Evaluating Known Zika Virus NS2B-NS3 Protease Inhibitor Scaffolds via In Silico Screening and Biochemical Assays
Abstract
:1. Introduction
2. Results and Discussion
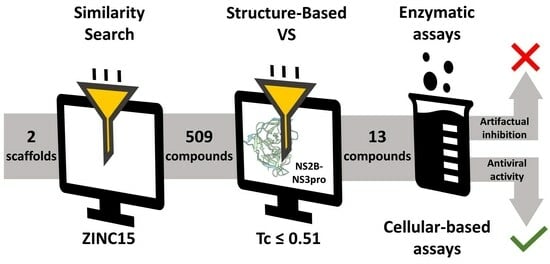
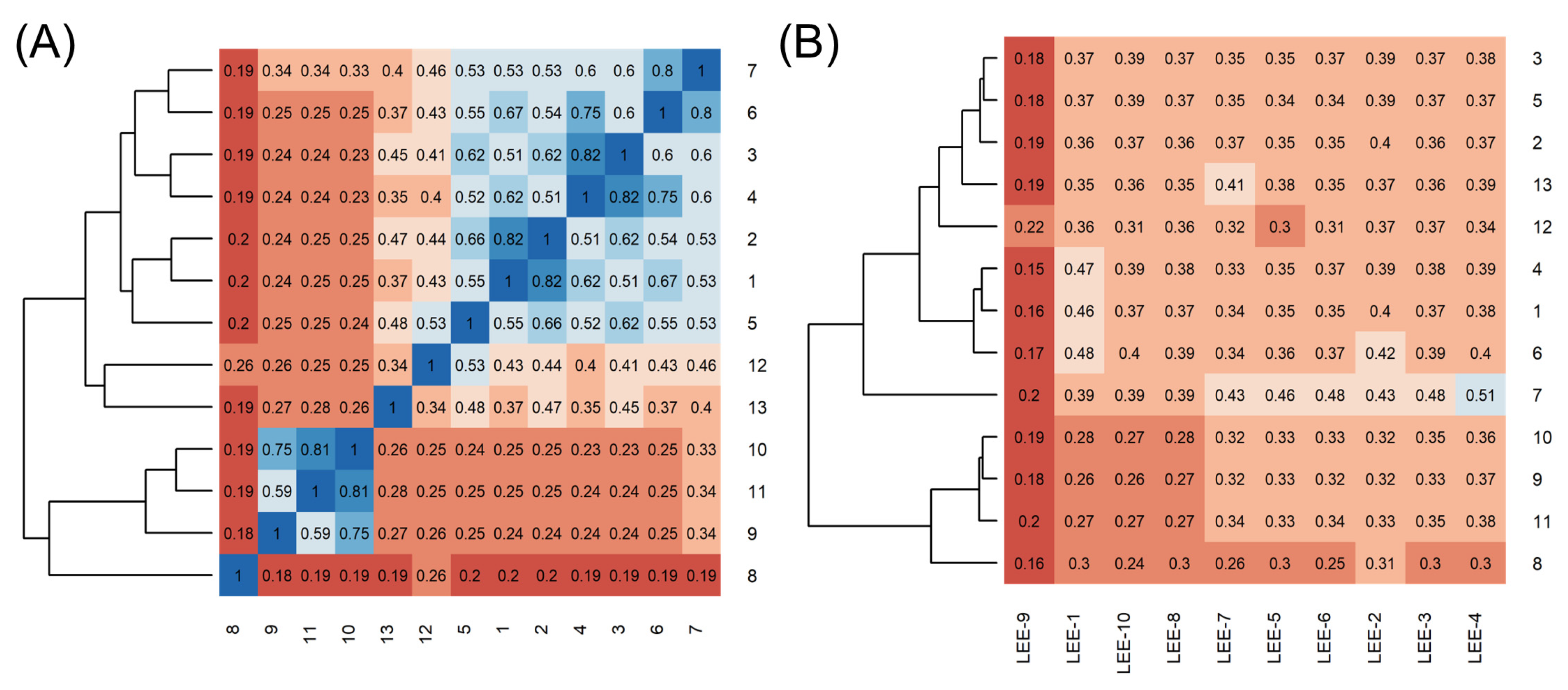
2.1. Using Similarity Search to Build a Compound Library for Virtual Screening
2.2. Virtual Screening of Compounds Based on Competitive Inhibitors
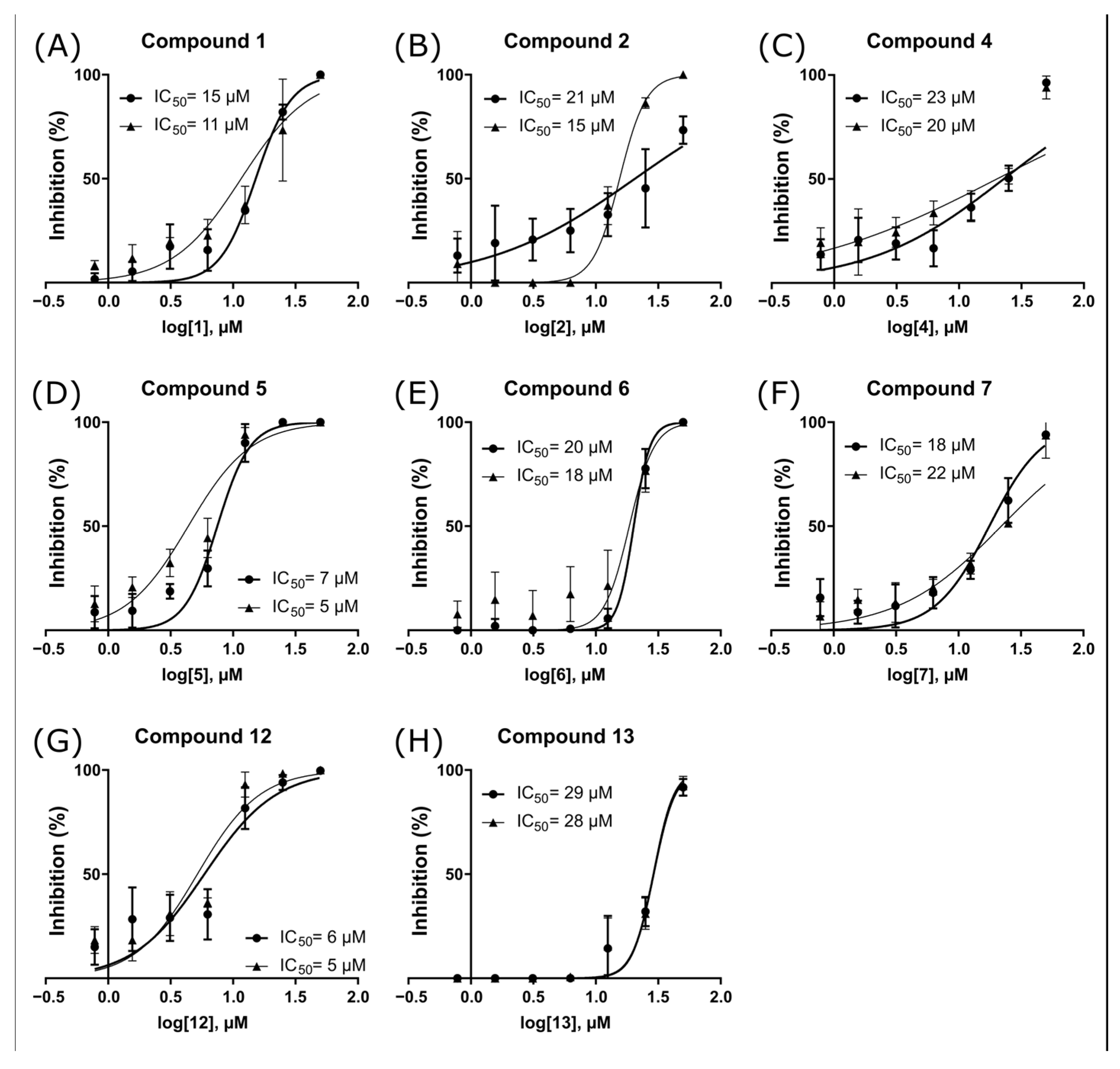
2.3. ZIKV NS2B-NS3pro Inhibitory Assays
2.4. Antiviral Activity Evaluation against ZIKV
3. Materials and Methods
3.1. Similarity Search Approach
3.2. Dataset Preparation
3.3. Virtual Screening by Molecular Docking
3.4. NS2B-NS3pro Expression and Purification
3.5. NS2B-NS3pro Enzyme Assays
3.6. Cruzain Enzyme Assays
3.7. Cell Lineage and Virus Strain
3.8. Viral Propagation
3.9. Viral Titration
3.10. Cytotoxicity Assay: 50% Cytotoxic Concentration (CC50)
3.11. Antiviral Activity Assay: 50% Effective Concentration (EC50)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dick, G.W.A. Zika Virus (I). Isolations and Serological Specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.-J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the Genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Mayor, S. Zika Infection in Pregnancy Is Linked to Range of Fetal Abnormalities, Data Indicate. BMJ 2016, 352, i1362. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, T.V.B.; Rodrigues, L.C.; de Alencar Ximenes, R.A.; de Barros Miranda-Filho, D.; Montarroyos, U.R.; de Melo, A.P.L.; Valongueiro, S.; de Albuquerque, M.d.F.P.M.; Souza, W.V.; Braga, C.; et al. Association between Zika Virus Infection and Microcephaly in Brazil, January to May, 2016: Preliminary Report of a Case-Control Study. Lancet Infect. Dis. 2016, 16, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Brasil, P.; Sequeira, P.C.; Freitas, A.D.A.; Zogbi, H.E.; Calvet, G.A.; De Souza, R.V.; Siqueira, A.M.; De Mendonca, M.C.L.; Nogueira, R.M.R.; De Filippis, A.M.B.; et al. Guillain-Barré Syndrome Associated with Zika Virus Infection. Lancet 2016, 387, 1482. [Google Scholar] [CrossRef] [PubMed]
- Gulland, A. Zika Virus Is a Global Public Health Emergency, Declares WHO. BMJ 2016, 352, i657. [Google Scholar] [CrossRef]
- World Health Organisation, 2022 Zika Epidemiology Update—February 2022. Available online: https://www.who.int/publications/m/item/zika-epidemiology-update---february-2022 (accessed on 20 June 2023).
- Pierson, T.C.; Diamond, M.S. The Continued Threat of Emerging Flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef]
- Mottin, M.; Borba, J.V.V.B.; Braga, R.C.; Torres, P.H.M.; Martini, M.C.; Proenca-Modena, J.L.; Judice, C.C.; Costa, F.T.M.; Ekins, S.; Perryman, A.L.; et al. The A–Z of Zika Drug Discovery. Drug Discov. Today 2018, 23, 1833–1847. [Google Scholar] [CrossRef]
- Chen, X.; Yang, K.; Wu, C.; Chen, C.; Hu, C.; Buzovetsky, O.; Wang, Z.; Ji, X.; Xiong, Y.; Yang, H. Mechanisms of Activation and Inhibition of Zika Virus NS2B-NS3 Protease. Cell Res. 2016, 26, 1260–1263. [Google Scholar] [CrossRef]
- Kang, C.B.; Keller, T.H.; Luo, D. Zika Virus Protease: An Antiviral Drug Target. Trends Microbiol. 2017, 25, 797–808. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Cortese, M.; Acosta, E.G.; Bartenschlager, R. Rewiring Cellular Networks by Members of the Flaviviridae Family. Nat. Rev. Microbiol. 2018, 16, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Phoo, W.W.; Zhang, Z.; Wirawan, M.; Chew, E.J.C.; Chew, A.B.L.; Kouretova, J.; Steinmetzer, T.; Luo, D. Structures of Zika Virus NS2B-NS3 Protease in Complex with Peptidomimetic Inhibitors. Antivir. Res. 2018, 160, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Phoo, W.W.; Li, Y.; Zhang, Z.; Lee, M.Y.; Loh, Y.R.; Tan, Y.B.; Ng, E.Y.; Lescar, J.; Kang, C.; Luo, D. Structure of the NS2B-NS3 Protease from Zika Virus after Self-Cleavage. Nat. Commun. 2016, 7, 13410. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C. Proteases from Dengue, West Nile and Zika Viruses as Drug Targets. Biophys. Rev. 2019, 11, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The Flavivirus NS2B-NS3 Protease-Helicase as a Target for Antiviral Drug Development. Antivir. Res. 2015, 118, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Wang, W.; Liu, S.; Chen, M.W.; Hung, A.W.; Keller, T.H.; Luo, D.; et al. Structural Dynamics of Zika Virus NS2B-NS3 Protease Binding to Dipeptide Inhibitors. Structure 2017, 25, 1242–1250. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Li, R.; Yang, H.Y.; Jansson, A.E.; Hill, J.; Keller, T.H.; Nacro, K.; et al. Structural Insights into the Inhibition of Zika Virus NS2B-NS3 Protease by a Small-Molecule Inhibitor. Structure 2018, 26, 555–564. [Google Scholar] [CrossRef]
- Lei, J.; Hansen, G.; Nitsche, C.; Klein, C.D.; Zhang, L.; Hilgenfeld, R. Crystal Structure of Zika Virus Ns2b-Ns3 Protease in Complex with a Boronate Inhibitor. Science 2016, 353, 503–505. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.B.; Luo, D. Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef]
- Voss, S.; Nitsche, C. Inhibitors of the Zika Virus Protease NS2B-NS3. Bioorg. Med. Chem. Lett. 2020, 30, 126965. [Google Scholar] [CrossRef]
- Wang, L.; Liang, R.; Gao, Y.; Li, Y.; Deng, X.; Xiang, R.; Zhang, Y.; Ying, T.; Jiang, S.; Yu, F. Development of Small-Molecule Inhibitors Against Zika Virus Infection. Front. Microbiol. 2019, 10, 2725. [Google Scholar] [CrossRef] [PubMed]
- Mottin, M.; De Paula Sousa, B.K.; De Moraes Roso Mesquita, N.C.; De Oliveira, K.I.Z.; Noske, G.D.; Sartori, G.R.; De Oliveira Albuquerque, A.; Urbina, F.; Puhl, A.C.; Moreira-Filho, J.T.; et al. Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika. J. Chem. Inf. Model. 2022, 62, 6825–6843. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, J.F.W.; den-Haan, H.; Chik, K.K.H.; Zhang, A.J.; Chan, C.C.S.; Poon, V.K.M.; Yip, C.C.Y.; Mak, W.W.N.; Zhu, Z.; et al. Structure-Based Discovery of Clinically Approved Drugs as Zika Virus NS2B-NS3 Protease Inhibitors That Potently Inhibit Zika Virus Infection in Vitro and in Vivo. Antivir. Res. 2017, 145, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Retallack, H.; Di Lullo, E.; Arias, C.; Knopp, K.A.; Laurie, M.T.; Sandoval-Espinosa, C.; Leon, W.R.M.; Krencik, R.; Ullian, E.M.; Spatazza, J.; et al. Zika Virus Cell Tropism in the Developing Human Brain and Inhibition by Azithromycin. Proc. Natl. Acad. Sci. USA 2016, 113, 14408–14413. [Google Scholar] [CrossRef]
- Bullard-Feibelman, K.M.; Govero, J.; Zhu, Z.; Salazar, V.; Veselinovic, M.; Diamond, M.S.; Geiss, B.J. The FDA-Approved Drug Sofosbuvir Inhibits Zika Virus Infection. Antivir. Res. 2017, 137, 134–140. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Powell, S.T.; Prasanth, K.R.; Schott-Lerner, G.; Soto-Acosta, R.; Galarza-Muñoz, G.; McGrath, E.L.; Urrabaz-Garza, R.; Gao, J.; et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 2016, 20, 259–270. [Google Scholar] [CrossRef]
- Santos, F.R.S.; Nunes, D.A.F.; Lima, W.G.; Davyt, D.; Santos, L.L.; Taranto, A.G.; Ferreira, M.S.J. Identification of Zika Virus NS2B-NS3 Protease Inhibitors by Structure-Based Virtual Screening and Drug Repurposing Approaches. J. Chem. Inf. Model. 2020, 60, 731–737. [Google Scholar] [CrossRef]
- Nitsche, C.; Passioura, T.; Varava, P.; Mahawaththa, M.C.; Leuthold, M.M.; Klein, C.D.; Suga, H.; Otting, G. De Novo Discovery of Nonstandard Macrocyclic Peptides as Noncompetitive Inhibitors of the Zika Virus NS2B-NS3 Protease. ACS Med. Chem. Lett. 2019, 10, 168–174. [Google Scholar] [CrossRef]
- da Silva-Júnior, E.F.; de Araújo-Júnior, J.X. Peptide Derivatives as Inhibitors of NS2B-NS3 Protease from Dengue, West Nile, and Zika Flaviviruses. Bioorg. Med. Chem. 2019, 27, 3963–3978. [Google Scholar] [CrossRef]
- Ji, M.; Zhu, T.; Xing, M.; Luan, N.; Mwangi, J.; Yan, X.; Mo, G.; Rong, M.; Li, B.; Lai, R.; et al. An Antiviral Peptide from Alopecosa Nagpag Spider Targets NS2B-NS3 Protease of Flaviviruses. Toxins 2019, 11, 584. [Google Scholar] [CrossRef]
- Rassias, G.; Zogali, V.; Swarbrick, C.M.D.; Ki Chan, K.W.; Chan, S.A.; Gwee, C.P.; Wang, S.; Kaplanai, E.; Canko, A.; Kiousis, D.; et al. Cell-Active Carbazole Derivatives as Inhibitors of the Zika Virus Protease. Eur. J. Med. Chem. 2019, 180, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ren, J.; Nocadello, S.; Rice, A.J.; Ojeda, I.; Light, S.; Minasov, G.; Vargas, J.; Nagarathnam, D.; Anderson, W.F.; et al. Identification of Novel Small Molecule Inhibitors against NS2B/NS3 Serine Protease from Zika Virus. Antivir. Res. 2017, 139, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Huo, T.; Lin, Y.L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef] [PubMed]
- Millies, B.; Von Hammerstein, F.; Gellert, A.; Hammerschmidt, S.; Barthels, F.; Göppel, U.; Immerheiser, M.; Elgner, F.; Jung, N.; Basic, M.; et al. Proline-Based Allosteric Inhibitors of Zika and Dengue Virus NS2B/NS3 Proteases. J. Med. Chem. 2019, 62, 11359–11382. [Google Scholar] [CrossRef] [PubMed]
- Brecher, M.; Li, Z.; Liu, B.; Zhang, J.; Koetzner, C.A.; Alifarag, A.; Jones, S.A.; Lin, Q.; Kramer, L.D.; Li, H. A Conformational Switch High-Throughput Screening Assay and Allosteric Inhibition of the Flavivirus NS2B-NS3 Protease. PLoS Pathog. 2017, 13, e1006411. [Google Scholar] [CrossRef] [PubMed]
- Willett, P.; Barnard, J.M.; Downs, G.M. Chemical Similarity Searching. J. Chem. Inf. Comput. Sci. 1998, 38, 983–996. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Guha, R.; Cherto, M.R. Rcdk: Integrating the CDK with R. Chem. Inform. Funct. R 2017, 1–22. [Google Scholar]
- Tanimoto, T.T. An Elementary Mathematical Theory of Classification and Prediction; International Business Machine Corperation: New York, NY, USA, 1958; pp. 1–11. [Google Scholar]
- Santos, L.H.; Caffarena, E.R.; Ferreira, R.S. PH and Non-Covalent Ligand Binding Modulate Zika Virus NS2B/NS3 Protease Binding Site Residues: Discoveries from MD and Constant PH MD Simulations. J. Biomol. Struct. Dyn. 2022, 40, 10359–10372. [Google Scholar] [CrossRef]
- Mukherjee, S.; Balius, T.E.; Rizzo, R.C. Docking Validation Resources: Protein Family and Ligand Flexibility Experiments. J. Chem. Inf. Model. 2010, 50, 1986–2000. [Google Scholar] [CrossRef]
- Balius, T.E.; Allen, W.J.; Mukherjee, S.; Rizzo, R.C. Grid-Based Molecular Footprint Comparison Method for Docking and de Novo Design: Application to HIVgp41. J. Comput. Chem. 2013, 34, 1226–1240. [Google Scholar] [CrossRef]
- Hilgenfeld, R.; Lei, J.; Zhang, L. The Structure of the Zika Virus Protease, NS2B/NS3pro. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; Volume 1062, pp. 131–145. 3p. [Google Scholar]
- Campos, D.M.O.; Bezerra, K.S.; Esmaile, S.C.; Fulco, U.L.; Albuquerque, E.L.; Oliveira, J.I.N. Intermolecular Interactions of Cn-716 and Acyl-KR-Aldehyde Dipeptide Inhibitors against Zika Virus. Phys. Chem. Chem. Phys. 2020, 22, 15683–15695. [Google Scholar] [CrossRef]
- Fischer, A.; Smieško, M.; Sellner, M.; Lill, M.A. Decision Making in Structure-Based Drug Discovery: Visual Inspection of Docking Results. J. Med. Chem. 2021, 64, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Bock, S.; Snitko, M.; Berger, T.; Weidner, T.; Holloway, S.; Kanitz, M.; Diederich, W.E.; Steuber, H.; Walter, C.; et al. Novel Dengue Virus NS2B/NS3 Protease Inhibitors. Antimicrob. Agents Chemother. 2015, 59, 1100–1109. [Google Scholar] [CrossRef]
- Hue, B.T.B.; Nguyen, P.H.; De, T.Q.; Van Hieu, M.; Jo, E.; Van Tuan, N.; Thoa, T.T.; Anh, L.D.; Son, N.H.; La Duc Thanh, D.; et al. Benzimidazole Derivatives as Novel Zika Virus Inhibitors. ChemMedChem 2020, 15, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Dibrov, S.M.; Parsons, J.; Carnevali, M.; Zhou, S.; Rynearson, K.D.; Ding, K.; Garcia Sega, E.; Brunn, N.D.; Boerneke, M.A.; Castaldi, M.P.; et al. Hepatitis C Virus Translation Inhibitors Targeting the Internal Ribosomal Entry Site. J. Med. Chem. 2014, 57, 1694–1707. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, A.; Ferreira, R.S.; Klumpp, C.; Mott, B.T.; Austin, C.P.; Inglese, J.; Thomas, C.J.; Maloney, D.J.; Shoichet, B.K.; Simeonov, A. Quantitative Analyses of Aggregation, Autofluorescence, and Reactivity Artifacts in a Screen for Inhibitors of a Thiol Protease. J. Med. Chem. 2010, 53, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Shoichet, B.K. Screening in a Spirit Haunted World. Drug Discov. Today 2006, 11, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Babaoglu, K.; Simconov, A.; Irwin, J.J.; Nelson, M.E.; Feng, B.; Thomas, C.J.; Cancian, L.; Costi, M.P.; Maltby, D.A.; Jadhav, A.; et al. Comprehensive Mechanistic Analysis of Hits from High-Throughput and Docking Screens against β-Lactamase. J. Med. Chem. 2008, 51, 2502–2511. [Google Scholar] [CrossRef]
- Feng, B.Y.; Shoichet, B.K. A Detergent-Based Assay for the Detection of Promiscuous Inhibitors. Nat. Protoc. 2006, 1, 550–553. [Google Scholar] [CrossRef]
- Dias, R.F.C.; Ribeiro, B.M.R.M.; Cassani, N.M.; Farago, D.N.; Antoniucci, G.A.; Rocha, R.E.O.; de Souza, F.O. Discovery and Structural Optimization of a Novel N-Acyl-2-Aminobenzothiazole Series as Potent Inhibitors of Zika Virus; Instituto de Química, Universidade Federal de Uberlândia: Uberlândia, Brazil, 2023; (submitted for publication). [Google Scholar]
- Santos, V.C.; Leite, P.G.; Santos, L.H.; Pascutti, P.G.; Kolb, P.; Machado, F.S.; Ferreira, R.S. Structure-Based Discovery of Novel Cruzain Inhibitors with Distinct Trypanocidal Activity Profiles. Eur. J. Med. Chem. 2023, 257, 115498. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Guo, C.; Hansell, E.; Doyle, P.S.; Caffrey, C.R.; Holler, T.P.; McKerrow, J.H.; Cohen, F.E. Synthesis and Structure-Activity Relationship Study of Potent Trypanocidal Thio Semicarbazone Inhibitors of the Trypanosomal Cysteine Protease Cruzain. J. Med. Chem. 2002, 45, 2695–2707. [Google Scholar] [CrossRef] [PubMed]
- Barbosa Da Silva, E.; Rocha, D.A.; Fortes, I.S.; Yang, W.; Monti, L.; Siqueira-Neto, J.L.; Caffrey, C.R.; McKerrow, J.; Andrade, S.F.; Ferreira, R.S. Structure-Based Optimization of Quinazolines as Cruzain and TbrCATL Inhibitors. J. Med. Chem. 2021, 64, 13054–13071. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.S.; Bryant, C.; Ang, K.K.H.; McKerrow, J.H.; Shoichet, B.K.; Renslo, A.R. Divergent Modes of Enzyme Inhibition in a Homologous Structure-Activity Series. J. Med. Chem. 2009, 52, 5005–5008. [Google Scholar] [CrossRef]
- Stork, C.; Chen, Y.; Šícho, M.; Kirchmair, J. Hit Dexter 2.0: Machine-Learning Models for the Prediction of Frequent Hitters. J. Chem. Inf. Model. 2019, 59, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Duan, D.; Torosyan, H.; Doak, A.K.; Ziebart, K.T.; Sterling, T.; Tumanian, G.; Shoichet, B.K. An Aggregation Advisor for Ligand Discovery. J. Med. Chem. 2015, 58, 7076–7087. [Google Scholar] [CrossRef]
- Seidler, J.; McGovern, S.L.; Doman, T.N.; Shoichet, B.K. Identification and Prediction of Promiscuous Aggregating Inhibitors among Known Drugs. J. Med. Chem. 2003, 46, 4477–4486. [Google Scholar] [CrossRef]
- Nunes, D.A.D.F.; Santos, F.R.D.S.; da Fonseca, S.T.D.; de Lima, W.G.; Nizer, W.S.D.C.; Ferreira, J.M.S.; de Magalhaes, J.C. NS2B-NS3 Protease Inhibitors as Promising Compounds in the Development of Antivirals against Zika Virus: A Systematic Review. J. Med. Virol. 2022, 94, 442–453. [Google Scholar] [CrossRef]
- Mirza, M.U.; Alanko, I.; Vanmeert, M.; Muzzarelli, K.M.; Salo-Ahen, O.M.H.; Abdullah, I.; Kovari, I.A.; Claes, S.; De Jonghe, S.; Schols, D.; et al. The Discovery of Zika Virus NS2B-NS3 Inhibitors with Antiviral Activity via an Integrated Virtual Screening Approach. Eur. J. Pharm. Sci. 2022, 175, 106220. [Google Scholar] [CrossRef]
- Serafim, M.S.M.; Kronenberger, T.; de Oliveira, R.B.; Kroon, E.G.; Abrahão, J.S.; Mota, B.E.F.; Maltarollo, V.G. Synthetic Curcumin Analogues Present Antiflavivirus Activity In Vitro with Potential Multiflavivirus Activity from a Thiazolylhydrazone Moiety. Future Pharmacol. 2023, 3, 364–378. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Bajusz, D.; Rácz, A.; Héberger, K. Why Is Tanimoto Index an Appropriate Choice for Fingerprint-Based Similarity Calculations? J. Cheminform. 2015, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. An Accessory Software Package For Molecular Mechanical Calculations. J. Am. Chem. Soc 2001, 222, U403. [Google Scholar]
- Allen, W.J.; Balius, T.E.; Mukherjee, S.; Brozell, S.R.; Moustakas, D.T.; Lang, P.T.; Case, D.A.; Kuntz, I.D.; Rizzo, R.C. DOCK 6: Impact of New Features and Current Docking Performance. J. Comput. Chem. 2015, 36, 1132–1156. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- do Valle Moreira, T.; Martins, L.C.; Diniz, L.A.; Bernardes, T.C.D.; de Oliveira, R.B.; Ferreira, R.S. Screening the Pathogen Box to Discover and Characterize New Cruzain and TbrCatL Inhibitors. Pathogens 2023, 12, 251. [Google Scholar] [CrossRef]
- Fonseca, N.C.; Da Cruz, L.F.; Da Silva Villela, F.; Do Nascimento Pereira, G.A.; De Siqueira-Neto, J.L.; Kellar, D.; Suzuki, B.M.; Ray, D.; De Souza, T.B.; Alves, R.J.; et al. Synthesis of a Sugar-Based Thiosemicarbazone Series and Structure-Activity Relationship versus the Parasite Cysteine Proteases Rhodesain, Cruzain, and Schistosoma Mansoni Cathepsin B1. Antimicrob. Agents Chemother. 2015, 59, 2666–2677. [Google Scholar] [CrossRef]
- Donald, C.L.; Brennan, B.; Cumberworth, S.L.; Rezelj, V.V.; Clark, J.J.; Cordeiro, M.T.; Freitas de Oliveira França, R.; Pena, L.J.; Wilkie, G.S.; Da Silva Filipe, A.; et al. Full Genome Sequence and SfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. PLoS Negl. Trop. Dis. 2016, 10, e0005048. [Google Scholar] [CrossRef]
- Coelho, S.V.A.; Neris, R.L.S.; Papa, M.P.; Schnellrath, L.C.; Meuren, L.M.; Tschoeke, D.A.; Leomil, L.; Verçoza, B.R.F.; Miranda, M.; Thompson, F.L.; et al. Development of Standard Methods for Zika Virus Propagation, Titration, and Purification. J. Virol. Methods 2017, 246, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]

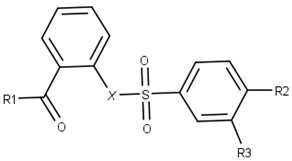 | ||||
| Compound | R1 | R2 | R3 | X |
| 1 | 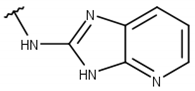 | 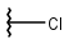 | 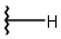 |  |
| 2 |  | 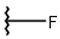 | 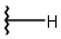 |  |
| 3 | 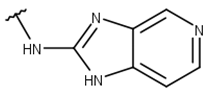 | 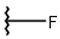 | 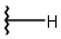 |  |
| 4 |  | 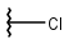 | 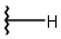 |  |
| 5 |  | 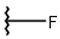 | 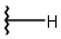 |  |
| 6 | 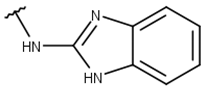 | 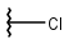 | 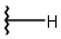 |  |
| 7 |  | 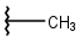 | 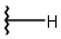 |  |
| 8 | 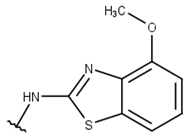 | 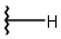 | 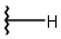 | 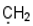 |
| 9 | 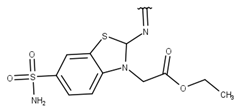 | 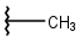 | 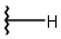 |  |
| 10 | 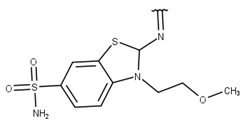 | 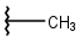 | 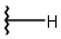 |  |
| 11 | 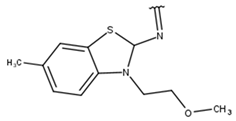 | 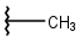 | 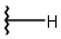 |  |
| 12 |  | 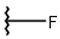 | 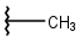 |  |
| 13 | 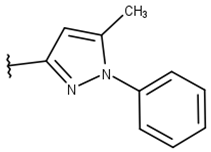 | 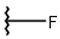 | 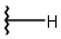 |  |
| % ZIKV NS2B/NS3pro Inhibition (100 µM) | ZIKV NS2B/NS3pro IC50 (µM) | % ZIKV NS2B/NS3pro Inhibition | % Cruzain Inhibition b | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Compound | With Preincubation | Without Preincubation | 0.001% Triton | 0.01% Triton | BSA | [Enzyme] = 0.2 nM b | [Enzyme] = 2 nM b | ||
| 1 | 100 ± 0 | 100 ± 0 | 93 ± 8 | 47 ± 34 | 69 ± 6 | 13 ± 3 | 45 ± 0.1 | 26 ± 7 | 97 ± 7 |
| 2 | 100 ± 0 | 100 ± 0 | 94 ± 7 | 30 ± 19 | 32 ± 2 | 18 ± 4 | 30 ± 0.1 | 28 ± 7 | 42 ± 11 |
| 3 | 57 ± 22 | 56 ± 11 | ND | ND | ND | ND | ND | ND | ND |
| 4 | 100 ± 0 | 100 ± 0 | 93 ± 8 | 25 ± 15 | 45 ± 4 | 21 ± 2 | 44 ± 0.2 | 29 ± 8 | −12 ± 9.1 c |
| 5 | 100 ± 0 | 100 ± 0 | 93 ± 3 | 6 ± 8 | 94 ± 8 | 5 ± 3 | 62 ± 0.1 | 23 ± 8 | 69 ± 24 |
| 6 | 100 ± 0 | 99 ± 2 | 89 ± 6 | 7 ± 9 | 55 ± 20 | 19 ± 2 | 99 ± 0.4 | 66 ± 21 | ND |
| 7 | 100 ± 0 | 90 ± 8 | 88 ± 5 | 12 ± 17 | 89 ± 7 | 19 ± 4 | 87 ± 0.04 | 27 ± 6 | ND |
| 8 a | 8 ± 8 | 15 ± 11 | ND | ND | ND | ND | ND | ND | ND |
| 9 a | 62 ± 8 | 58 ± 8 | 90 ± 3 | 12 ± 7 | 0 | ND | ND | ND | ND |
| 10 a | 42 ± 3 | 30 ± 12 | 87 ± 3 | 8 ± 3 | 0 | ND | ND | ND | ND |
| 11 a | 13 ± 4 | 9 ± 7 | ND | ND | ND | ND | ND | ND | ND |
| 12 | 100 ± 0 | 91 ± 9 | 90 ± 5 | 5 ± 7 | 87 ± 18 | 5 ± 1 | 94 ± 0.06 | 30 ± 10 | ND |
| 13 | 95 ± 2 | 100 ± 1 | 100 ± 0 | 45 ± 9 | 57 ± 15 | 28 ± 1 | 43 ± 0.1 | 26 ± 10 | ND |
| Compound | CC50 (µM) | EC50 (µM) |
|---|---|---|
| 1 | 22 ± 0.79 | NA |
| 2 | 32.47 ± 1.95 | NA |
| 3 | 119.79 ± 3.68 | NA |
| 4 | 57.11 ± 1.86 | NA |
| 5 | <12.5 | NA |
| 6 | 38.36 ± 2.18 | NA |
| 7 | 32.76 ± 1.55 | NA |
| 8 | 23.57 ± 1 | NA |
| 9 | 100.95 ± 4.23 | 50 |
| 10 | 76.21 ± 3.81 | NA |
| 11 | 73.36 ± 4.02 | 50 |
| 12 | <12.5 | NA |
| 13 | <12.5 | NA |
| Ribavirin a | >100 | 4.1 ± 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, L.H.; Rocha, R.E.O.; Dias, D.L.; Ribeiro, B.M.R.M.; Serafim, M.S.M.; Abrahão, J.S.; Ferreira, R.S. Evaluating Known Zika Virus NS2B-NS3 Protease Inhibitor Scaffolds via In Silico Screening and Biochemical Assays. Pharmaceuticals 2023, 16, 1319. https://doi.org/10.3390/ph16091319
Santos LH, Rocha REO, Dias DL, Ribeiro BMRM, Serafim MSM, Abrahão JS, Ferreira RS. Evaluating Known Zika Virus NS2B-NS3 Protease Inhibitor Scaffolds via In Silico Screening and Biochemical Assays. Pharmaceuticals. 2023; 16(9):1319. https://doi.org/10.3390/ph16091319
Chicago/Turabian StyleSantos, Lucianna H., Rafael E. O. Rocha, Diego L. Dias, Beatriz M. R. M. Ribeiro, Mateus Sá M. Serafim, Jônatas S. Abrahão, and Rafaela S. Ferreira. 2023. "Evaluating Known Zika Virus NS2B-NS3 Protease Inhibitor Scaffolds via In Silico Screening and Biochemical Assays" Pharmaceuticals 16, no. 9: 1319. https://doi.org/10.3390/ph16091319
APA StyleSantos, L. H., Rocha, R. E. O., Dias, D. L., Ribeiro, B. M. R. M., Serafim, M. S. M., Abrahão, J. S., & Ferreira, R. S. (2023). Evaluating Known Zika Virus NS2B-NS3 Protease Inhibitor Scaffolds via In Silico Screening and Biochemical Assays. Pharmaceuticals, 16(9), 1319. https://doi.org/10.3390/ph16091319