Endogenous Lipid Carriers—Bench-to-Bedside Roadblocks in Production and Drug Loading of Exosomes
Abstract
:1. Introduction
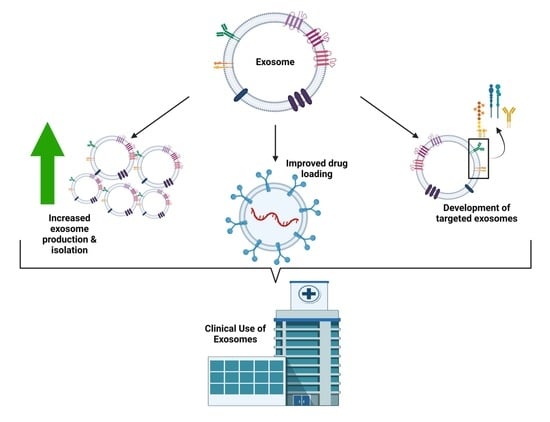
2. Exosomal Drug Delivery: Challenges
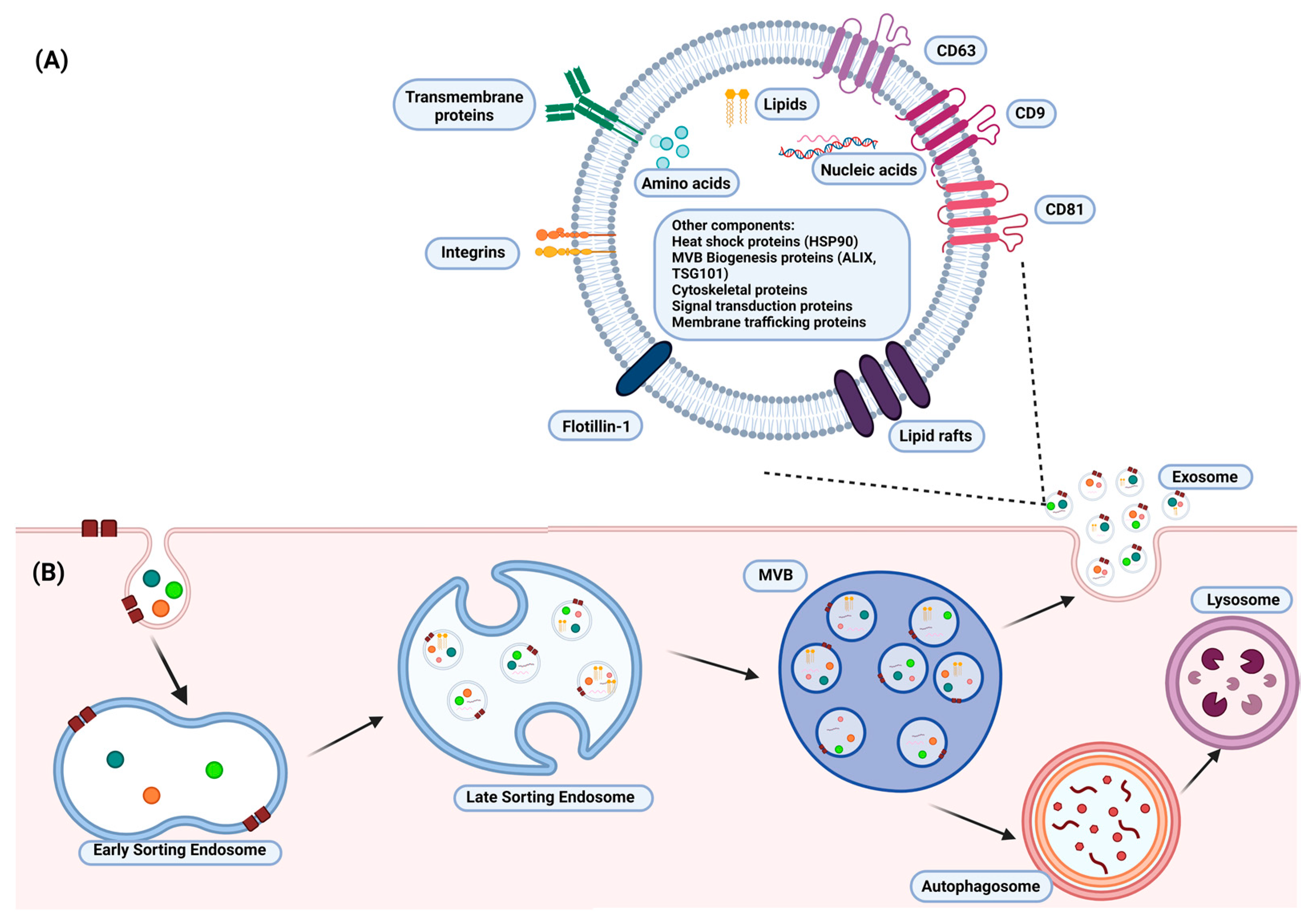
2.1. Exosome Production and Isolation
2.2. Exosome Drug Loading
3. Exosomal Drug Delivery: Solutions
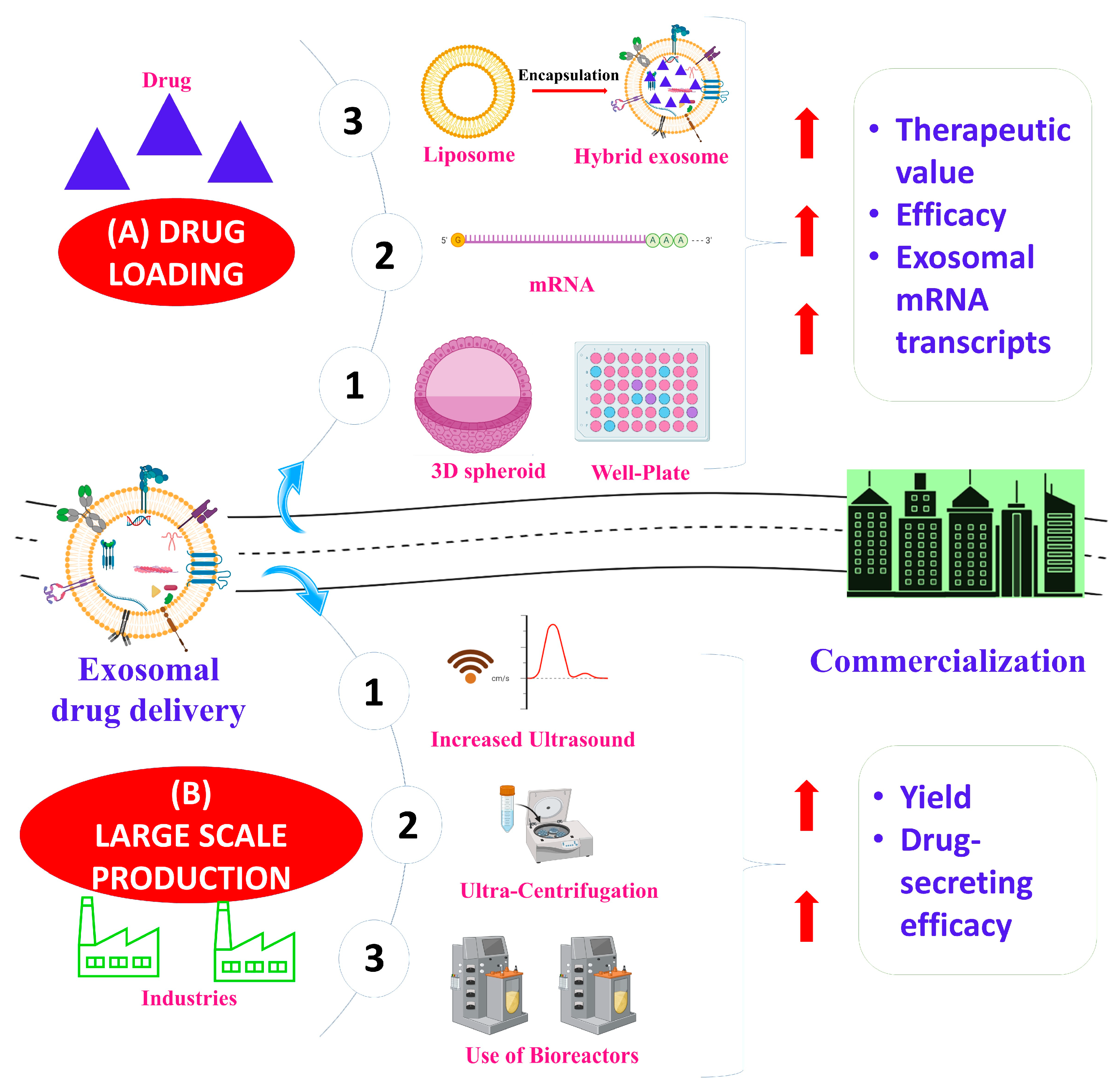
3.1. Exosome Production and Isolation
3.1.1. Source Selection
3.1.2. Upstream Modifications
Soluble Factors
Chemical and Physical Stimulation
3D Culture
Biomaterials
3.1.3. Downstream Modifications
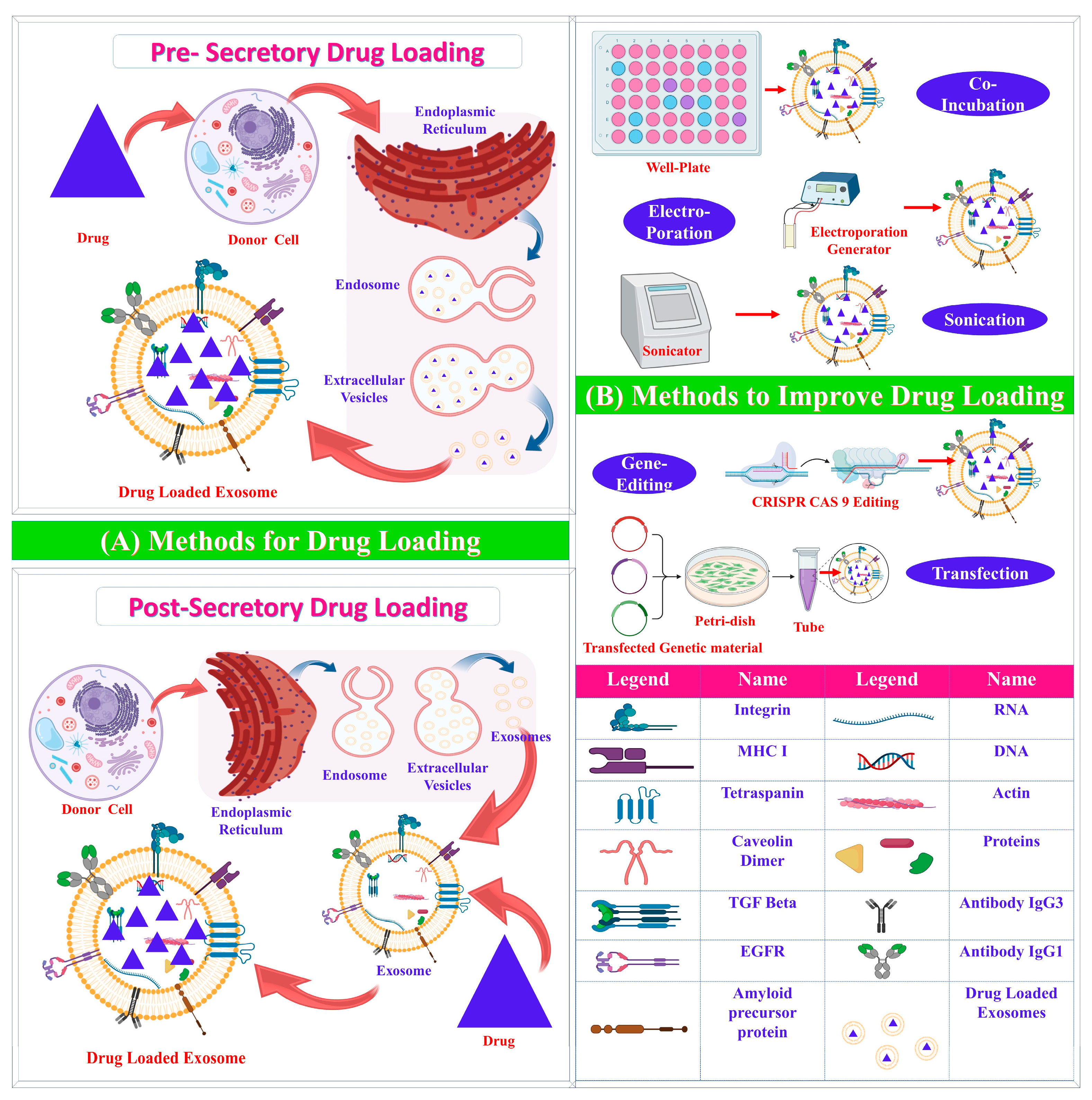
3.2. Exosome Drug Loading
3.2.1. Pre-Secretory Drug Loading
3.2.2. Post-Secretory Drug Loading
3.3. Targeted Exosome Delivery
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR/CAS9 | Clustered regularly interspaced short palindromic repeats-associated protein 9 |
| ESCRT | Endosomal sorting complex required for transport |
| ESE | Early-sorting endosome |
| EXOSD | Exosome separation and detection |
| EXOtic | Exosomal transfer into cells |
| EXPLOR | Exosome system via optically reversible protein–protein interactions |
| HIFα | Hypoxia-induced factor α |
| HUR | Human antigen R |
| ILV | Intraluminal vesicle |
| LPS | Lipopolysaccharide |
| LSE | Late-sorting endosome |
| MVB | Multivesicular body |
| NDFIP1 | Nedd4 family interacting protein 1 |
| NEF | Negative regulatory factor |
| PEG | Polyethylene glycol |
| PLGA | Poly(lactic-co-glycolic acid) |
| PTEN | Phosphatase and TENsin homolog deleted on chromosome 10 |
| STEAP3 | Six-transmembrane epithelial antigen of prostate 3 |
| TRAIL | Tumor necrosis factor-related apoptosis-induced ligand |
References
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as Therapeutic Drug Carriers and Delivery Vehicles across Biological Membranes: Current Perspectives and Future Challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as Drug Delivery Vehicles for Parkinson’s Disease Therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular Vesicles as Drug Delivery Systems: Why and How? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef]
- An, Y.; Lin, S.; Tan, X.; Zhu, S.; Nie, F.; Zhen, Y.; Gu, L.; Zhang, C.; Wang, B.; Wei, W.; et al. Exosomes from Adipose-derived Stem Cells and Application to Skin Wound Healing. Cell Prolif. 2021, 54, e12993. [Google Scholar] [CrossRef]
- Kumar, D.N.; Chaudhuri, A.; Aqil, F.; Dehari, D.; Munagala, R.; Singh, S.; Gupta, R.C.; Agrawal, A.K. Exosomes as Emerging Drug Delivery and Diagnostic Modality for Breast Cancer: Recent Advances in Isolation and Application. Cancers 2022, 14, 1435. [Google Scholar] [CrossRef]
- Hussen, B.M.; Faraj, G.S.H.; Rasul, M.F.; Hidayat, H.J.; Salihi, A.; Baniahmad, A.; Taheri, M.; Ghafouri-Frad, S. Strategies to Overcome the Main Challenges of the Use of Exosomes as Drug Carrier for Cancer Therapy. Cancer Cell Int. 2022, 22, 323. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Gao, W.; Xie, N. Exosomes as Anticancer Drug Delivery Vehicles: Prospects and Challenges. Front. Biosci.-Landmark 2022, 27, 293. [Google Scholar] [CrossRef]
- Ramirez, M.I.; Amorim, M.G.; Gadelha, C.; Milic, I.; Welsh, J.A.; Freitas, V.M.; Nawaz, M.; Akbar, N.; Couch, Y.; Makin, L.; et al. Technical Challenges of Working with Extracellular Vesicles. Nanoscale 2018, 10, 881–906. [Google Scholar] [CrossRef] [Green Version]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Wong, D.K.; Hong, K.Y.; Raffai, R.L. Cushioned-Density Gradient Ultracentrifugation (C-DGUC): A Refined and High Performance Method for the Isolation, Characterization, and Use of Exosomes. Methods Mol. Biol. 2018, 1740, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Onódi, Z.; Pelyhe, C.; Terézia Nagy, C.; Brenner, G.B.; Almási, L.; Kittel, Á.; Manček-Keber, M.; Ferdinandy, P.; Buzás, E.I.; Giricz, Z. Isolation of High-Purity Extracellular Vesicles by the Combination of Iodixanol Density Gradient Ultracentrifugation and Bind-Elute Chromatography from Blood Plasma. Front. Physiol. 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rider, M.A.; Hurwitz, S.N.; Meckes, D.G. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci. Rep. 2016, 6, 23978. [Google Scholar] [CrossRef] [PubMed]
- Emam, S.E.; Ando, H.; Lila, A.S.A.; Shimizu, T.; Ukawa, M.; Okuhira, K.; Ishima, Y.; Mahdy, M.A.; Ghazy, F.S.; Ishida, T. A Novel Strategy to Increase the Yield of Exosomes (Extracellular Vesicles) for an Expansion of Basic Research. Biol. Pharm. Bull. 2018, 41, 733–742. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Borg, E.G.F.; Liaci, A.M.; Vos, H.R.; Stoorvogel, W. A Novel Three Step Protocol to Isolate Extracellular Vesicles from Plasma or Cell Culture Medium with Both High Yield and Purity. J. Extracell. Vesicles 2020, 9, 1791450. [Google Scholar] [CrossRef]
- Benedikter, B.J.; Bouwman, F.G.; Vajen, T.; Heinzmann, A.C.A.; Grauls, G.; Mariman, E.C.; Wouters, E.F.M.; Savelkoul, P.H.; Lopez-Iglesias, C.; Koenen, R.R.; et al. Ultrafiltration Combined with Size Exclusion Chromatography Efficiently Isolates Extracellular Vesicles from Cell Culture Media for Compositional and Functional Studies. Sci. Rep. 2017, 7, 15297. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, E.M.; Vestad, B.; Steffensen, L.A.; Aass, H.C.D.; Saeed, M.; Øvstebø, R.; Costea, D.E.; Galtung, H.K.; Søland, T.M. Efficient Extracellular Vesicle Isolation by Combining Cell Media Modifications, Ultrafiltration, and Size-Exclusion Chromatography. PLoS ONE 2018, 13, e0204276. [Google Scholar] [CrossRef] [Green Version]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-Based Isolation Minimally Alters Extracellular Vesicles’ Characteristics Compared to Precipitating Agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-Based Isolation of Melanoma Cell-Derived Exosomes from Plasma of Patients with Melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef]
- Filipović, L.; Spasojević, M.; Prodanović, R.; Korać, A.; Matijaševic, S.; Brajušković, G.; de Marco, A.; Popović, M. Affinity-Based Isolation of Extracellular Vesicles by Means of Single-Domain Antibodies Bound to Macroporous Methacrylate-Based Copolymer. New Biotechnol. 2022, 69, 36–48. [Google Scholar] [CrossRef]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for Exosome Isolation and Analysis: Enabling Liquid Biopsy for Personalized Medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talebjedi, B.; Tasnim, N.; Hoorfar, M.; Mastromonaco, G.F.; De Almeida Monteiro Melo Ferraz, M. Exploiting Microfluidics for Extracellular Vesicle Isolation and Characterization: Potential Use for Standardized Embryo Quality Assessment. Front. Vet. Sci. 2021, 7, 620809. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Han, S.; Ma, L.; Zhang, H.; Zhan, Z.; Aguilar, H.A.; Zhang, H.; Xiao, K.; Gu, Y.; Gu, Z.; et al. Synergistically Bifunctional Paramagnetic Separation Enables Efficient Isolation of Urine Extracellular Vesicles and Downstream Phosphoproteomic Analysis. ACS Appl. Mater. Interfaces 2021, 13, 3622–3630. [Google Scholar] [CrossRef]
- Jiawei, S.; Zhi, C.; Kewei, T.; Xiaoping, L. Magnetic Bead-Based Adsorption Strategy for Exosome Isolation. Front. Bioeng. Biotechnol. 2022, 10, 942077. [Google Scholar] [CrossRef] [PubMed]
- Paolini, L.; Monguió-Tortajada, M.; Costa, M.; Antenucci, F.; Barilani, M.; Clos-Sansalvador, M.; Andrade, A.C.; Driedonks, T.A.P.; Giancaterino, S.; Kronstadt, S.M.; et al. Large-Scale Production of Extracellular Vesicles: Report on the “MassivEVs” ISEV Workshop. J. Extracell. Biol. 2022, 1, e63. [Google Scholar] [CrossRef]
- Jeyaram, A.; Jay, S.M. Preservation and Storage Stability of Extracellular Vesicles for Therapeutic Applications. AAPS J. 2017, 20, 1. [Google Scholar] [CrossRef]
- Lőrincz, Á.M.; Timár, C.I.; Marosvári, K.A.; Veres, D.S.; Otrokocsi, L.; Kittel, Á.; Ligeti, E. Effect of Storage on Physical and Functional Properties of Extracellular Vesicles Derived from Neutrophilic Granulocytes. J. Extracell. Vesicles 2014, 3, 25465. [Google Scholar] [CrossRef]
- Maroto, R.; Zhao, Y.; Jamaluddin, M.; Popov, V.L.; Wang, H.; Kalubowilage, M.; Zhang, Y.; Luisi, J.; Sun, H.; Culbertson, C.T.; et al. Effects of Storage Temperature on Airway Exosome Integrity for Diagnostic and Functional Analyses. J. Extracell. Vesicles 2017, 6, 1359478. [Google Scholar] [CrossRef] [Green Version]
- Bunggulawa, E.J.; Wang, W.; Yin, T.; Wang, N.; Durkan, C.; Wang, Y.; Wang, G. Recent Advancements in the Use of Exosomes as Drug Delivery Systems. J. Nanobiotechnol. 2018, 16, 81. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, D.; Moreira, J.N.; Rodrigues, L.R. New Advances in Exosome-Based Targeted Drug Delivery Systems. Crit. Rev. Oncol./Hematol. 2022, 172, 103628. [Google Scholar] [CrossRef]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel Is Incorporated by Mesenchymal Stromal Cells and Released in Exosomes That Inhibit in vitro Tumor Growth: A New Approach for Drug Delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Sterzenbach, U.; Putz, U.; Low, L.-H.; Silke, J.; Tan, S.-S.; Howitt, J. Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol. Ther. 2017, 25, 1269–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Zhang, W.; Li, Y.; Chang, J.; Tian, F.; Zhao, F.; Ma, Y.; Sun, J. Microfluidic Sonication to Assemble Exosome Membrane-Coated Nanoparticles for Immune Evasion-Mediated Targeting. Nano Lett. 2019, 19, 7836–7844. [Google Scholar] [CrossRef] [PubMed]
- Salarpour, S.; Forootanfar, H.; Pournamdari, M.; Ahmadi-Zeidabadi, M.; Esmaeeli, M.; Pardakhty, A. Paclitaxel Incorporated Exosomes Derived from Glioblastoma Cells: Comparative Study of Two Loading Techniques. Daru 2019, 27, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Morales, B.; Antunes-Ricardo, M.; González-Valdez, J. Exosome-Mediated Insulin Delivery for the Potential Treatment of Diabetes Mellitus. Pharmaceutics 2021, 13, 1870. [Google Scholar] [CrossRef]
- Tang, M.; Chen, Y.; Li, B.; Sugimoto, H.; Yang, S.; Yang, C.; LeBleu, V.S.; McAndrews, K.M.; Kalluri, R. Therapeutic Targeting of STAT3 with Small Interference RNAs and Antisense Oligonucleotides Embedded Exosomes in Liver Fibrosis. FASEB J. 2021, 35, e21557. [Google Scholar] [CrossRef]
- Deng, W.; Meng, Y.; Wang, B.; Wang, C.-X.; Hou, C.-X.; Zhu, Q.-H.; Tang, Y.-T.; Ye, J.-H. In Vitro Experimental Study on the Formation of MicroRNA-34a Loaded Exosomes and Their Inhibitory Effect in Oral Squamous Cell Carcinoma. Cell Cycle 2022, 21, 1775–1783. [Google Scholar] [CrossRef]
- González-Sarrías, A.; Iglesias-Aguirre, C.E.; Cortés-Martín, A.; Vallejo, F.; Cattivelli, A.; del Pozo-Acebo, L.; Del Saz, A.; López de las Hazas, M.C.; Dávalos, A.; Espín, J.C. Milk-Derived Exosomes as Nanocarriers to Deliver Curcumin and Resveratrol in Breast Tissue and Enhance Their Anticancer Activity. Int. J. Mol. Sci. 2022, 23, 2860. [Google Scholar] [CrossRef]
- Wang, Z.; Rich, J.; Hao, N.; Gu, Y.; Chen, C.; Yang, S.; Zhang, P.; Huang, T.J. Acoustofluidics for Simultaneous Nanoparticle-Based Drug Loading and Exosome Encapsulation. Microsyst. Nanoeng. 2022, 8, 45. [Google Scholar] [CrossRef]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering Hybrid Exosomes by Membrane Fusion with Liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Shi, G.; Guo, J.; Wang, C.; He, Y. Exosome-Encapsulated Antibiotic against Intracellular Infections of Methicillin-Resistant Staphylococcus Aureus. Int. J. Nanomed. 2018, 13, 8095–8104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, R.; Yu, Z.; Du, J.; Hu, S.; Yuan, C.; Guo, H.; Zhang, Y.; Yang, H. A High-Throughput Nanofluidic Device for Exosome Nanoporation to Develop Cargo Delivery Vehicles. Small 2021, 17, 2102150. [Google Scholar] [CrossRef]
- Fuhrmann, G.; Chandrawati, R.; Parmar, P.A.; Keane, T.J.; Maynard, S.A.; Bertazzo, S.; Stevens, M.M. Engineering Extracellular Vesicles with the Tools of Enzyme Prodrug Therapy. Adv. Mater. 2018, 30, 1706616. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Lee, C.-S.; Kim, S.; Chen, C.; Aghaloo, T.; Lee, M. Generation of Small RNA-Modulated Exosome Mimetics for Bone Regeneration. ACS Nano 2020, 14, 11973–11984. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Che, S.P.Y.; LeBleu, V.S.; Kalluri, R. Effective Delivery of STING Agonist Using Exosomes Suppresses Tumor Growth and Enhances Antitumor Immunity. J. Biol. Chem. 2021, 296, 100523. [Google Scholar] [CrossRef]
- Wei, H.; Chen, J.; Wang, S.; Fu, F.; Zhu, X.; Wu, C.; Liu, Z.; Zhong, G.; Lin, J. A Nanodrug Consisting of Doxorubicin and Exosome Derived from Mesenchymal Stem Cells for Osteosarcoma Treatment in vitro. Int. J. Nanomed. 2019, 14, 8603–8610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [Green Version]
- Yim, N.; Ryu, S.-W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, W.; Park, J.-H.; et al. Exosome Engineering for Efficient Intracellular Delivery of Soluble Proteins Using Optically Reversible Protein–Protein Interaction Module. Nat. Commun. 2016, 7, 12277. [Google Scholar] [CrossRef] [Green Version]
- Burrello, J.; Monticone, S.; Gai, C.; Gomez, Y.; Kholia, S.; Camussi, G. Stem Cell-Derived Extracellular Vesicles and Immune-Modulation. Front. Cell Dev. Biol. 2016, 4, 83. [Google Scholar] [CrossRef] [Green Version]
- Ishiy, C.S.R.A.; Ormanji, M.S.; Maquigussa, E.; Ribeiro, R.S.; da Silva Novaes, A.; Boim, M.A. Comparison of the Effects of Mesenchymal Stem Cells with Their Extracellular Vesicles on the Treatment of Kidney Damage Induced by Chronic Renal Artery Stenosis. Stem. Cells Int. 2020, 2020, 8814574. [Google Scholar] [CrossRef]
- Fernández-Francos, S.; Eiro, N.; Costa, L.A.; Escudero-Cernuda, S.; Fernández-Sánchez, M.L.; Vizoso, F.J. Mesenchymal Stem Cells as a Cornerstone in a Galaxy of Intercellular Signals: Basis for a New Era of Medicine. Int. J. Mol. Sci. 2021, 22, 3576. [Google Scholar] [CrossRef] [PubMed]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.-C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol. Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Defourny, K.A.Y.; Smid, E.J.; Abee, T. Gram-Positive Bacterial Extracellular Vesicles and Their Impact on Health and Disease. Front. Microbiol. 2018, 9, 1502. [Google Scholar] [CrossRef] [Green Version]
- Bitto, N.J.; Zavan, L.; Johnston, E.L.; Stinear, T.P.; Hill, A.F.; Kaparakis-Liaskos, M. Considerations for the Analysis of Bacterial Membrane Vesicles: Methods of Vesicle Production and Quantification Can Influence Biological and Experimental Outcomes. Microbiol. Spectr. 2021, 9, e01273-21. [Google Scholar] [CrossRef]
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-Derived Exosomes for Oral Delivery of Paclitaxel. Nanomedicine 2017, 13, 1627–1636. [Google Scholar] [CrossRef]
- Marcilla, A.; Trelis, M.; Cortés, A.; Sotillo, J.; Cantalapiedra, F.; Minguez, M.T.; Valero, M.L.; del Pino, M.M.S.; Muñoz-Antoli, C.; Toledo, R.; et al. Extracellular Vesicles from Parasitic Helminths Contain Specific Excretory/Secretory Proteins and Are Internalized in Intestinal Host Cells. PLoS ONE 2012, 7, e45974. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host. Microbe 2018, 24, 637–652.e8. [Google Scholar] [CrossRef] [Green Version]
- Adamo, G.; Fierli, D.; Romancino, D.P.; Picciotto, S.; Barone, M.E.; Aranyos, A.; Božič, D.; Morsbach, S.; Raccosta, S.; Stanly, C.; et al. Nanoalgosomes: Introducing Extracellular Vesicles Produced by Microalgae. J. Extracell. Vesicles 2021, 10, e12081. [Google Scholar] [CrossRef]
- Picciotto, S.; Barone, M.E.; Fierli, D.; Aranyos, A.; Adamo, G.; Božič, D.; Romancino, D.P.; Stanly, C.; Parkes, R.; Morsbach, S.; et al. Isolation of Extracellular Vesicles from Microalgae: Towards the Production of Sustainable and Natural Nanocarriers of Bioactive Compounds. Biomater. Sci. 2021, 9, 2917–2930. [Google Scholar] [CrossRef]
- Petousis-Harris, H. Impact of Meningococcal Group B OMV Vaccines, beyond Their Brief. Hum. Vaccines Immunother. 2018, 14, 1058–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ti, D.; Hao, H.; Tong, C.; Liu, J.; Dong, L.; Zheng, J.; Zhao, Y.; Liu, H.; Fu, X.; Han, W. LPS-Preconditioned Mesenchymal Stromal Cells Modify Macrophage Polarization for Resolution of Chronic Inflammation via Exosome-Shuttled Let-7b. J. Transl. Med. 2015, 13, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Bonacquisti, E.E.; Brown, A.D.; Nguyen, J. Boosting the Biogenesis and Secretion of Mesenchymal Stem Cell-Derived Exosomes. Cells 2020, 9, 660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glebov, K.; Löchner, M.; Jabs, R.; Lau, T.; Merkel, O.; Schloss, P.; Steinhäuser, C.; Walter, J. Serotonin Stimulates Secretion of Exosomes from Microglia Cells: Serotonin Stimulates Microglial Exosome Release. Glia 2015, 63, 626–634. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kita, S.; Tanaka, Y.; Fukuda, S.; Obata, Y.; Okita, T.; Nishida, H.; Takahashi, Y.; Kawachi, Y.; Tsugawa-Shimizu, Y.; et al. Adiponectin Stimulates Exosome Release to Enhance Mesenchymal Stem-Cell-Driven Therapy of Heart Failure in Mice. Mol. Ther. 2020, 28, 2203–2219. [Google Scholar] [CrossRef]
- Drago, F.; Lombardi, M.; Prada, I.; Gabrielli, M.; Joshi, P.; Cojoc, D.; Franck, J.; Fournier, I.; Vizioli, J.; Verderio, C. ATP Modifies the Proteome of Extracellular Vesicles Released by Microglia and Influences Their Action on Astrocytes. Front. Pharmacol. 2017, 8, 910. [Google Scholar] [CrossRef] [Green Version]
- Hooper, C.; Sainz-Fuertes, R.; Lynham, S.; Hye, A.; Killick, R.; Warley, A.; Bolondi, C.; Pocock, J.; Lovestone, S. Wnt3a Induces Exosome Secretion from Primary Cultured Rat Microglia. BMC Neurosci. 2012, 13, 144. [Google Scholar] [CrossRef] [Green Version]
- Yuyama, K.; Takahashi, K.; Usuki, S.; Mikami, D.; Sun, H.; Hanamatsu, H.; Furukawa, J.; Mukai, K.; Igarashi, Y. Plant Sphingolipids Promote Extracellular Vesicle Release and Alleviate Amyloid-β Pathologies in a Mouse Model of Alzheimer’s Disease. Sci. Rep. 2019, 9, 16827. [Google Scholar] [CrossRef] [Green Version]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.-E.; El-Baba, M.D.; Saxena, P.; Ausländer, S.; Tan, K.R.; Fussenegger, M. Designer Exosomes Produced by Implanted Cells Intracerebrally Deliver Therapeutic Cargo for Parkinson’s Disease Treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and Opportunities in Exosome Research—Perspectives from Biology, Engineering, and Cancer Therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-King, H.; García, N.A.; Ontoria-Oviedo, I.; Ciria, M.; Montero, J.A.; Sepúlveda, P. Hypoxia Inducible Factor-1α Potentiates Jagged 1-Mediated Angiogenesis by Mesenchymal Stem Cell-Derived Exosomes. Stem Cells 2017, 35, 1747–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhou, X.; Yao, Q.; Liu, Y.; Zhang, H.; Dong, Z. HIF-1-Mediated Production of Exosomes during Hypoxia Is Protective in Renal Tubular Cells. Am. J. Physiol. Ren. Physiol. 2017, 313, F906–F913. [Google Scholar] [CrossRef] [Green Version]
- Böker, K.O.; Lemus-Diaz, N.; Ferreira, R.R.; Schiller, L.; Schneider, S.; Gruber, J. The Impact of the CD9 Tetraspanin on Lentivirus Infectivity and Exosome Secretion. Mol. Ther. 2018, 26, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, G.-H.; Wu, J.; Mou, F.-F.; Xie, W.-H.; Wang, F.-B.; Wang, Q.-L.; Fang, J.; Xu, Y.-W.; Dong, Y.-R.; Liu, J.-R.; et al. Exosomes Derived from Hypoxia-Preconditioned Mesenchymal Stromal Cells Ameliorate Cognitive Decline by Rescuing Synaptic Dysfunction and Regulating Inflammatory Responses in APP/PS1 Mice. FASEB J. 2018, 32, 654–668. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Zhao, H.; Zhang, M.; He, Y.; Li, X.; Xu, Y.; Liu, X. Hypoxia Induced Changes of Exosome Cargo and Subsequent Biological Effects. Front. Immunol. 2022, 13, 824188. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.A.; Ontoria-Oviedo, I.; González-King, H.; Diez-Juan, A.; Sepúlveda, P. Glucose Starvation in Cardiomyocytes Enhances Exosome Secretion and Promotes Angiogenesis in Endothelial Cells. PLoS ONE 2015, 10, e0138849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Wang, J.; Xiao, Y.; Li, F.; Niu, L.; Liu, X.; Meng, L.; Zheng, H. Ultrasound-Mediated Augmented Exosome Release from Astrocytes Alleviates Amyloid-β-Induced Neurotoxicity. Theranostics 2021, 11, 4351–4362. [Google Scholar] [CrossRef]
- Guo, Y.; Wan, Z.; Zhao, P.; Wei, M.; Liu, Y.; Bu, T.; Sun, W.; Li, Z.; Yuan, L. Ultrasound Triggered Topical Delivery of Bmp7 MRNA for White Fat Browning Induction via Engineered Smart Exosomes. J. Nanobiotechnol. 2021, 19, 402. [Google Scholar] [CrossRef] [PubMed]
- Ambattu, L.A.; Ramesan, S.; Dekiwadia, C.; Hanssen, E.; Li, H.; Yeo, L.Y. High Frequency Acoustic Cell Stimulation Promotes Exosome Generation Regulated by a Calcium-Dependent Mechanism. Commun. Biol. 2020, 3, 553. [Google Scholar] [CrossRef]
- Kim, M.; Yun, H.-W.; Park, D.Y.; Choi, B.H.; Min, B.-H. Three-Dimensional Spheroid Culture Increases Exosome Secretion from Mesenchymal Stem Cells. Tissue Eng. Regen. Med. 2018, 15, 427–436. [Google Scholar] [CrossRef]
- Cao, J.; Wang, B.; Tang, T.; Lv, L.; Ding, Z.; Li, Z.; Hu, R.; Wei, Q.; Shen, A.; Fu, Y.; et al. Three-Dimensional Culture of MSCs Produces Exosomes with Improved Yield and Enhanced Therapeutic Efficacy for Cisplatin-Induced Acute Kidney Injury. Stem Cell Res. Ther. 2020, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- Koh, B.; Sulaiman, N.; Fauzi, M.B.; Law, J.X.; Ng, M.H.; Idrus, R.B.H.; Yazid, M.D. Three Dimensional Microcarrier System in Mesenchymal Stem Cell Culture: A Systematic Review. Cell Biosci. 2020, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhao, J.; Li, Q.; Hou, L.; Wang, Y.; Li, S.; Jiang, F.; Zhu, Z.; Tian, L. Exosomes Derived from Three-Dimensional Cultured Human Umbilical Cord Mesenchymal Stem Cells Ameliorate Pulmonary Fibrosis in a Mouse Silicosis Model. Stem Cell Res. Ther. 2020, 11, 503. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.B.; Luthers, C.R.; Lerman, M.J.; Fisher, J.P.; Jay, S.M. Enhanced Extracellular Vesicle Production and Ethanol-Mediated Vascularization Bioactivity via a 3D-Printed Scaffold-Perfusion Bioreactor System. Acta Biomater. 2019, 95, 236–244. [Google Scholar] [CrossRef]
- Burns, A.B.; Doris, C.; Vehar, K.; Saxena, V.; Bardliving, C.; Shamlou, P.A.; Phillips, M.I. Novel Low Shear 3D Bioreactor for High Purity Mesenchymal Stem Cell Production. PLoS ONE 2021, 16, e0252575. [Google Scholar] [CrossRef]
- Du, W.; Zhang, K.; Zhang, S.; Wang, R.; Nie, Y.; Tao, H.; Han, Z.; Liang, L.; Wang, D.; Liu, J.; et al. Enhanced Proangiogenic Potential of Mesenchymal Stem Cell-Derived Exosomes Stimulated by a Nitric Oxide Releasing Polymer. Biomaterials 2017, 133, 70–81. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Y.; Feng, C.; Chang, J.; Fu, R.; Wu, T.; Yu, F.; Wang, X.; Xia, L.; Wu, C.; et al. Lithium-Containing Biomaterials Stimulate Bone Marrow Stromal Cell-Derived Exosomal MiR-130a Secretion to Promote Angiogenesis. Biomaterials 2019, 192, 523–536. [Google Scholar] [CrossRef]
- Park, D.J.; Yun, W.S.; Kim, W.C.; Park, J.-E.; Lee, S.H.; Ha, S.; Choi, J.S.; Key, J.; Seo, Y.J. Improvement of Stem Cell-Derived Exosome Release Efficiency by Surface-Modified Nanoparticles. J. Nanobiotechnol. 2020, 18, 178. [Google Scholar] [CrossRef]
- Wu, Z.; He, D.; Li, H. Bioglass Enhances the Production of Exosomes and Improves Their Capability of Promoting Vascularization. Bioact. Mater. 2021, 6, 823–835. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Dubuke, M.L.; Rockwell, H.E.; Coles, A.H.; Sapp, E.; Didiot, M.-C.; Echeverria, D.; Stoppato, M.; Sere, Y.Y.; et al. Serum Deprivation of Mesenchymal Stem Cells Improves Exosome Activity and Alters Lipid and Protein Composition. iScience 2019, 16, 230–241. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Rawat, S.; Arora, V.; Kottarath, S.K.; Dinda, A.K.; Vaishnav, P.K.; Nayak, B.; Mohanty, S. An Improvised One-Step Sucrose Cushion Ultracentrifugation Method for Exosome Isolation from Culture Supernatants of Mesenchymal Stem Cells. Stem Cell Res. Ther. 2018, 9, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.T.; Wunsch, B.H.; Dogra, N.; Ahsen, M.E.; Lee, K.; Yadav, K.K.; Weil, R.; Pereira, M.A.; Patel, J.V.; Duch, E.A.; et al. Integrated Nanoscale Deterministic Lateral Displacement Arrays for Separation of Extracellular Vesicles from Clinically-Relevant Volumes of Biological Samples. Lab Chip 2018, 18, 3913–3925. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lin, S.; Xia, F.; Liu, Y.; Zhang, D.; Wang, F.; Wang, Y.; Li, Q.; Niu, J.; Cao, C.; et al. ExoSD Chips for High-Purity Immunomagnetic Separation and High-Sensitivity Detection of Gastric Cancer Cell-Derived Exosomes. Biosens. Bioelectron. 2021, 194, 113594. [Google Scholar] [CrossRef] [PubMed]
- Heath, N.; Grant, L.; De Oliveira, T.M.; Rowlinson, R.; Osteikoetxea, X.; Dekker, N.; Overman, R. Rapid Isolation and Enrichment of Extracellular Vesicle Preparations Using Anion Exchange Chromatography. Sci. Rep. 2018, 8, 5730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Mao, J.; Barrero, R.A.; Wang, P.; Zhang, F.; Wang, T. Development of a CD63 Aptamer for Efficient Cancer Immunochemistry and Immunoaffinity-Based Exosome Isolation. Molecules 2020, 25, 5585. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, Y.; Qian, X. Target-Specific Exosome Isolation through Aptamer-Based Microfluidics. Biosensors 2022, 12, 257. [Google Scholar] [CrossRef]
- Zhang, K.; Yue, Y.; Wu, S.; Liu, W.; Shi, J.; Zhang, Z. Rapid Capture and Nondestructive Release of Extracellular Vesicles Using Aptamer-Based Magnetic Isolation. ACS Sens. 2019, 4, 1245–1251. [Google Scholar] [CrossRef]
- Kimiz-Gebologlu, I.; Oncel, S.S. Exosomes: Large-Scale Production, Isolation, Drug Loading Efficiency, and Biodistribution and Uptake. J. Control. Release 2022, 347, 533–543. [Google Scholar] [CrossRef]
- Gebeyehu, A.; Kommineni, N.; Bagde, A.; Meckes, D.G.; Sachdeva, M.S. Role of Exosomes for Delivery of Chemotherapeutic Drugs. Crit. Rev. Ther. Drug Carrier Syst. 2021, 38, 53–97. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, H.; Ning, T.; Liu, D.; Deng, T.; Liu, R.; Bai, M.; Zhu, K.; Li, J.; Fan, Q.; et al. Exosome-Delivered c-Met SiRNA Could Reverse Chemoresistance to Cisplatin in Gastric Cancer. Int. J. Nanomed. 2020, 15, 2323–2335. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.; Kolluri, K.K.; Gowers, K.H.C.; Janes, S.M. TRAIL Delivery by MSC-Derived Extracellular Vesicles Is an Effective Anticancer Therapy. J. Extracell. Vesicles 2017, 6, 1265291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.F.; O’Driscoll, L. MiR-134 in Extracellular Vesicles Reduces Triple-Negative Breast Cancer Aggression and Increases Drug Sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Wang, Q.; Cao, F.; Han, B.; Xu, L. MiRNA-134 Suppresses Esophageal Squamous Cell Carcinoma Progression by Targeting FOXM1. Int. J. Clin. Exp. Pathol. 2019, 12, 2130–2138. [Google Scholar] [PubMed]
- Yang, Z.; Shi, J.; Xie, J.; Wang, Y.; Sun, J.; Liu, T.; Zhao, Y.; Zhao, X.; Wang, X.; Ma, Y.; et al. Large-Scale Generation of Functional MRNA-Encapsulating Exosomes via Cellular Nanoporation. Nat. Biomed. Eng. 2020, 4, 69–83. [Google Scholar] [CrossRef]
- Nawaz, M.; Heydarkhan-Hagvall, S.; Tangruksa, B.; González-King Garibotti, H.; Jing, Y.; Maugeri, M.; Kohl, F.; Hultin, L.; Reyahi, A.; Camponeschi, A.; et al. Lipid Nanoparticles Deliver the Therapeutic VEGFA MRNA In Vitro and In Vivo and Transform Extracellular Vesicles for Their Functional Extensions. Adv. Sci. 2023, 2206187. [Google Scholar] [CrossRef]
- Wu, M.; Ozcelik, A.; Rufo, J.; Wang, Z.; Fang, R.; Jun Huang, T. Acoustofluidic Separation of Cells and Particles. Microsyst. Nanoeng. 2019, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, F.; Rufo, J.; Chen, C.; Yang, S.; Li, L.; Zhang, J.; Cheng, J.; Kim, Y.; Wu, M.; et al. Acoustofluidic Salivary Exosome Isolation. J. Mol. Diagn. 2020, 22, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Wang, X.; Ren, J.; Lin, F.; Wu, J. Recent Advances in Acoustofluidic Separation Technology in Biology. Microsyst. Nanoeng. 2022, 8, 94. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; He, D.; Guo, Q.; Zhang, Z.; Ru, D.; Wang, L.; Gong, K.; Liu, F.; Duan, Y.; Li, H. Exosome-Liposome Hybrid Nanoparticle Codelivery of TP and MiR497 Conspicuously Overcomes Chemoresistant Ovarian Cancer. J. Nanobiotechnol. 2022, 20, 50. [Google Scholar] [CrossRef]
- Osteikoetxea, X.; Silva, A.; Lázaro-Ibáñez, E.; Salmond, N.; Shatnyeva, O.; Stein, J.; Schick, J.; Wren, S.; Lindgren, J.; Firth, M.; et al. Engineered Cas9 Extracellular Vesicles as a Novel Gene Editing Tool. J. Extracell. Vesicles 2022, 11, e12225. [Google Scholar] [CrossRef] [PubMed]
- Jafari, D.; Shajari, S.; Jafari, R.; Mardi, N.; Gomari, H.; Ganji, F.; Forouzandeh Moghadam, M.; Samadikuchaksaraei, A. Designer Exosomes: A New Platform for Biotechnology Therapeutics. BioDrugs 2020, 34, 567–586. [Google Scholar] [CrossRef] [PubMed]
- Sutaria, D.S.; Jiang, J.; Elgamal, O.A.; Pomeroy, S.M.; Badawi, M.; Zhu, X.; Pavlovicz, R.; Azevedo-Pouly, A.C.P.; Chalmers, J.; Li, C.; et al. Low Active Loading of Cargo into Engineered Extracellular Vesicles Results in Inefficient MiRNA Mimic Delivery. J. Extracell. Vesicles 2017, 6, 1333882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Z, L.; X, Z.; M, W.; X, G.; L, Z.; R, S.; W, S.; Y, D.; G, Y.; L, Y. In Vitro and in Vivo RNA Inhibition by CD9-HuR Functionalized Exosomes Encapsulated with MiRNA or CRISPR/DCas9. Nano Lett. 2019, 19, 19–28. [Google Scholar] [CrossRef]
- Antes, T.J.; Middleton, R.C.; Luther, K.M.; Ijichi, T.; Peck, K.A.; Liu, W.J.; Valle, J.; Echavez, A.K.; Marbán, E. Targeting Extracellular Vesicles to Injured Tissue Using Membrane Cloaking and Surface Display. J. Nanobiotechnol. 2018, 16, 61. [Google Scholar] [CrossRef] [PubMed]
- Martins, Á.M.; Ramos, C.C.; Freitas, D.; Reis, C.A. Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications. Cells 2021, 10, 109. [Google Scholar] [CrossRef]
- Royo, F.; Cossío, U.; de Angulo, A.R.; Llop, J.; Falcon-Perez, J.M. Modification of the Glycosylation of Extracellular Vesicles Alters Their Biodistribution in Mice. Nanoscale 2019, 11, 1531–1537. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Wang, F.; Hu, Y.; Luo, Y.; Wei, Y.; Xu, K.; Zhang, H.; Liu, H.; Bo, L.; Lv, S.; et al. Exosome-Based Bone-Targeting Drug Delivery Alleviates Impaired Osteoblastic Bone Formation and Bone Loss in Inflammatory Bowel Diseases. Cell Rep. Med. 2023, 4, 100881. [Google Scholar] [CrossRef]
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward Tailored Exosomes: The Exosomal Tetraspanin Web Contributes to Target Cell Selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- Zou, J.; Shi, M.; Liu, X.; Jin, C.; Xing, X.; Qiu, L.; Tan, W. Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy. Anal. Chem. 2019, 91, 2425–2430. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered Exosomes for Targeted Co-Delivery of MiR-21 Inhibitor and Chemotherapeutics to Reverse Drug Resistance in Colon Cancer. J. Nanobiotechnol. 2020, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liang, Y.; Li, X.; Ouyang, K.; Wang, M.; Cao, T.; Li, W.; Liu, J.; Xiong, J.; Li, B.; et al. Exosome-Mediated Delivery of Kartogenin for Chondrogenesis of Synovial Fluid-Derived Mesenchymal Stem Cells and Cartilage Regeneration. Biomaterials 2021, 269, 120539. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Han, Y.; An, Y.; Ding, Y.; He, C.; Wang, X.; Tang, Q. NRP-1 Targeted and Cargo-Loaded Exosomes Facilitate Simultaneous Imaging and Therapy of Glioma in vitro and in vivo. Biomaterials 2018, 178, 302–316. [Google Scholar] [CrossRef] [PubMed]
| Isolation Technique | Principle | Recovery (%) | Pros | Cons | References |
|---|---|---|---|---|---|
| Ultracentrifugation | Sedimentation rate | 5–20 | High sample capacity and low cost | Time-consuming and low purity | [9,10] |
| Density gradient ultracentrifugation | Density, size, shape | 10–40 | High purity and protein concentration | Long run time and low yield | [9,11,12] |
| Polymer-based precipitation | Sedimentation rate | 90+ | High yield | Low purity | [13,14,15] |
| Ultrafiltration | Size | 30 | Maintains integrity; simple and low-cost | Moderate purity; low yield due to exosome trapping in filter pores | [9,16,17] |
| Size-exclusion chromatography | Size | 40–80 | High purity, integrity, and functionality; reduction of exosome aggregation | Low extraction volume | [9,18] |
| Immunoaffinity chromatography | Surface marker | 90+ | Maintain integrity | Low capacity and low yield | [9,19,20] |
| Microfluidics | Surface marker | 40–90 | Low cost and low input sample required | Low sample capacity; cargo may be modified | [9,21,22] |
| Magnetic bead isolation | Surface marker | 80+ | Maintain integrity | Possible impurities | [23,24] |
| Methods | Principle | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Pre-secretory Drug Loading | ||||
| Co-incubation | Drug incubated with parent cell | Easy; effective in hydrophobic drugs | Low loading efficacy; possible drug toxicity | [31] |
| Gene editing | Editing of genes | Overexpression of specific molecules | Low loading efficacy; possible toxicity | [32] |
| Post-Secretory Drug Loading | ||||
| Sonication | Mechanical shear force decreases membrane integrity | Large amount of drug loaded | Possible damage to intracellular components and integrity | [3,33,34] |
| Electroporation | High-voltage electric charge decreases membrane integrity | Effective loading of hydrophilic drugs and nucleic acids | Possible aggregation; low loading efficacy | [35] |
| Passive incubation | Passive diffusion | Effective loading of hydrophobic drugs; does not affect exosome integrity | Not useful for hydrophilic drugs; low drug-loading capacity | [3,34,36,37,38,39] |
| Freeze–thaw | Repeated freeze–thaw cycles to decrease membrane integrity | Easy process | Low loading efficacy; possible aggregation and inactivation | [3,40] |
| Nanoporation | Nanosecond electrical pulse decreases membrane integrity | Effective loading of small molecules | Possible aggregation | [41,42] |
| Saponin treatment | Formation of porous structure on exosome membrane | Increased loading capacity compared to electroporation | May cause hemolysis in vivo; requires further purification | [3,43] |
| Extrusion | Mechanical stress decreases membrane integrity | Provides uniform distribution | May damage membrane; possible drug leakage | [3,44] |
| Upstream Modifications | Fold Increase | Alterations and Effects | References |
|---|---|---|---|
| Soluble Factors | |||
| Lipopolysaccharide (LPS) | 1.37 | Upregulation of let-7b increased immunotherapeutic effect | [62] |
| N-methyldopamine and norepinephrine | 3 | No significant change | [63] |
| Serotonin and calcium | 2–2.5 | - | [64] |
| Adiponectin | 3 | Present in exosomes | [65] |
| ATP | 4 | No significant change | [66] |
| Wnt3a | - | Present in exosomes; increased neuroprotective abilities | [67] |
| Plant ceramide | 2.5 | - | [68] |
| Chemical/physical stimulation | |||
| Hypoxia | 1.5 | Dependent on cell type; increased expression of nucleic acids and proteins | [71,72,74,75] |
| Serum deprivation | Varies | Decreased exosome protein content | [52,76] |
| Flow/stretch | 37 | Over 200 proteins expressed differently from typical exosomes | [77,78] |
| High-frequency ultrasound | 8–10 | Increased exosome protein content | [79] |
| 3D cultivation | |||
| 3D spheroid culture | 2–3 | - | [80] |
| Microcarrier-based suspension | 20; 140 with tangential flow system | No significant change | [52,81,82,83] |
| 3D print fibrillar scaffold with perfusion system | 100 | Decreased exosome protein content | [84] |
| Low-shear unsubmerged 3D-printed polylactic acid lattice matrix | 2 | Maintained protein expression | [85] |
| Biomaterials | |||
| Nitric oxide-releasing polymer | Not significant | Enhanced pro-angiogenic activity | [86] |
| Lithium-incorporated bioactive glass ceramic | Not significant | Enhanced pro-angiogenic activity | [87] |
| Iron-oxide coated poly-lactic-co-glycosidic acid (PLGA) nanoparticle | 2 | Increased antioxidant or tissue regeneration factors | [88] |
| Bioglass | 2 | Modulation of cargo through altered expression of microRNA; enhanced ability to promote vascularization | [89] |
| EXOtic | ~6.8 | - | [69] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richards, T.; Patel, H.; Patel, K.; Schanne, F. Endogenous Lipid Carriers—Bench-to-Bedside Roadblocks in Production and Drug Loading of Exosomes. Pharmaceuticals 2023, 16, 421. https://doi.org/10.3390/ph16030421
Richards T, Patel H, Patel K, Schanne F. Endogenous Lipid Carriers—Bench-to-Bedside Roadblocks in Production and Drug Loading of Exosomes. Pharmaceuticals. 2023; 16(3):421. https://doi.org/10.3390/ph16030421
Chicago/Turabian StyleRichards, Terjahna, Himaxi Patel, Ketan Patel, and Frank Schanne. 2023. "Endogenous Lipid Carriers—Bench-to-Bedside Roadblocks in Production and Drug Loading of Exosomes" Pharmaceuticals 16, no. 3: 421. https://doi.org/10.3390/ph16030421
APA StyleRichards, T., Patel, H., Patel, K., & Schanne, F. (2023). Endogenous Lipid Carriers—Bench-to-Bedside Roadblocks in Production and Drug Loading of Exosomes. Pharmaceuticals, 16(3), 421. https://doi.org/10.3390/ph16030421