry 2018, 10, 485. [Google Scholar] [CrossRef]
- Ahmed, S.; Dera, D.; Hassan, S.U.; Bouaynaya, N.; Rasool, G. Failure detection in deep neural networks for medical imaging. Front. Med. Technol. 2022, 4, 919046. [Google Scholar] [CrossRef]
- Dera, D.; Ahmed, S.; Bouaynaya, N.C.; Rasool, G. TRustworthy Uncertainty Propagation for Sequential Time-Series Analysis in RNNs. IEEE Trans. Knowl. Data Eng. 2023, 36, 882–896. [Google Scholar] [CrossRef]
- Waqas, A.; Farooq, H.; Bouaynaya, N.C.; Rasool, G. Exploring Robust Architectures for Deep Artificial Neural Networks. Commun. Eng. 2022, 1, 46. [Google Scholar] [CrossRef]
- Benedum, C.M.; Sondhi, A.; Fidyk, E.; Cohen, A.B.; Nemeth, S.; Adamson, B.; Estévez, M.; Bozkurt, S. Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning. Cancers 2023, 15, 1853. [Google Scholar] [CrossRef]
- Specht, D.S.; Waqas, A.; Rasool, G.; Clifford, C.; Bouaynaya, N. Intelligent Helipad Detection and (Grad-Cam) Estimation Using Satellite Imagery. Transp. Res. Board 2021, TRBAM-21-01973. Available online: https://annualmeeting.mytrb.org/OnlineProgram/Details/15715 (accessed on 25 January 2024).
- Congress, U.S. Health Insurance Portability and Accountability Act of 1996. 1996. Available online: https://www.govinfo.gov/content/pkg/PLAW-104publ191/pdf/PLAW-104publ191.pdf (accessed on 1 December 2023).
- Oh, S.R.; Seo, Y.D.; Lee, E.; Kim, Y.G. A comprehensive survey on security and privacy for electronic health data. Int. J. Environ. Res. Public Health 2021, 18, 9668. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. CCG’s Genome Characterization Pipeline. Available online: https://www.cancer.gov/ccg/research/genome-characterization-pipeline (accessed on 18 June 2023).
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Hinkson, I.V.; Davidsen, T.M.; Klemm, J.D.; Chandramouliswaran, I.; Kerlavage, A.R.; Kibbe, W.A. A Comprehensive Infrastructure for Big Data in Cancer Research: Accelerating Cancer Research and Precision Medicine. Front. Cell Dev. Biol. 2017, 5, 83. [Google Scholar] [CrossRef]
- Vesteghem, C.; Brøndum, R.F.; Sønderkær, M.; Sommer, M.; Schmitz, A.; Bødker, J.S.; Dybkær, K.; El-Galaly, T.C.; Bøgsted, M. Implementing the FAIR Data Principles in precision oncology: Review of supporting initiatives. Brief. Bioinform. 2020, 21, 936–945. [Google Scholar] [CrossRef]
- Kuhn, K. The cancer biomedical informatics grid (caBIG™): Infrastructure and applications for a worldwide research community. Medinfo 2007, 1, 330. [Google Scholar]
- Scheufele, E.; Aronzon, D.; Coopersmith, R.; McDuffie, M.T.; Kapoor, M.; Uhrich, C.A.; Avitabile, J.E.; Liu, J.; Housman, D.; Palchuk, M.B. tranSMART: An open source knowledge management and high content data analytics platform. AMIA Summits Transl. Sci. Proc. 2014, 2014, 96. [Google Scholar]
- Murphy, S.N.; Weber, G.; Mendis, M.; Gainer, V.; Chueh, H.C.; Churchill, S.; Kohane, I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J. Am. Med. Inform. Assoc. 2010, 17, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Messiou, C.; Lee, R.; Salto-Tellez, M. Multimodal analysis and the oncology patient: Creating a hospital system for integrated diagnostics and discovery. Comput. Struct. Biotechnol. J. 2023, 21, 4536–4539. [Google Scholar] [CrossRef] [PubMed]
- Santaolalla, A.; Hulsen, T.; Davis, J.; Ahmed, H.U.; Moore, C.M.; Punwani, S.; Attard, G.; McCartan, N.; Emberton, M.; Coolen, A.; et al. The ReIMAGINE multimodal warehouse: Using artificial intelligence for accurate risk stratification of prostate cancer. Front. Artif. Intell. 2021, 4, 769582. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Longabaugh, W.; Pot, D.; Clunie, D.; Pieper, S.; Lewis, R.; Aerts, H.; Homeyer, A.; Herrmann, M.; Wagner, U.; et al. NCI Imaging Data Commons. Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, e101. [Google Scholar] [CrossRef]
- Thangudu, R.R.; Rudnick, P.A.; Holck, M.; Singhal, D.; MacCoss, M.J.; Edwards, N.J.; Ketchum, K.A.; Kinsinger, C.R.; Kim, E.; Basu, A. Abstract LB-242: Proteomic Data Commons: A resource for proteogenomic analysis. Cancer Res. 2020, 80, LB-242. [Google Scholar] [CrossRef]
- Asiimwe, R.; Lam, S.; Leung, S.; Wang, S.; Wan, R.; Tinker, A.; McAlpine, J.N.; Woo, M.M.; Huntsman, D.G.; Talhouk, A. From biobank and data silos into a data commons: Convergence to support translational medicine. J. Transl. Med. 2021, 19, 493. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Prabakaran, S. Big data in digital healthcare: Lessons learnt and recommendations for general practice. Heredity 2020, 124, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Lecaros, J.A. Biobanks for Biomedical Research: Evolution and Future. In Handbook of Bioethical Decisions. Volume I: Decisions at the Bench; Springer: Cham, Switzerland, 2023; pp. 295–323. [Google Scholar]
- Cancer Data Aggregator. Available online: https://datacommons.cancer.gov/cancer-data-aggregator (accessed on 15 June 2023).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Willems, S.M.; Abeln, S.; Feenstra, K.A.; de Bree, R.; van der Poel, E.F.; Baatenburg de Jong, R.J.; Heringa, J.; van den Brekel, M.W. The potential use of big data in oncology. Oral Oncol. 2019, 98, 8–12. [Google Scholar] [CrossRef]
- Nambiar, A.; Mundra, D. An Overview of Data Warehouse and Data Lake in Modern Enterprise Data Management. Big Data Cogn. Comput. 2022, 6, 132. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Dolin, R.H.; Alschuler, L.; Beebe, C.; Biron, P.V.; Boyer, S.L.; Essin, D.; Kimber, E.; Lincoln, T.; Mattison, J.E. The HL7 clinical document architecture. J. Am. Med. Inform. Assoc. 2001, 8, 552–569. [Google Scholar] [CrossRef]
- HL7 FHIR. Available online: https://www.hl7.org/fhir/ (accessed on 1 December 2023).
- Clinical Data Interchange Standards Consortium. Available online: https://www.cdisc.org/ (accessed on 1 December 2023).
- Babre, D.K. Clinical data interchange standards consortium: A bridge to overcome data standardisation. Perspect. Clin. Res. 2013, 4, 115–116. [Google Scholar] [CrossRef]
- Overview of SNOMED CT. National Library of Medicine. Available online: https://www.nlm.nih.gov/healthit/snomedct/snomed_overview.html (accessed on 1 December 2023).
- NCI Thesaurus. Available online: https://ncit.nci.nih.gov/ncitbrowser/ (accessed on 1 December 2023).
- Amazon Web Services. Amazon QuickSight. Available online: https://aws.amazon.com/quicksight/ (accessed on 1 March 2023).
- Amazon Web Services. Amazon S3. Available online: https://aws.amazon.com/s3/ (accessed on 1 March 2023).
- Amazon Web Services. AWS Lake Formation. Available online: https://aws.amazon.com/lake-formation/ (accessed on 1 March 2023).
- Amazon Web Services. Data Catalog and Crawlers in AWS Glue. Available online: https://docs.aws.amazon.com/glue/latest/dg/catalog-and-crawler.html (accessed on 1 March 2023).
- Amazon Web Services. Serverless Computing—AWS Lambda—Amazon Web Services. Available online: https://aws.amazon.com/lambda/ (accessed on 7 August 2023).
- Amazon Web Services. AWS Glue. Available online: https://aws.amazon.com/glue/ (accessed on 1 March 2023).
- Amazon Web Services. Amazon Redshift. Available online: https://aws.amazon.com/redshift/ (accessed on 1 March 2023).
- Amazon Web Services. Amazon Athena. Available online: https://aws.amazon.com/athena/ (accessed on 1 March 2023).
- Amazon Web Services. Encryption at Rest. Available online: https://docs.aws.amazon.com/redshift/latest/mgmt/security-server-side-encryption.html (accessed on 7 August 2023).
- Amazon Web Services. Security in AWS Glue. Available online: https://docs.aws.amazon.com/glue/latest/dg/security.html (accessed on 7 August 2023).
- Amazon Web Services. Amazon CloudWatch. Available online: https://aws.amazon.com/cloudwatch/ (accessed on 7 August 2023).
- Medical Imaging and Data Resource Center (MIDRIC). Available online: https://www.midrc.org/ (accessed on 28 November 2023).








| Data Source | Storage Size | # of Cases |
|---|---|---|
| MINDS | 25.85 MB | 41,499 |
| PDC | 36 TB | 3081 |
| GDC | 3.78 PB (17.68 TB public) | 86,962 |
| IDC | 40.96 TB | 63,788 |
| Program | # of Cases |
|---|---|
| Foundation Medicine (FM) | 18,004 |
| The Cancer Genome Atlas (TCGA) | 11,315 |
| Therapeutically Applicable Research to Generate Effective Treatments (TARGET) | 6542 |
| Clinical Proteomic Tumor Analysis Consortium (CPTAC) | 1526 |
| Multiple Myeloma Research Foundation (MMRF) | 995 |
| BEATAML1.0 | 756 |
| NCI Center for Cancer Research (NCICCR) | 481 |
| REBC | 440 |
| Cancer Genome Characterization Initiatives (CGCI) | 371 |
| Count Me In (CMI) | 296 |
| Human Cancer Model Initiative (HCMI) | 228 |
| West Coast Prostrate Cancer Dream Team (WCDT) | 99 |
| Applied Proteogenomics OrganizationaL Learning and Outcomes (APOLLO) | 87 |
| EXCEPTIONAL RESPONDERS | 84 |
| Oregon Health and Science University (OHSU) | 80 |
| The Molecular Profiling to Predict Response to Treatment (MP2PRT) | 52 |
| Environment And Genetics in Lung Cancer Etiology (EAGLE) | 50 |
| ORGANOID | 49 |
| Clinical Trials Sequencing Project (CTSP) | 44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripathi, A.; Waqas, A.; Venkatesan, K.; Yilmaz, Y.; Rasool, G. Building Flexible, Scalable, and Machine Learning-Ready Multimodal Oncology Datasets. Sensors 2024, 24, 1634. https://doi.org/10.3390/s24051634
Tripathi A, Waqas A, Venkatesan K, Yilmaz Y, Rasool G. Building Flexible, Scalable, and Machine Learning-Ready Multimodal Oncology Datasets. Sensors. 2024; 24(5):1634. https://doi.org/10.3390/s24051634
Chicago/Turabian StyleTripathi, Aakash, Asim Waqas, Kavya Venkatesan, Yasin Yilmaz, and Ghulam Rasool. 2024. "Building Flexible, Scalable, and Machine Learning-Ready Multimodal Oncology Datasets" Sensors 24, no. 5: 1634. https://doi.org/10.3390/s24051634







