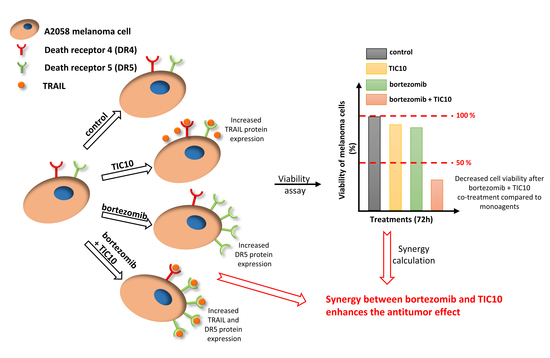
The Synergistic Activity of Bortezomib and TIC10 against A2058 Melanoma Cells
Abstract
:1. Introduction
2. Results
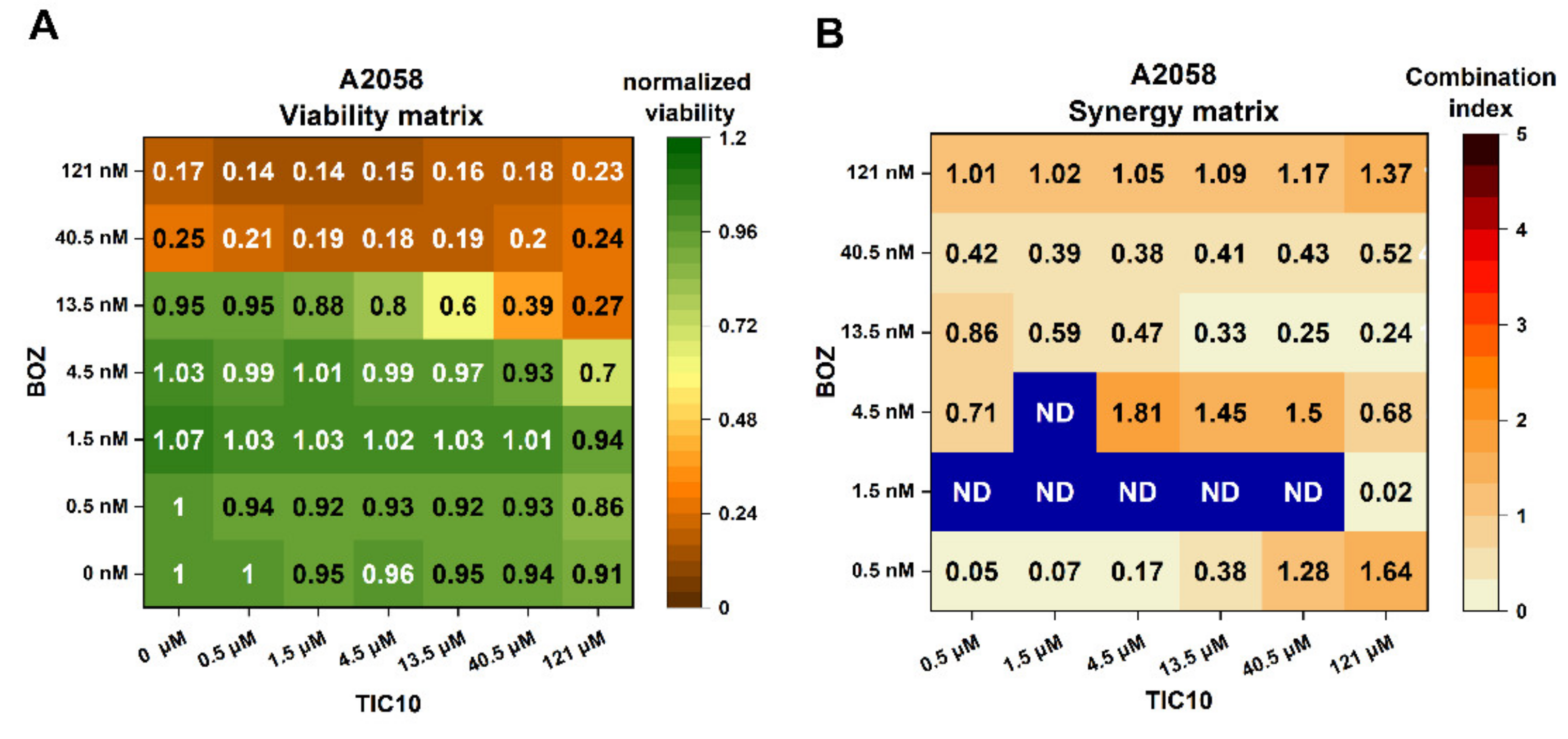
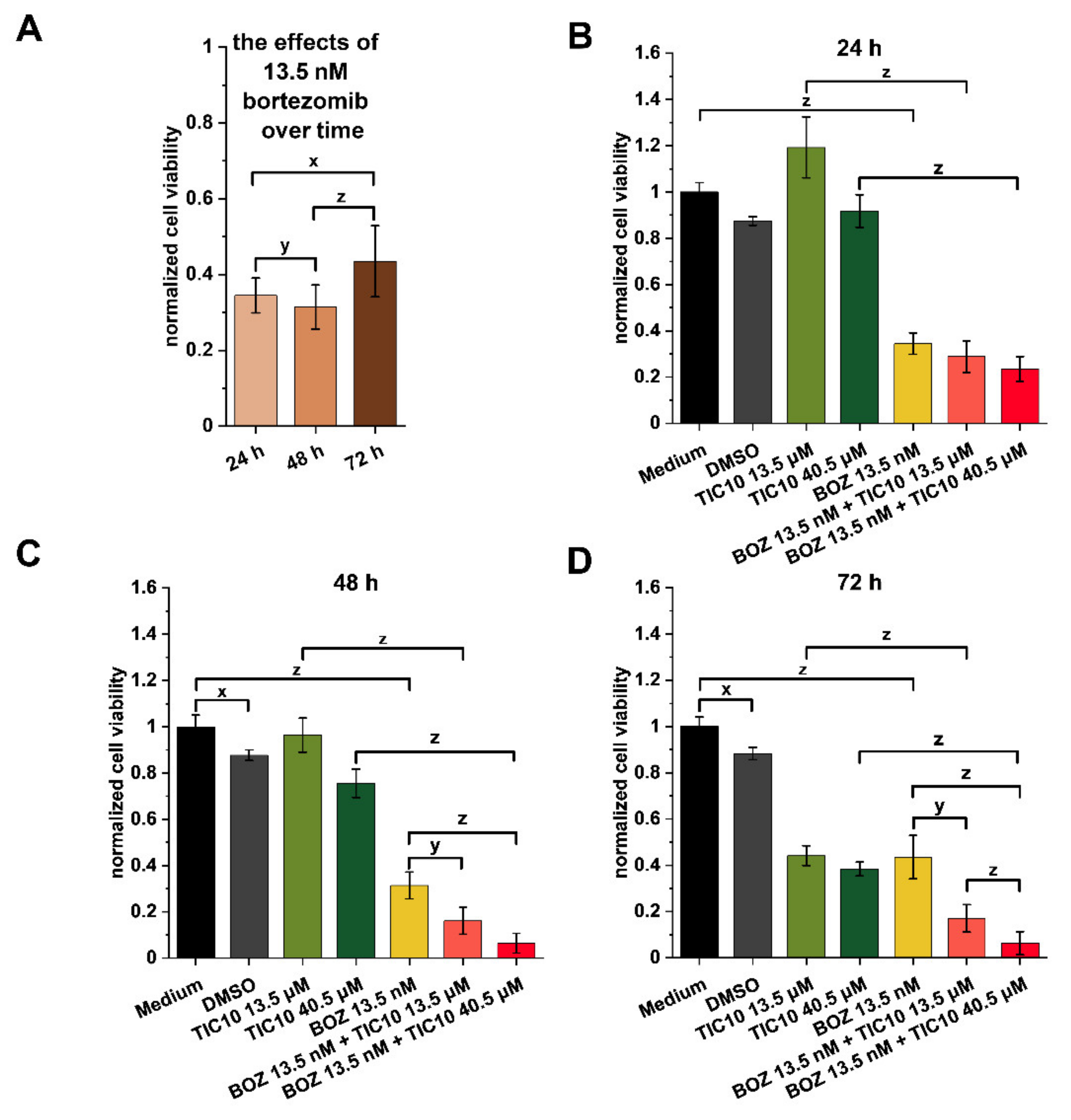
2.1. Influence of the Compounds on Cell Viability and Combination Index Assessments
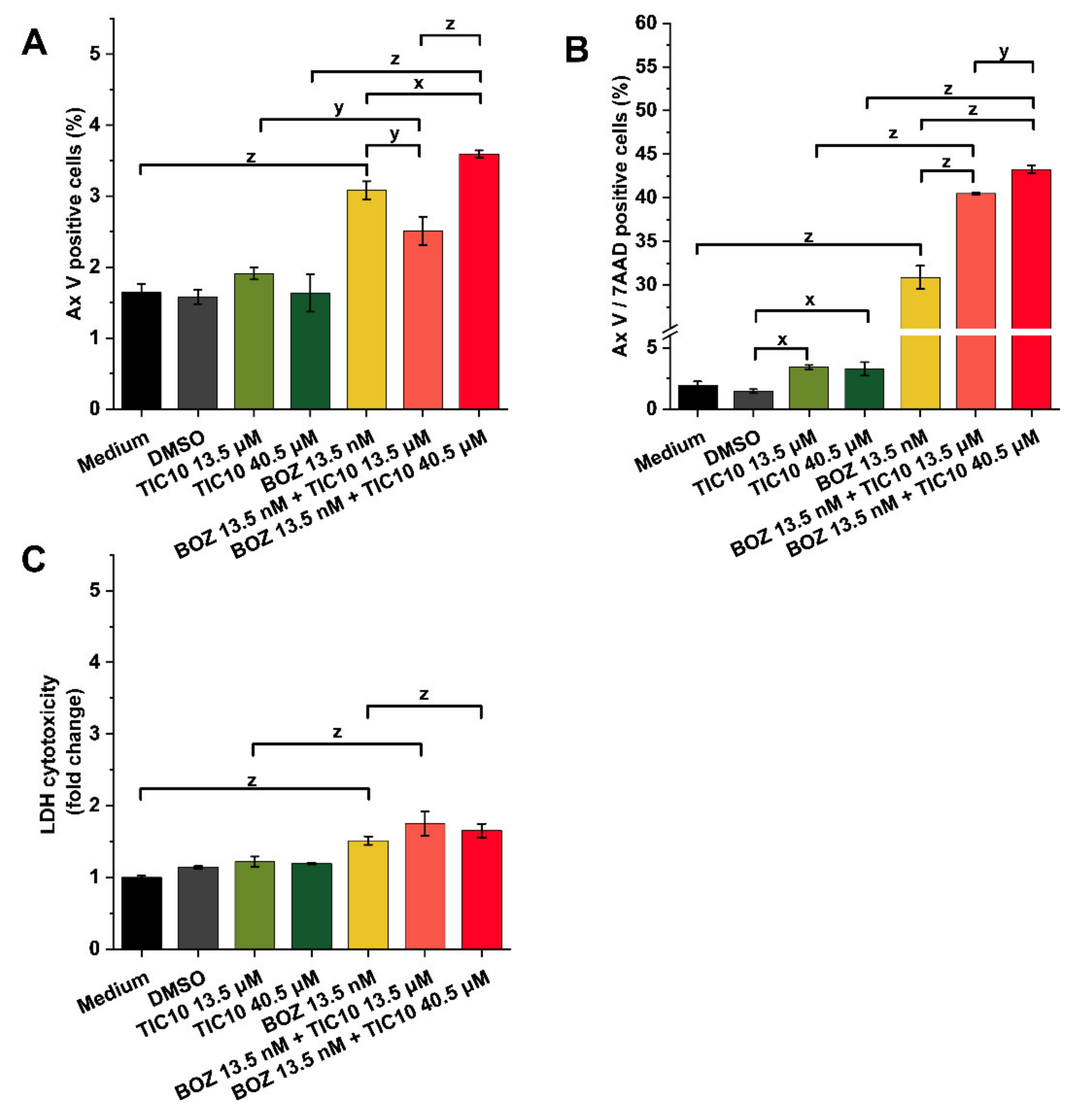
2.2. Effects on Cell Death and Cytotoxicity
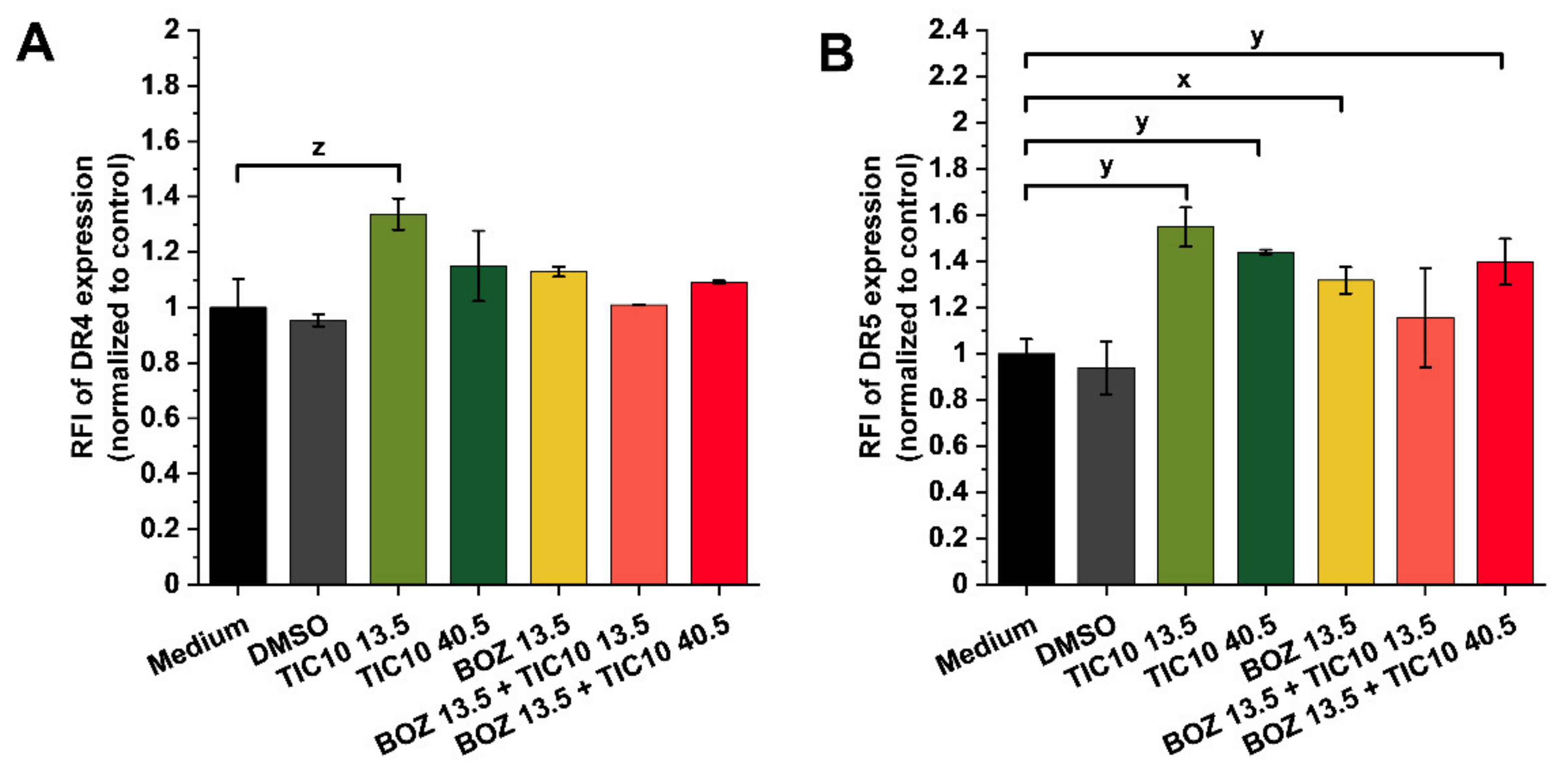
2.3. Association of Synergism with Death Receptor Expression at mRNA and Protein Level
3. Discussion
4. Materials and Methods
4.1. Substances
4.2. Cell Culturing
4.3. Endpoint Viability Assay
4.4. Synergy Quantification
4.5. Real-Time Viability Assay
4.6. Apoptosis Assay and Brightfield Images
4.7. Cytotoxicity Assay
4.8. RNA Isolation and Gene Expression Analysis of Death Receptors
4.9. Flow Cytometric Detection of Death Receptors
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 7-AAD | 7-Aminoactinomycin |
| ANOVA | one-way analysis of variance |
| Ax V | Annexin V |
| BOZ | bortezomib |
| CI | combination index |
| DISC | death-inducing signaling complex |
| DMSO | dimethyl sulfoxide |
| DR4 | death receptor 4 |
| DR5 | death receptor 5 |
| ECACC | European Collection of Authenticated Cell Cultures |
| FITC | fluorescein isothiocyanate |
| Fisher’s LSD | Fisher’s least significant difference |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| LDH | lactate dehydrogenase |
| MFI | mean fluorescence intensity |
| NTC | no template control |
| RNA | ribonucleic acid |
| PBS | phosphate buffer |
| PE | phycoerythrin |
| RFI | ratio of mean fluorescence intensity |
| RTCA | Real Time Cell Analyzer |
| SD | standard deviation |
| TIC10 | TRAIL-inducing compound |
| TNFRSF10A | TNF Receptor Superfamily Member 10a/death receptor 4 |
| TNFRSF10B | TNF Receptor Superfamily Member 10b/death receptor 5 |
| TRAIL | TNF-related apoptosis-inducing ligand |
| UV | ultraviolet |
References
- Pham, D.D.M.; Guhan, S.; Tsao, H. KIT and Melanoma: Biological Insights and Clinical Implications. Yonsei Med. J. 2020, 61, 562–571. [Google Scholar] [CrossRef]
- Bertrand, J.U.; Steingrimsson, E.; Jouenne, F.; Bressac-de Paillerets, B.; Larue, L. Melanoma Risk and Melanocyte Biology. Acta Derm. Venereol. 2020, 100, adv00139. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [Green Version]
- Rastrelli, M.; Tropea, S.; Rossi, C.R.; Alaibac, M. Melanoma: Epidemiology, Risk Factors, Pathogenesis, Diagnosis and Classification. In Vivo 2014, 28, 1005–1011. [Google Scholar] [PubMed]
- Simiczyjew, A.; Dratkiewicz, E.; Mazurkiewicz, J.; Ziętek, M.; Matkowski, R.; Nowak, D. The Influence of Tumor Microenvironment on Immune Escape of Melanoma. Int. J. Mol. Sci. 2020, 21, 8359. [Google Scholar] [CrossRef] [PubMed]
- Keilholz, U.; Ascierto, P.A.; Dummer, R.; Robert, C.; Lorigan, P.; van Akkooi, A.; Arance, A.; Blank, C.U.; Chiarion Sileni, V.; Donia, M.; et al. ESMO consensus conference recommendations on the management of metastatic melanoma: Under the auspices of the ESMO Guidelines Committee. Ann. Oncol. 2020, 31, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Kozar, I.; Margue, C.; Rothengatter, S.; Haan, C.; Kreis, S. Many ways to resistance: How melanoma cells evade targeted therapies. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Steininger, J.; Gellrich, F.F.; Schulz, A.; Westphal, D.; Beissert, S.; Meier, F. Systemic Therapy of Metastatic Melanoma: On the Road to Cure. Cancers 2021, 13, 1430. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Qasim, M.; Kim, J.H. Nanoparticle-Mediated Combination Therapy: Two-in-One Approach for Cancer. Int. J. Mol. Sci. 2018, 19, 3264. [Google Scholar] [CrossRef] [Green Version]
- Takács, A.; Lajkó, E.; Láng, O.; Istenes, I.; Kőhidai, L. Alpha-lipoic acid alters the antitumor effect of bortezomib in melanoma cells in vitro. Sci. Rep. 2020, 10, 14287. [Google Scholar] [CrossRef]
- Markowitz, J.; Luedke, E.A.; Grignol, V.P.; Hade, E.M.; Paul, B.K.; Mundy-Bosse, B.L.; Brooks, T.R.; Dao, T.V.; Kondalasula, S.V.; Lesinski, G.B.; et al. A phase I trial of bortezomib and interferon-α-2b in metastatic melanoma. J. Immunother. 2014, 37, 55–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starobova, H.; Vetter, I. Pathophysiology of Chemotherapy-Induced Peripheral Neuropathy. Front. Mol. Neurosci. 2017, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Bahleda, R.; Le Deley, M.C.; Bernard, A.; Chaturvedi, S.; Hanley, M.; Poterie, A.; Gazzah, A.; Varga, A.; Touat, M.; Deutsch, E.; et al. Phase I trial of bortezomib daily dose: Safety, pharmacokinetic profile, biological effects and early clinical evaluation in patients with advanced solid tumors. Investig. New Drugs 2018, 36, 619–628. [Google Scholar] [CrossRef]
- Bárány, P.; Oláh, R.S.; Kovács, I.; Czuczi, T.; Szabó, C.L.; Takács, A.; Lajkó, E.; Láng, O.; Kőhidai, L.; Schlosser, G.; et al. Ferrocene-Containing Impiridone (ONC201) Hybrids: Synthesis, DFT Modelling, In Vitro Evaluation, and Structure—Activity Relationships. Molecules 2018, 23, 2248. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Kho, D.; Xu, J.; Gajan, A.; Wu, K.; Wu, G.S. ONC201 activates ER stress to inhibit the growth of triple-negative breast cancer cells. Oncotarget 2017, 8, 21626–21638. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, V.V.; Morrow, S.; Rahman Kawakibi, A.; Zhou, L.; Ralff, M.; Ray, J.; Jhaveri, A.; Ferrarini, I.; Lee, Y.; Parker, C.; et al. ONC201 and imipridones: Anti-cancer compounds with clinical efficacy. Neoplasia 2020, 22, 725–744. [Google Scholar] [CrossRef]
- Karpel-Massler, G.; Siegelin, M.D. TIC10/ONC201-a potential therapeutic in glioblastoma. Transl. Cancer Res. 2017, 6, S1439–S1440. [Google Scholar] [CrossRef]
- Wang, S.; El-Deiry, W.S. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 2003, 22, 8628–8633. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.T.; Sun, S.Y. Regulation of Cancer Metastasis by TRAIL/Death Receptor Signaling. Biomolecules 2021, 11, 499. [Google Scholar] [CrossRef] [PubMed]
- Аrtykov, А.; Belov, D.A.; Shipunova, V.O.; Trushina, D.B.; Deyev, S.M.; Dolgikh, D.A.; Kirpichnikov, M.P.; Gasparian, M.E. Chemotherapeutic Agents Sensitize Resistant Cancer Cells to the DR5-Specific Variant DR5-B more Efficiently than to TRAIL by Modulating the Surface Expression of Death and Decoy Receptors. Cancers 2020, 12, 1129. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yue, P.; Chen, S.; Hu, L.; Lonial, S.; Khuri, F.R.; Sun, S.Y. The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res. 2007, 67, 4981–4988. [Google Scholar] [CrossRef] [Green Version]
- Greer, Y.E.; Lipkowitz, S. TIC10/ONC201: A bend in the road to clinical development. Oncoscience 2015, 2, 75–76. [Google Scholar] [CrossRef]
- Pruss, M.; Dwucet, A.; Tanriover, M.; Hlavac, M.; Kast, R.E.; Debatin, K.-M.; Wirtz, C.R.; Halatsch, M.-E.; Siegelin, M.D.; Westhoff, M.-A.; et al. Dual metabolic reprogramming by ONC201/TIC10 and 2-Deoxyglucose induces energy depletion and synergistic anti-cancer activity in glioblastoma. Br. J. Cancer 2020, 122, 1146–1157. [Google Scholar] [CrossRef] [Green Version]
- Kouroukis, T.C.; Baldassarre, F.G.; Haynes, A.E.; Imrie, K.; Reece, D.E.; Cheung, M.C. Bortezomib in multiple myeloma: Systematic review and clinical considerations. Curr. Oncol. 2014, 21, e573–e603. [Google Scholar] [CrossRef] [Green Version]
- Manasanch, E.E.; Orlowski, R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433. [Google Scholar] [CrossRef]
- Chou, T.C. Preclinical versus clinical drug combination studies. Leuk. Lymphoma 2008, 49, 2059–2080. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Thibeault, S. Effect of DMSO concentration, cell density and needle gauge on the viability of cryopreserved cells in three dimensional hyaluronan hydrogel. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; Volume 2013, pp. 6228–6231. [Google Scholar] [CrossRef] [Green Version]
- Yan, G.; Du, Q.; Wei, X.; Miozzi, J.; Kang, C.; Wang, J.; Han, X.; Pan, J.; Xie, H.; Chen, J.; et al. Application of Real-Time Cell Electronic Analysis System in Modern Pharmaceutical Evaluation and Analysis. Molecules 2018, 23, 3280. [Google Scholar] [CrossRef] [Green Version]
- Lü, S.; Wang, J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomark. Res. 2013, 1, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Politou, M.; Karadimitris, A.; Terpos, E.; Kotsianidis, I.; Apperley, J.F.; Rahemtulla, A. No evidence of mutations of the PSMB5 (beta-5 subunit of proteasome) in a case of myeloma with clinical resistance to Bortezomib. Leuk Res. 2006, 30, 240–241. [Google Scholar] [CrossRef] [PubMed]
- De Wilt, L.H.; Jansen, G.; Assaraf, Y.G.; van Meerloo, J.; Cloos, J.; Schimmer, A.D.; Chan, E.T.; Kirk, C.J.; Peters, G.J.; Kruyt, F.A. Proteasome-based mechanisms of intrinsic and acquired bortezomib resistance in non-small cell lung cancer. Biochem. Pharm. 2012, 83, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, V.V.; Talekar, M.K.; Lulla, A.R.; Kline, C.L.B.; Zhou, L.; Hall, J.; Van den Heuvel, A.P.J.; Dicker, D.T.; Babar, J.; Grupp, S.A.; et al. Single agent and synergistic combinatorial efficacy of first-in-class small molecule imipridone ONC201 in hematological malignancies. Cell Cycle 2018, 17, 468–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizawa, J.; Kojima, K.; Chachad, D.; Ruvolo, P.; Ruvolo, V.; Jacamo, R.O.; Borthakur, G.; Mu, H.; Zeng, Z.; Tabe, Y.; et al. ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci. Signal 2016, 9, ra17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, J.E.; Krigsfeld, G.; Mayes, P.A.; Patel, L.; Dicker, D.T.; Patel, A.S.; Dolloff, N.G.; Messaris, E.; Scata, K.A.; Wang, W.; et al. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci. Transl. Med. 2013, 5, 171ra117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, J.E.; Krigsfeld, G.; Patel, L.; Mayes, P.A.; Dicker, D.T.; Wu, G.S.; El-Deiry, W.S. Identification of TRAIL-inducing compounds highlights small molecule ONC201/TIC10 as a unique anti-cancer agent that activates the TRAIL pathway. Mol. Cancer 2015, 14, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpel-Massler, G.; Bâ, M.; Shu, C.; Halatsch, M.E.; Westhoff, M.A.; Bruce, J.N.; Canoll, P.; Siegelin, M.D. TIC10/ONC201 synergizes with Bcl-2/Bcl-xL inhibition in glioblastoma by suppression of Mcl-1 and its binding partners in vitro and in vivo. Oncotarget 2015, 6, 36456–36471. [Google Scholar] [CrossRef] [Green Version]
- Pan, G.; O‘Rourke, K.; Chinnaiyan, A.M.; Gentz, R.; Ebner, R.; Ni, J.; Dixit, V.M. The receptor for the cytotoxic ligand TRAIL. Science 1997, 276, 111–113. [Google Scholar] [CrossRef]
- Sträter, J.; Hinz, U.; Walczak, H.; Mechtersheimer, G.; Koretz, K.; Herfarth, C.; Möller, P.; Lehnert, T. Expression of TRAIL and TRAIL receptors in colon carcinoma: TRAIL-R1 is an independent prognostic parameter. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2002, 8, 3734–3740. [Google Scholar]
- Bychkov, M.L.; Gasparian, M.E.; Dolgikh, D.A.; Kirpichnikov, M.P. Combination of TRAIL with bortezomib shifted apoptotic signaling from DR4 to DR5 death receptor by selective internalization and degradation of DR4. PLoS ONE 2014, 9, e109756. [Google Scholar] [CrossRef]
- Chou, T.-C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the Lactate Dehydrogenase Assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef]
| Drug | A2058 | U266 |
|---|---|---|
| bortezomib | 23.07 ± 2.41 nM | 1.45 ± 0.06 nM |
| TIC10 | ND | 0.97 ± 0.26 μM |
| Treatment | DR4 | DR5 |
|---|---|---|
| DMSO | 0.49 ± 0.04 | 0.32 ± 0.04 |
| TIC10 13.5 μM | 0.08 ± 0.15 | 0.28 ± 0.15 |
| TIC10 40.5 μM | 0.07 ± 0.05 | 0.46 ± 0.08 |
| BOZ 13.5 nM | 1.20 ± 0.11 | 0.15 ± 0.11 |
| BOZ 13.5 nM + TIC10 13.5 μM | 1.39 ± 0.02 | 0.42 ± 0.02 |
| BOZ 13.5 nM + TIC10 40.5 μM | 1.56 ± 0.06 | 0.47 ± 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takács, A.; Szász, Z.; Kalabay, M.; Bárány, P.; Csámpai, A.; Hegyesi, H.; Láng, O.; Lajkó, E.; Kőhidai, L. The Synergistic Activity of Bortezomib and TIC10 against A2058 Melanoma Cells. Pharmaceuticals 2021, 14, 820. https://doi.org/10.3390/ph14080820
Takács A, Szász Z, Kalabay M, Bárány P, Csámpai A, Hegyesi H, Láng O, Lajkó E, Kőhidai L. The Synergistic Activity of Bortezomib and TIC10 against A2058 Melanoma Cells. Pharmaceuticals. 2021; 14(8):820. https://doi.org/10.3390/ph14080820
Chicago/Turabian StyleTakács, Angéla, Zsófia Szász, Márton Kalabay, Péter Bárány, Antal Csámpai, Hargita Hegyesi, Orsolya Láng, Eszter Lajkó, and László Kőhidai. 2021. "The Synergistic Activity of Bortezomib and TIC10 against A2058 Melanoma Cells" Pharmaceuticals 14, no. 8: 820. https://doi.org/10.3390/ph14080820