Eltrombopag as an Allosteric Inhibitor of the METTL3-14 Complex Affecting the m6A Methylation of RNA in Acute Myeloid Leukemia Cells
Abstract
:1. Introduction
2. Results and Discussion
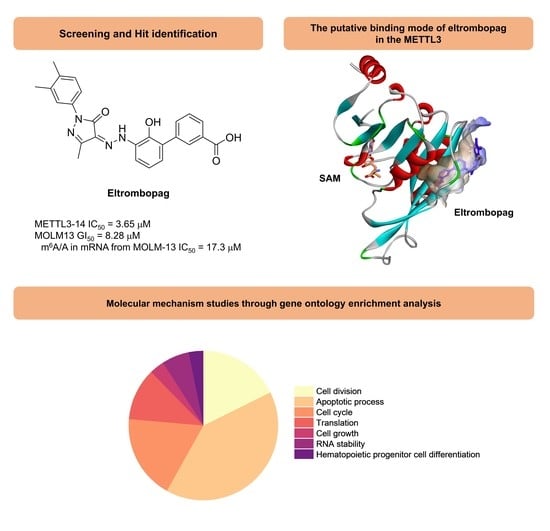
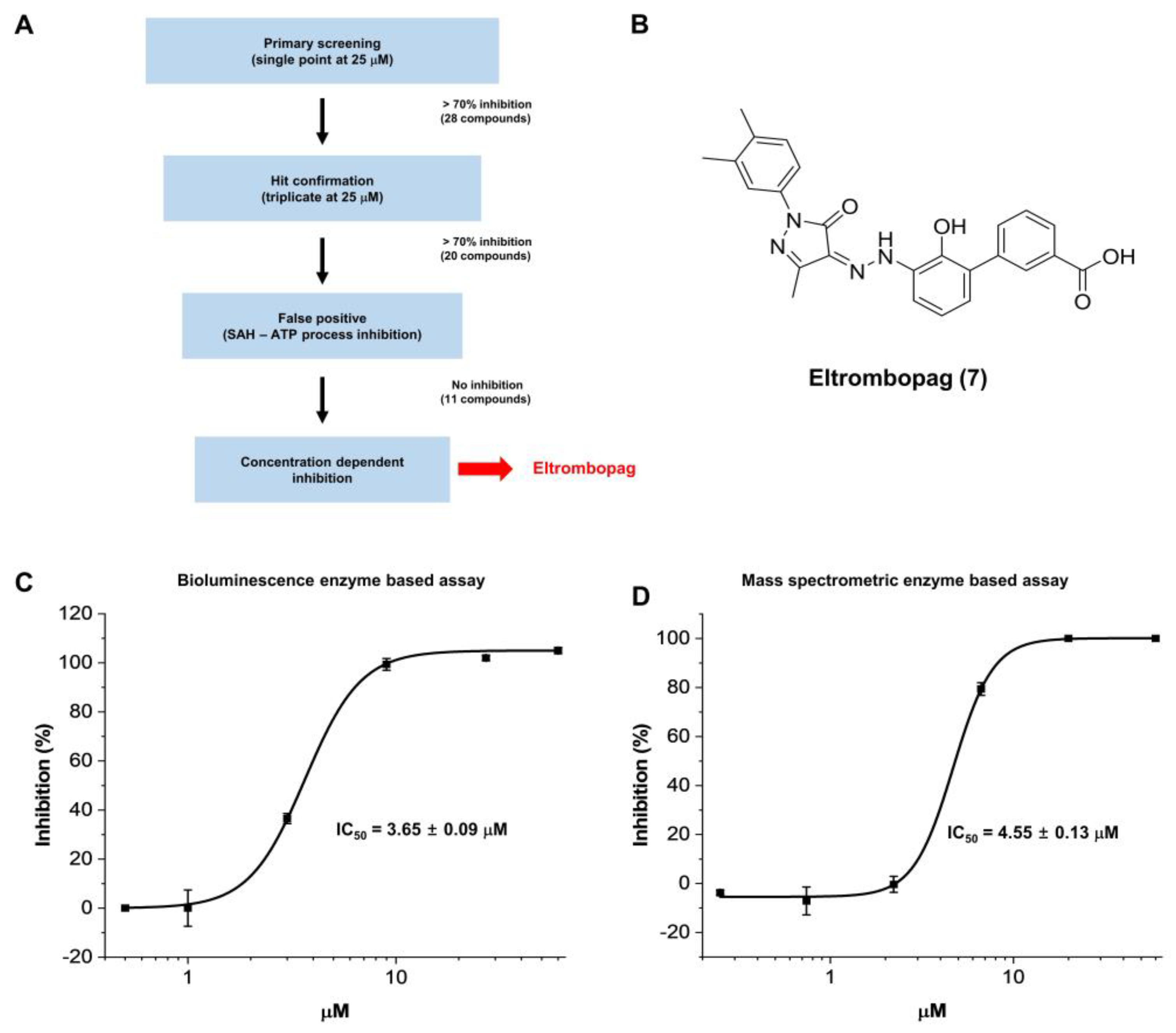
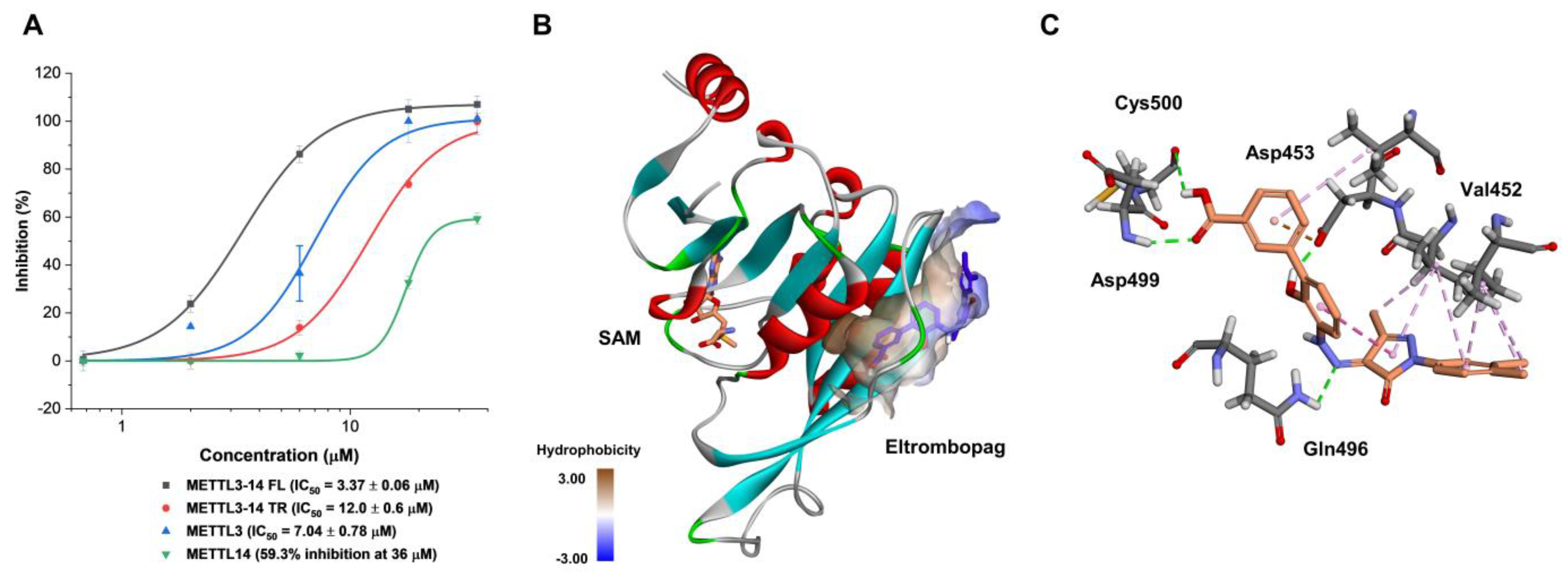
2.1. Enzyme Assay and Hit Identification of Eltrombopag as a METTL3-14 Inhibitor
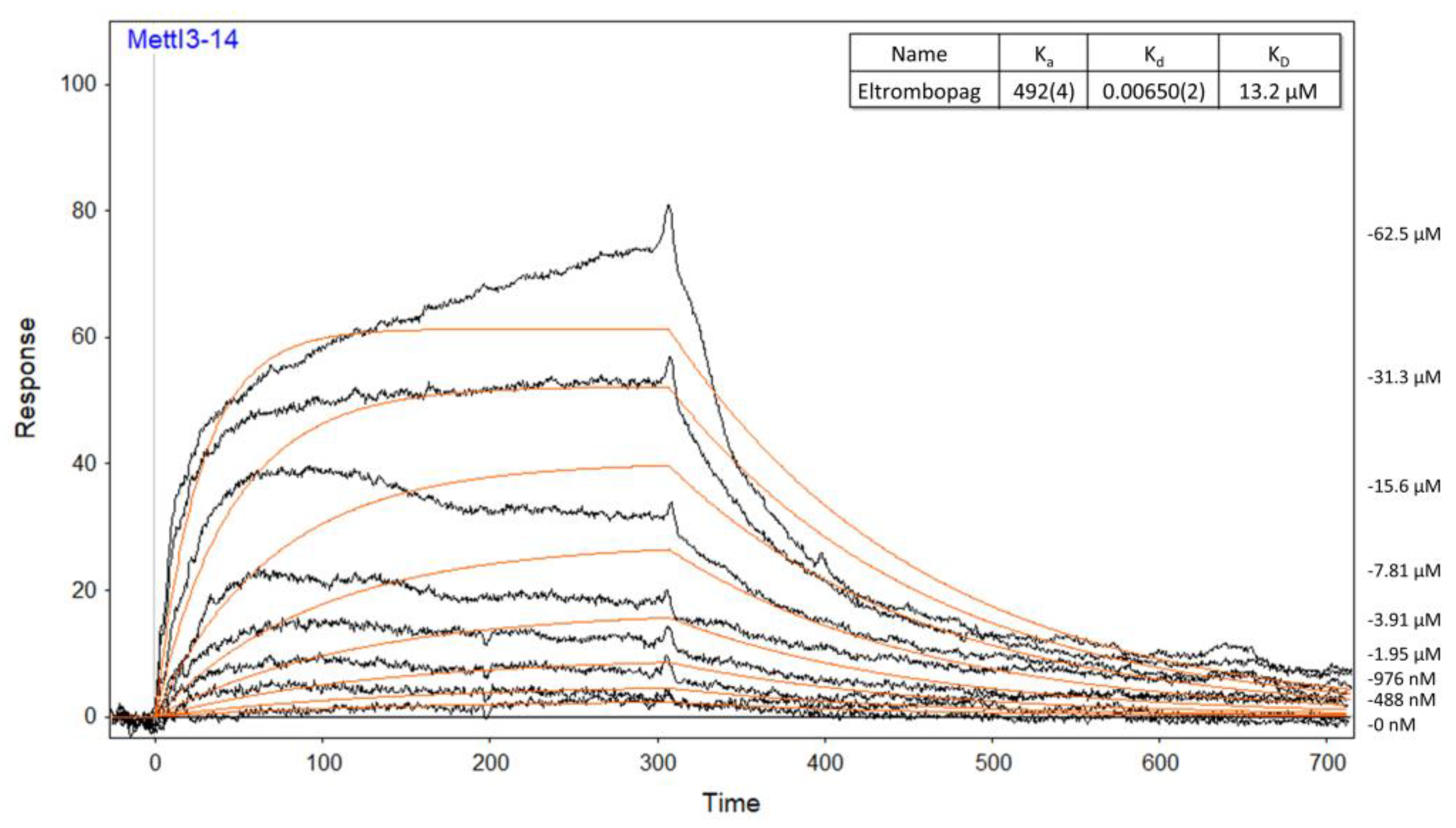
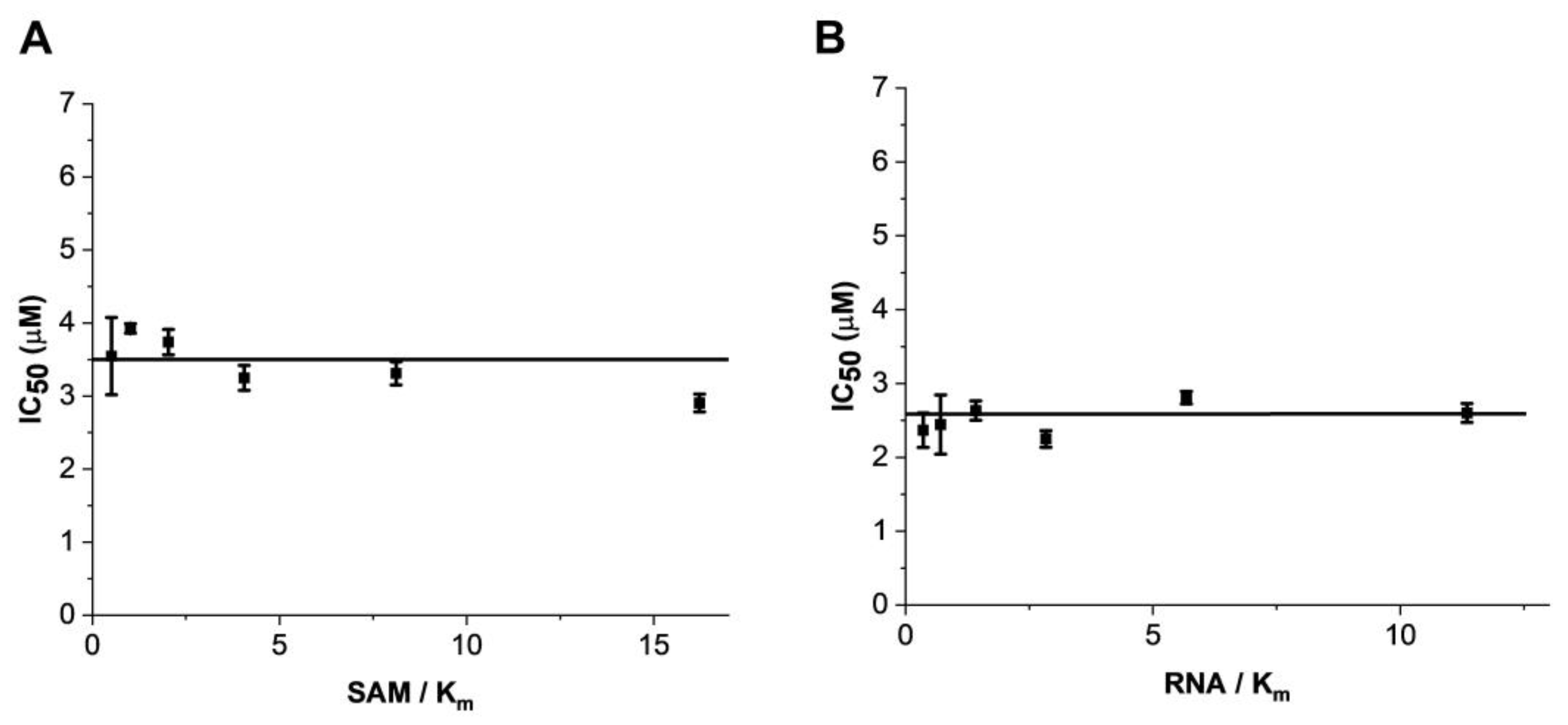
2.2. Mode of Enzyme Inhibition and Predicted Binding Mode of Eltrombopag in METTL3-14
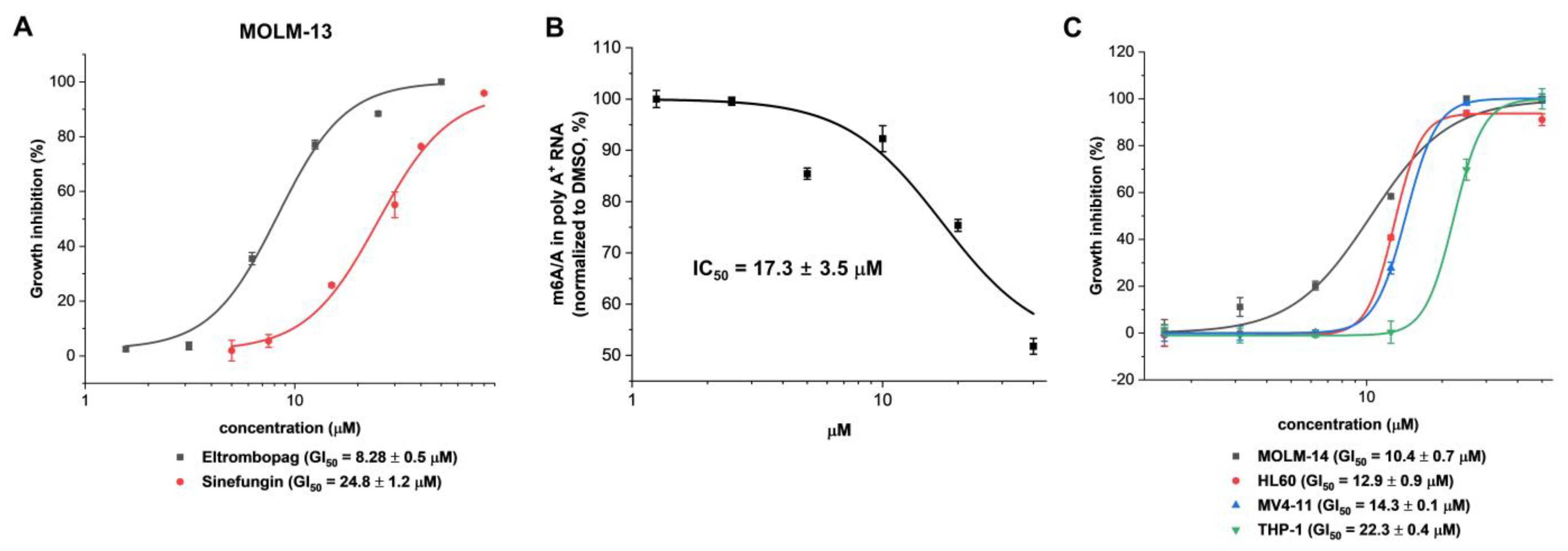
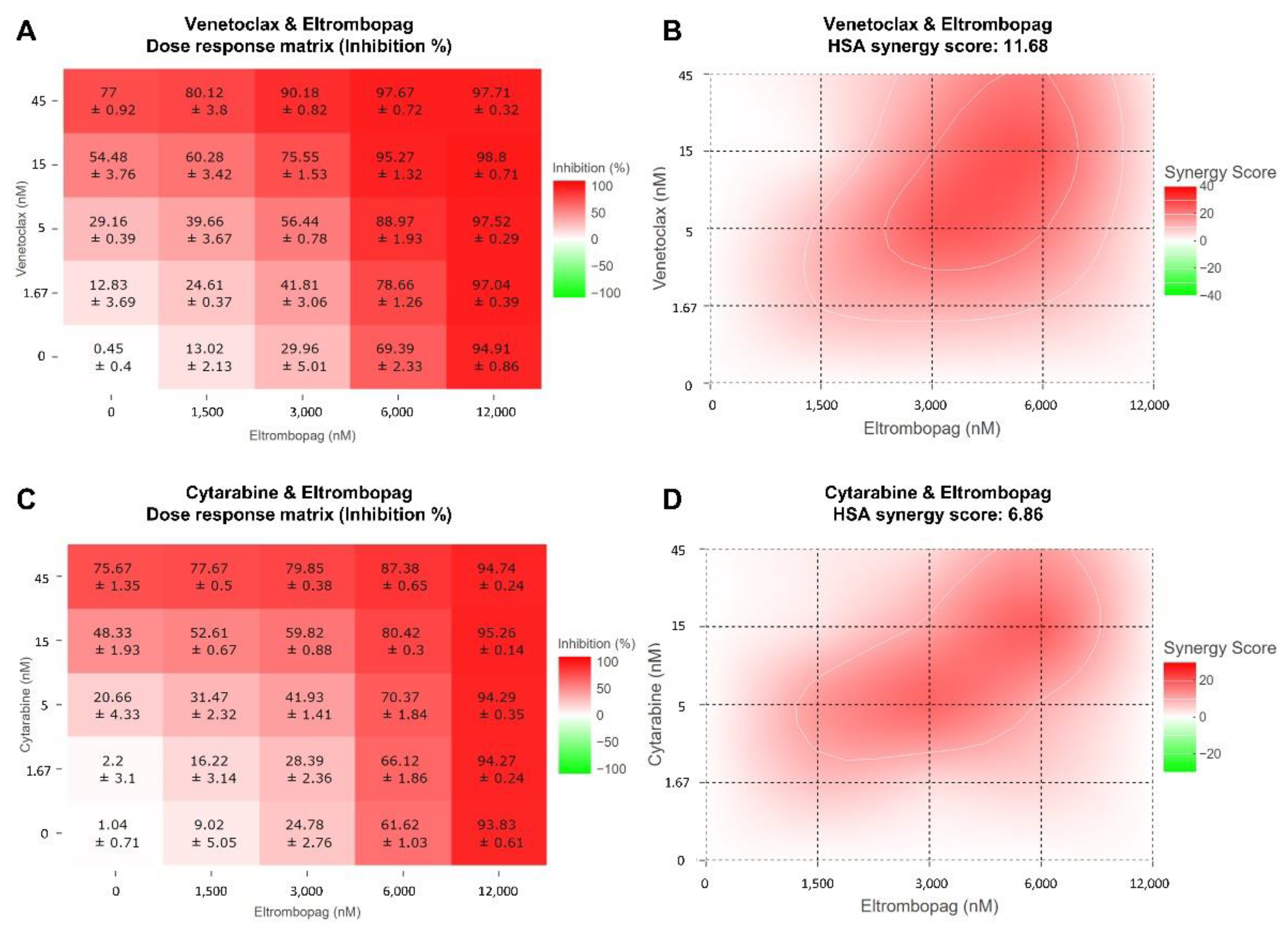
2.3. Cellular Activity Evaluation of Eltrombopag on Acute Myeloid Leukemia Cell Lines
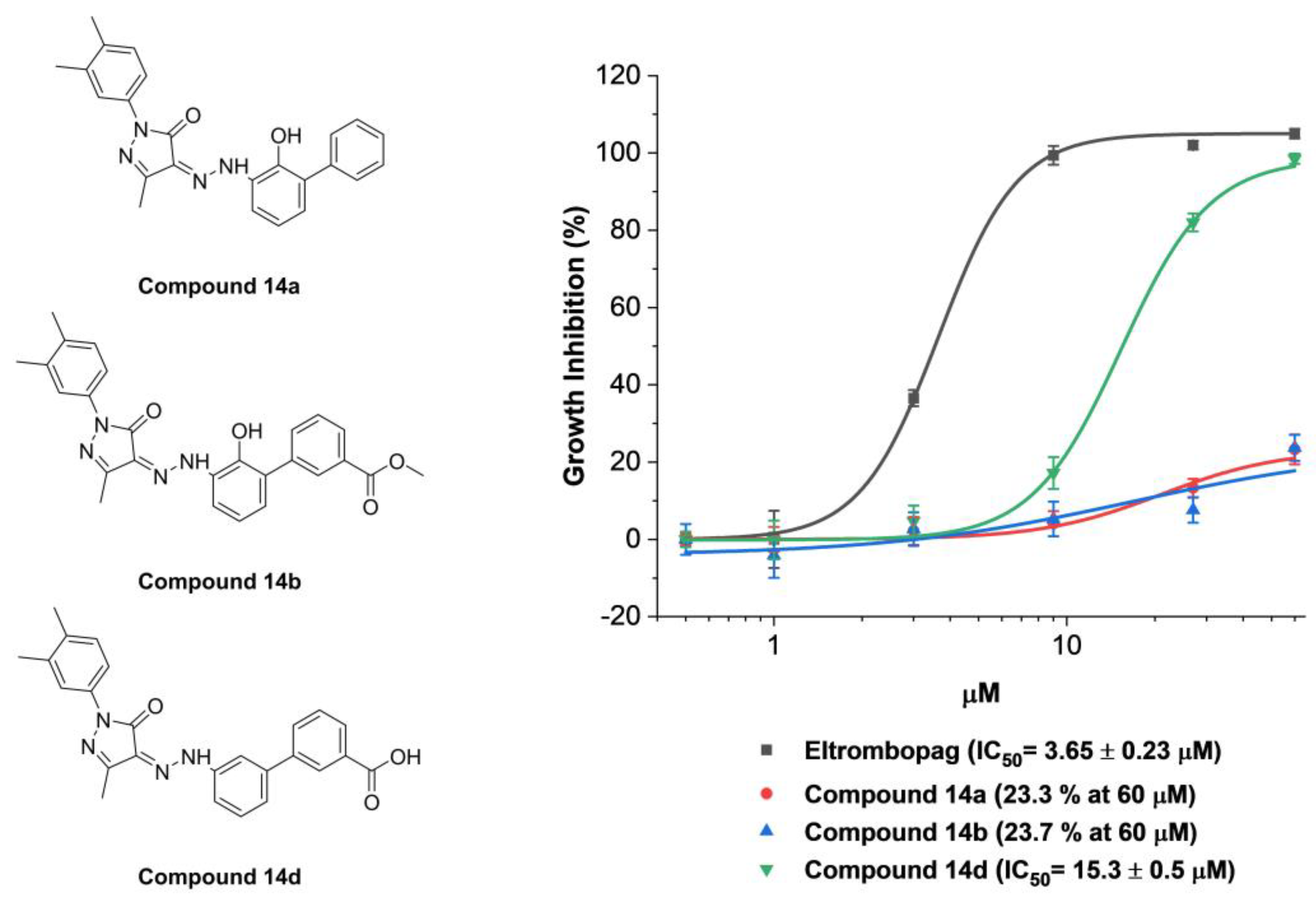
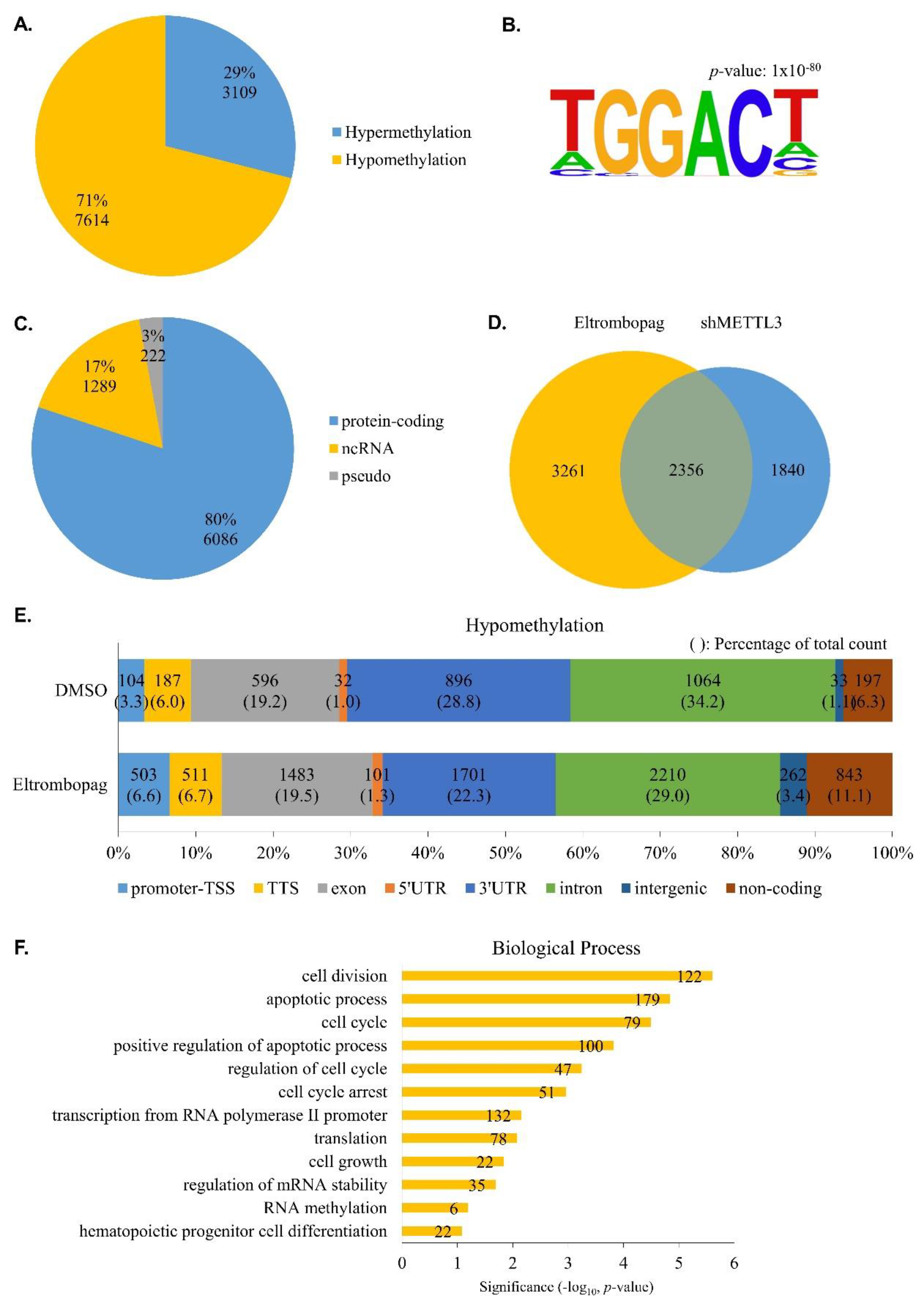
2.4. Identification of Anti-Leukemia Potential of Eltrombopag at Molecular Level
3. Materials and Methods
3.1. General Methods for Chemistry
3.2. Cloning, Expression and Purification of METTL3 and METTL14
3.3. METTL3-14 Enzyme-Based Bioluminescence Assay (Screening Assay)
3.4. METTL3-14 Enzyme-Based Bioluminescence Assay (False Positive Response Experiment)
3.5. Mass Spectrometry Based METTL3-14 Enzyme Based Assay
3.6. Surface Plasmon Resonance
3.7. Selectivity Profiling
3.8. Allosteric Binding Pocket Prediction and Molecular Docking
3.9. Cell Culture and shMETTL3 Knockdown
3.10. Anti-Proliferative Assay Protocol
3.11. Combinatorial Analysis of AML Drugs with Eltrombopag
3.12. Quantitative Analysis of m6A Level by Mass Spectrometry
3.13. N6-Methyladenosine-Sequencing (m6A-seq) and Sequencing Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desrosiers, R.C.; Friderici, K.H.; Rottman, F.M. Characterization of Novikoff Hepatoma MRNA Methylation and Heterogeneity in the Methylated 5’ Terminus. Biochemistry 1975, 14, 4367–4374. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6A RNA Methylomes Revealed by M6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.E.; Miceli, S.M.; Roberts, R.J.; Manley, J.L. Sequence Specificity of the Human MRNA N6-Adenosine Methylase in Vitro. Nucleic Acids Res. 1990, 18, 5735–5741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Fustin, J.-M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-Dependent Demethylation of N6-Methyladenosine Regulates MRNA Splicing and Is Required for Adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-Transcriptional Gene Regulation by MRNA Modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Yue, M.; Wang, J.; Kumar, S.; Wechsler-Reya, R.J.; Zhang, Z.; Ogawa, Y.; Kellis, M.; Duester, G.; et al. N6-Methyladenosine RNA Modification Regulates Embryonic Neural Stem Cell Self-Renewal through Histone Modifications. Nat. Neurosci. 2018, 21, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Sui, X.; Hu, Y.; Ren, C.; Cao, Q.; Zhou, S.; Cao, Y.; Li, M.; Shu, W.; Huo, R. METTL3-Mediated m6A Is Required for Murine Oocyte Maturation and Maternal-to-Zygotic Transition. Cell Cycle 2020, 19, 391–404. [Google Scholar] [CrossRef]
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging Role of m6 A RNA Methylation in Nutritional Physiology and Metabolism. Obes. Rev. 2020, 21, e12942. [Google Scholar] [CrossRef]
- Dorn, L.E.; Lasman, L.; Chen, J.; Xu, X.; Hund, T.J.; Medvedovic, M.; Hanna, J.H.; van Berlo, J.H.; Accornero, F. The N6-Methyladenosine MRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019, 139, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmiedel, J.; Friedman, N.; Le-Trilling, V.T.K.; et al. M6A Modification Controls the Innate Immune Response to Infection by Targeting Type I Interferons. Nat. Immunol. 2019, 20, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhan, Y.; Zhuo, L.; Zhang, Q.; Li, G.; Li, Q.; Qi, S.; Zhu, J.; Lv, Q.; Shen, Y.; et al. Biological Functions of M6A Methyltransferases. Cell Biosci. 2021, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Roignant, J.-Y.; Soller, M. M 6 A in MRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic M6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, Z.; Liu, Y.; Zhao, Y.; Yao, Y.; Wu, R.; Liu, Q.; Wang, Y.; Wang, X. A Dynamic Reversible RNA N6-methyladenosine Modification: Current Status and Perspectives. J. Cell Physiol. 2019, 234, 7948–7956. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of M6A Writers Reveals Two Distinct Classes of MRNA Methylation at Internal and 5′ Sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA M6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-Methyladenosine Modification Destabilizes Developmental Regulators in Embryonic Stem Cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural Basis of N6-Adenosine Methylation by the METTL3–METTL14 Complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Śledź, P.; Jinek, M. Structural Insights into the Molecular Mechanism of the M6A Writer Complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Zhang, J.; Zhu, J.-S. The Role of M6A RNA Methylation in Human Cancer. Mol. Cancer 2019, 18, 103. [Google Scholar] [CrossRef] [Green Version]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA M6A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in Cancer: Mechanisms and Therapeutic Targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef]
- De Kouchkovsky, I.; Abdul-Hay, M. Acute Myeloid Leukemia: A Comprehensive Review and 2016 Update. Blood Cancer J. 2016, 6, e441. [Google Scholar] [CrossRef]
- Kwok, C.-T.; Marshall, A.D.; Rasko, J.E.J.; Wong, J.J.L. Genetic Alterations of M6A Regulators Predict Poorer Survival in Acute Myeloid Leukemia. J. Hematol. Oncol. 2017, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via MRNA M6A Modification. Cell Stem. Cell 2018, 22, 191–205.e9. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N6-Methyladenosine (M6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of Normal Hematopoietic and Leukemia Cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-Bound METTL3 Maintains Myeloid Leukaemia by M6A-Dependent Translation Control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Kennedy, S.; Hajian, T.; Gibson, E.; Seitova, A.; Xu, C.; Arrowsmith, C.H.; Vedadi, M. A Radioactivity-Based Assay for Screening Human M6A-RNA Methyltransferase, METTL3-METTL14 Complex, and Demethylase ALKBH5. J. Biomol. Screen 2016, 21, 290–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buker, S.M.; Gurard-Levin, Z.A.; Wheeler, B.D.; Scholle, M.D.; Case, A.W.; Hirsch, J.L.; Ribich, S.; Copeland, R.A.; Boriack-Sjodin, P.A. A Mass Spectrometric Assay of METTL3/METTL14 Methyltransferase Activity. SLAS Discov. Adv. Sci. Drug Discov. 2020, 25, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Bedi, R.K.; Huang, D.; Eberle, S.A.; Wiedmer, L.; Śledź, P.; Caflisch, A. Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem 2020, 15, 744–748. [Google Scholar] [CrossRef]
- Moroz-Omori, E.V.; Huang, D.; Bedi, R.K.; Cheriyamkunnel, S.J.; Elena, B.; Aymeric, D.; Rzeczkowski, M.D.; Lars, W.; Paweł, Ś.; Amedeo, C. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. ChemMedchem 2020, 16, 3035–3043. [Google Scholar] [CrossRef]
- Dolbois, A.; Bedi, R.K.; Bochenkova, E.; Müller, A.; Moroz-Omori, E.V.; Huang, D.; Caflisch, A. 1,4,9-Triazaspiro[5.5]Undecan-2-One Derivatives as Potent and Selective METTL3 Inhibitors. J. Med. Chem. 2021, 64, 12738–12760. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small Molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Ianniello, Z.; Paiardini, A.; Fatica, A. N6-Methyladenosine (M6A): A Promising New Molecular Target in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 251. [Google Scholar] [CrossRef] [Green Version]
- Malone, T.; Blumenthal, R.M.; Cheng, X. Structure-Guided Analysis Reveals Nine Sequence Motifs Conserved among DNA Amino-Methyl-Transferases, and Suggests a Catalytic Mechanism for These Enzymes. J. Mol. Biol. 1995, 253, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Huang, M.; Wei, Y. Diversity of the Reaction Mechanisms of SAM-Dependent Enzymes. Acta Pharm. Sin. B 2021, 11, 632–650. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, S.; Jin, M.S.; Kim, Y. Discovery of Substituted Indole Derivatives as Allosteric Inhibitors of m6A-RNA Methyltransferase, METTL3-14 Complex. Drug Dev. Res. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Erickson-Miller, C.L.; DeLorme, E.; Tian, S.-S.; Hopson, C.B.; Stark, K.; Giampa, L.; Valoret, E.I.; Duffy, K.J.; Luengo, J.L.; Rosen, J.; et al. Discovery and Characterization of a Selective, Nonpeptidyl Thrombopoietin Receptor Agonist. Exp. Hematol. 2005, 33, 85–93. [Google Scholar] [CrossRef]
- Townsley, D.M.; Scheinberg, P.; Winkler, T.; Desmond, R.; Dumitriu, B.; Rios, O.; Weinstein, B.; Valdez, J.; Lotter, J.; Feng, X.; et al. Eltrombopag Added to Standard Immunosuppression for Aplastic Anemia. N. Engl. J. Med. 2017, 376, 1540–1550. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Dai, M.; Fu, Q.; Chen, S. Eltrombopag for the Treatment of Refractory Thrombocytopenia Associated with Connective Tissue Disease. Sci. Rep. 2021, 11, 5459. [Google Scholar] [CrossRef]
- Lu, S.; He, X.; Ni, D.; Zhang, J. Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J. Med. Chem. 2019, 62, 6405–6421. [Google Scholar] [CrossRef]
- Huang, W.; Lu, S.; Huang, Z.; Liu, X.; Mou, L.; Luo, Y.; Zhao, Y.; Liu, Y.; Chen, Z.; Hou, T.; et al. Allosite: A Method for Predicting Allosteric Sites. Bioinformatics 2013, 29, 2357–2359. [Google Scholar] [CrossRef] [Green Version]
- Jayagopal, B.; Murugesh, S. QBD-Driven HPLC Method of Eltrombopag Olamine: Degradation Pathway Proposal, Structure Elucidation, and in Silico Toxicity Prediction. J. Pharm. Biomed. Anal. 2021, 203, 114231. [Google Scholar] [CrossRef]
- Roth, M.; Will, B.; Simkin, G.; Narayanagari, S.; Barreyro, L.; Bartholdy, B.; Tamari, R.; Mitsiades, C.S.; Verma, A.; Steidl, U. Eltrombopag Inhibits the Proliferation of Leukemia Cells via Reduction of Intracellular Iron and Induction of Differentiation. Blood 2012, 120, 386–394. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genom. Proteom. Bioinform. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Argenziano, M.; Tortora, C.; Paola, A.D.; Pota, E.; Martino, M.D.; Pinto, D.D.; Leva, C.D.; Rossi, F. Eltrombopag and Its Iron Chelating Properties in Pediatric Acute Myeloid Leukemia. Oncotarget 2021, 12, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, M.; Cherif, H.; Fenaux, P.; Mittelman, M.; Verma, A.; Portella, M.S.O.; Burgess, P.; Ramos, P.M.; Choi, J.; Platzbecker, U. Azacitidine with or without Eltrombopag for First-Line Treatment of Intermediate- or High-Risk MDS with Thrombocytopenia. Blood 2018, 132, 2629–2638. [Google Scholar] [CrossRef] [PubMed]
- Basood, M.; Oster, H.S.; Mittelman, M. Thrombocytopenia in patients with myelodysplastic syndromes—Still an unsolved problem. Mediterr. J. Hematol. Infect. Dis. 2018, 10, e2018046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, J.; Renaud, Y.; Albrecht, S.; Ghavi-Helm, Y.; Roignant, J.-Y.; Silies, M.; Junion, G. M6A RNA Methylation Regulates Promoter- Proximal Pausing of RNA Polymerase II. Mol. Cell 2021, 81, 3356–3367.e6. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Meng, J.; Cui, X.; Rao, M.K.; Chen, Y.; Huang, Y. Exome-Based Analysis for RNA Epigenome Sequencing Data. Bioinformatics 2013, 29, 1565–1567. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
| Methyltransferase | Remaining Activity (%) 1 | Control IC50 (μM) 2 | Control Compound |
|---|---|---|---|
| DOT1L | 90.0 ± 2.8 | 0.217 | Chaetocin |
| G9a | 99.0 ± 3.3 | 0.762 | SAH |
| MLL4 complex | 70.9 ± 2.4 | 5.09 | SAH |
| PRDM9 | 130 ± 4 | 4.16 | SAH |
| PRMT1 | 96.1 ± 3.9 | 0.637 | SAH |
| SETD2 | 97.3 ± 4.1 | 10.8 | SAH |
| SMYD3 | 98.7 ± 5.7 | 35.3 | SAH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-H.; Choi, N.; Kim, S.; Jin, M.S.; Shen, H.; Kim, Y.-C. Eltrombopag as an Allosteric Inhibitor of the METTL3-14 Complex Affecting the m6A Methylation of RNA in Acute Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 440. https://doi.org/10.3390/ph15040440
Lee J-H, Choi N, Kim S, Jin MS, Shen H, Kim Y-C. Eltrombopag as an Allosteric Inhibitor of the METTL3-14 Complex Affecting the m6A Methylation of RNA in Acute Myeloid Leukemia Cells. Pharmaceuticals. 2022; 15(4):440. https://doi.org/10.3390/ph15040440
Chicago/Turabian StyleLee, Je-Heon, Namjeong Choi, Subin Kim, Mi Sun Jin, Haihong Shen, and Yong-Chul Kim. 2022. "Eltrombopag as an Allosteric Inhibitor of the METTL3-14 Complex Affecting the m6A Methylation of RNA in Acute Myeloid Leukemia Cells" Pharmaceuticals 15, no. 4: 440. https://doi.org/10.3390/ph15040440
APA StyleLee, J.-H., Choi, N., Kim, S., Jin, M. S., Shen, H., & Kim, Y.-C. (2022). Eltrombopag as an Allosteric Inhibitor of the METTL3-14 Complex Affecting the m6A Methylation of RNA in Acute Myeloid Leukemia Cells. Pharmaceuticals, 15(4), 440. https://doi.org/10.3390/ph15040440