Unite and Conquer: Association of Two G-Quadruplex Aptamers Provides Antiproliferative and Antimigration Activity for Cells from High-Grade Glioma Patients
Abstract
:1. Introduction
2. Results
2.1. Assessment of Bi-(AID-1-T) Specificity to the Target Protein
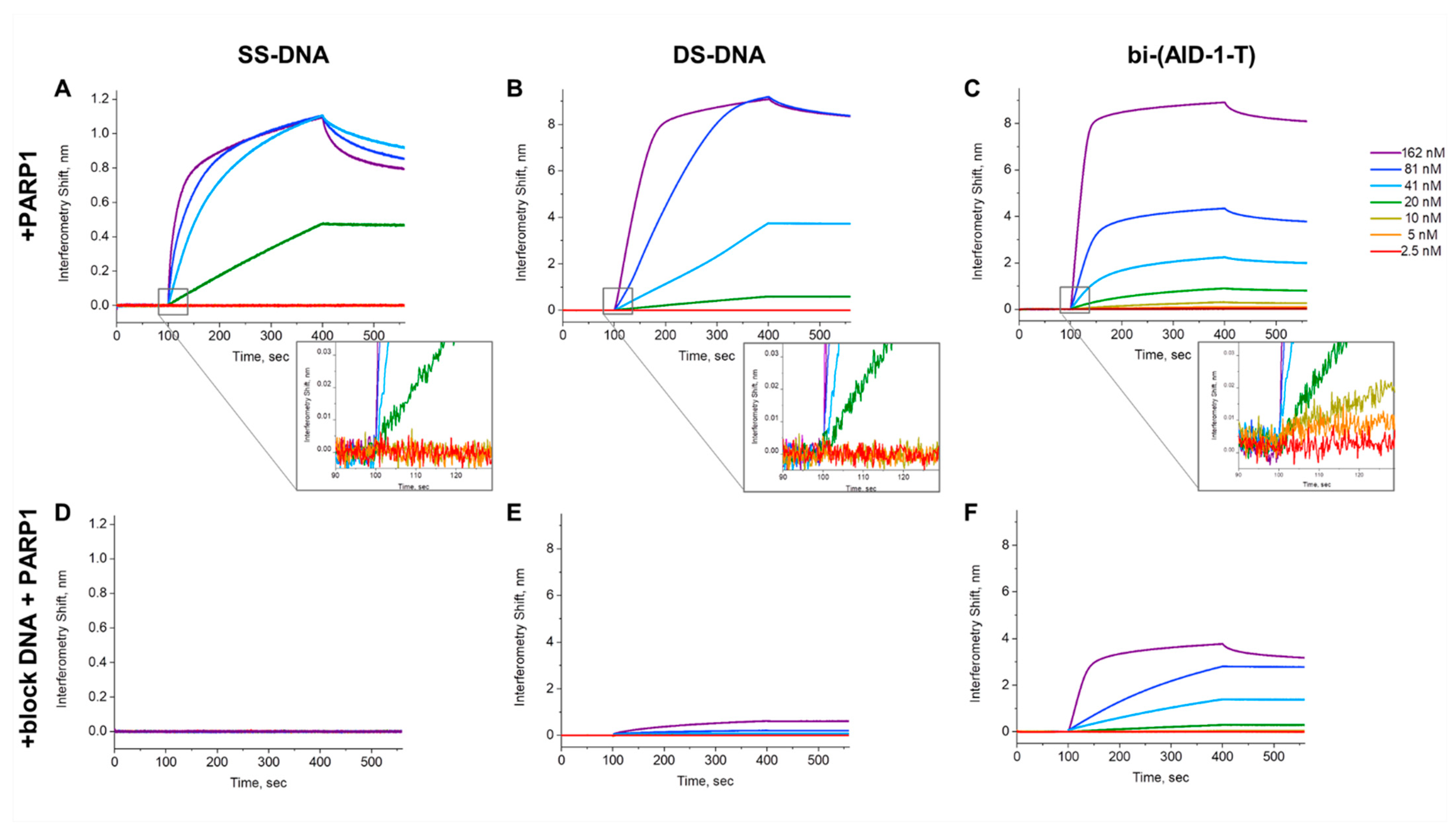
2.2. Assessment of PARP1 Affinity to Bi-(AID-1)-T
2.3. G-Quadruplex Bi-(AID-1-C) as an Analog of Bi-(AID-1-T)
2.4. Evaluation of Bi-(AID-1-C) Specificity to the Target Protein
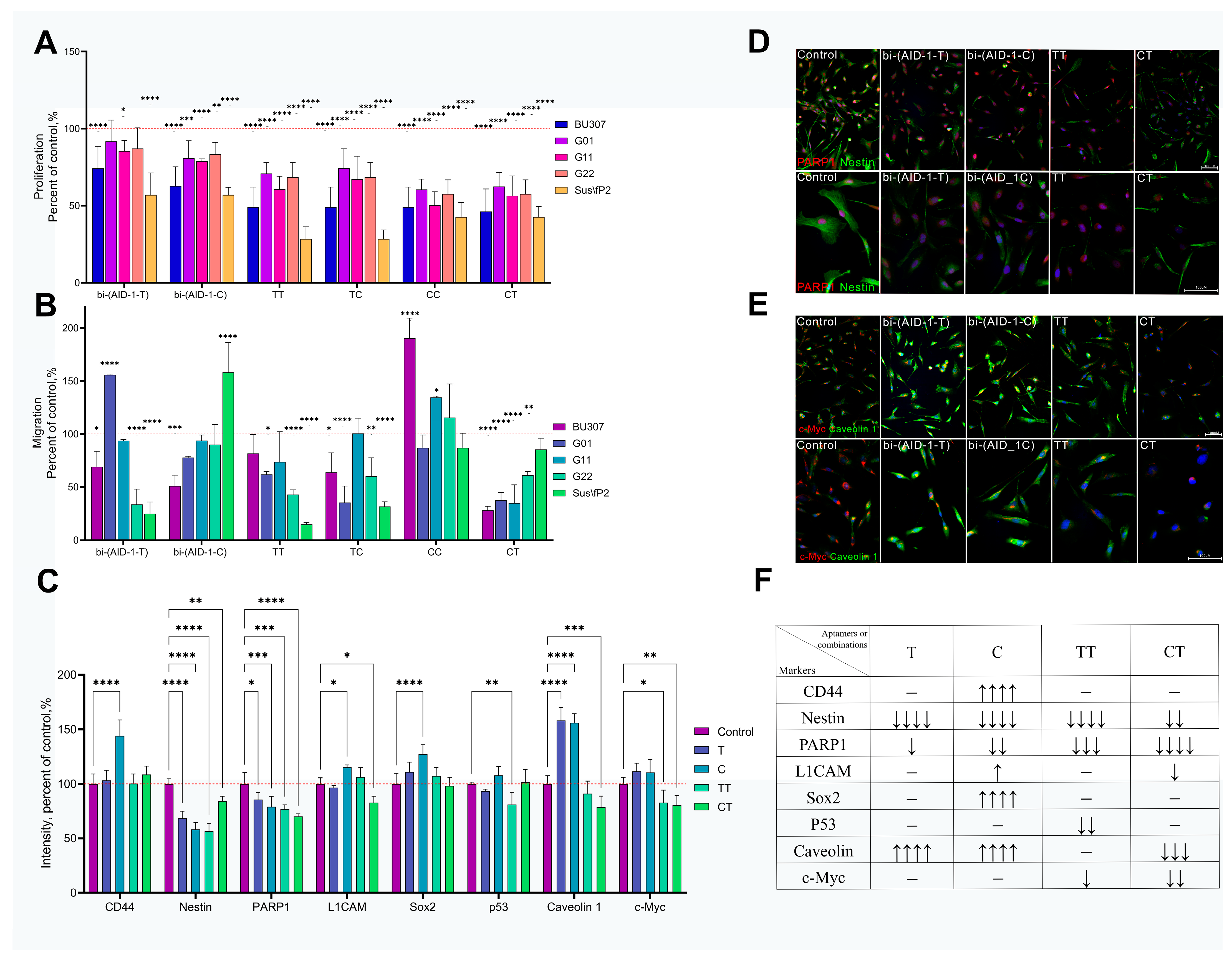
2.5. Complex Effects of Bi-(AID-1-T) and Bi-(AID-1-C) Aptamers on Human GBM Tumor Cells
3. Discussion
4. Material and Methods
4.1. Cell Cultures
4.2. Assessment of Bi-(AID-1-T) Specificity to the Target Protein
4.2.1. Lysate Preparation and Pull-Down
4.2.2. Chromatography–Mass Spectrometry Analysis
4.3. Assessment of PARP1 Affinity to Bi-(AID-1-T)
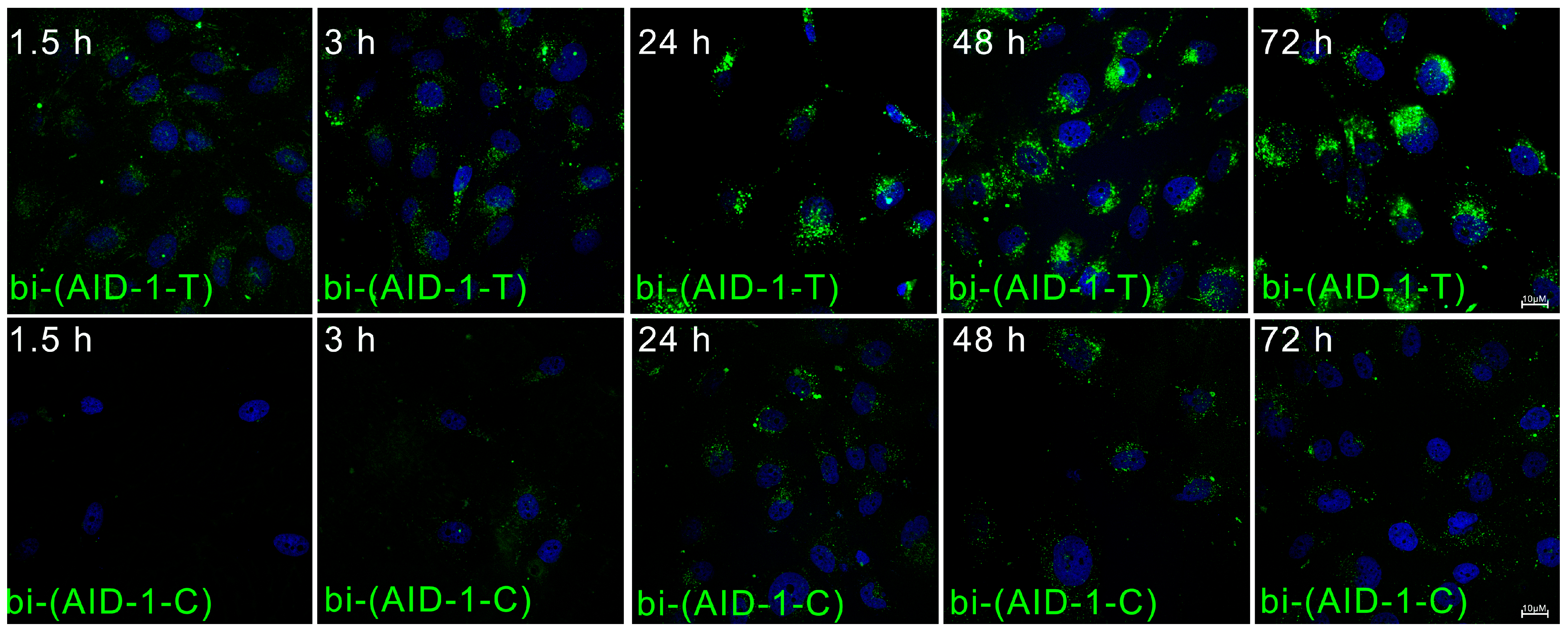
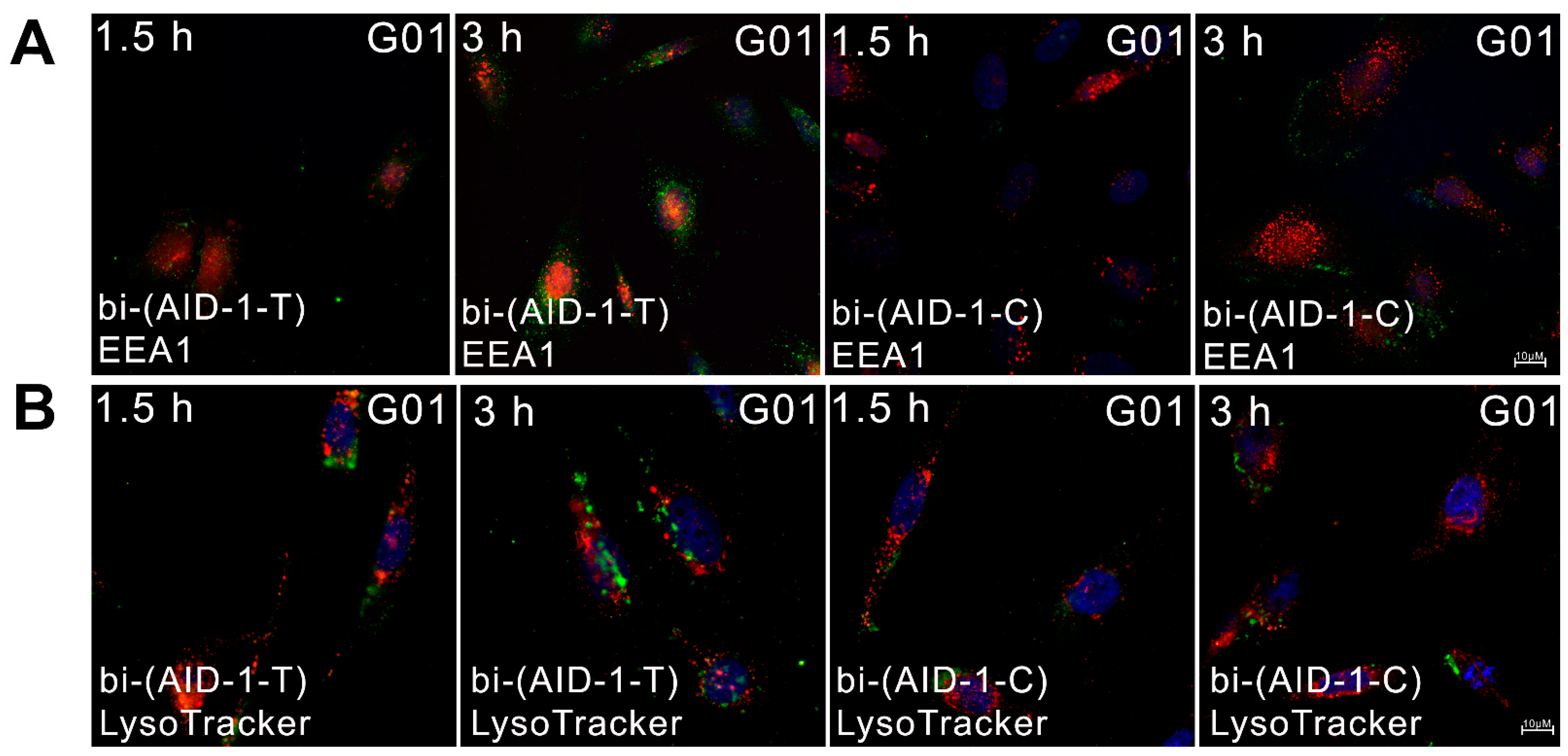
4.4. Cellular Uptake and Localization of Aptamers
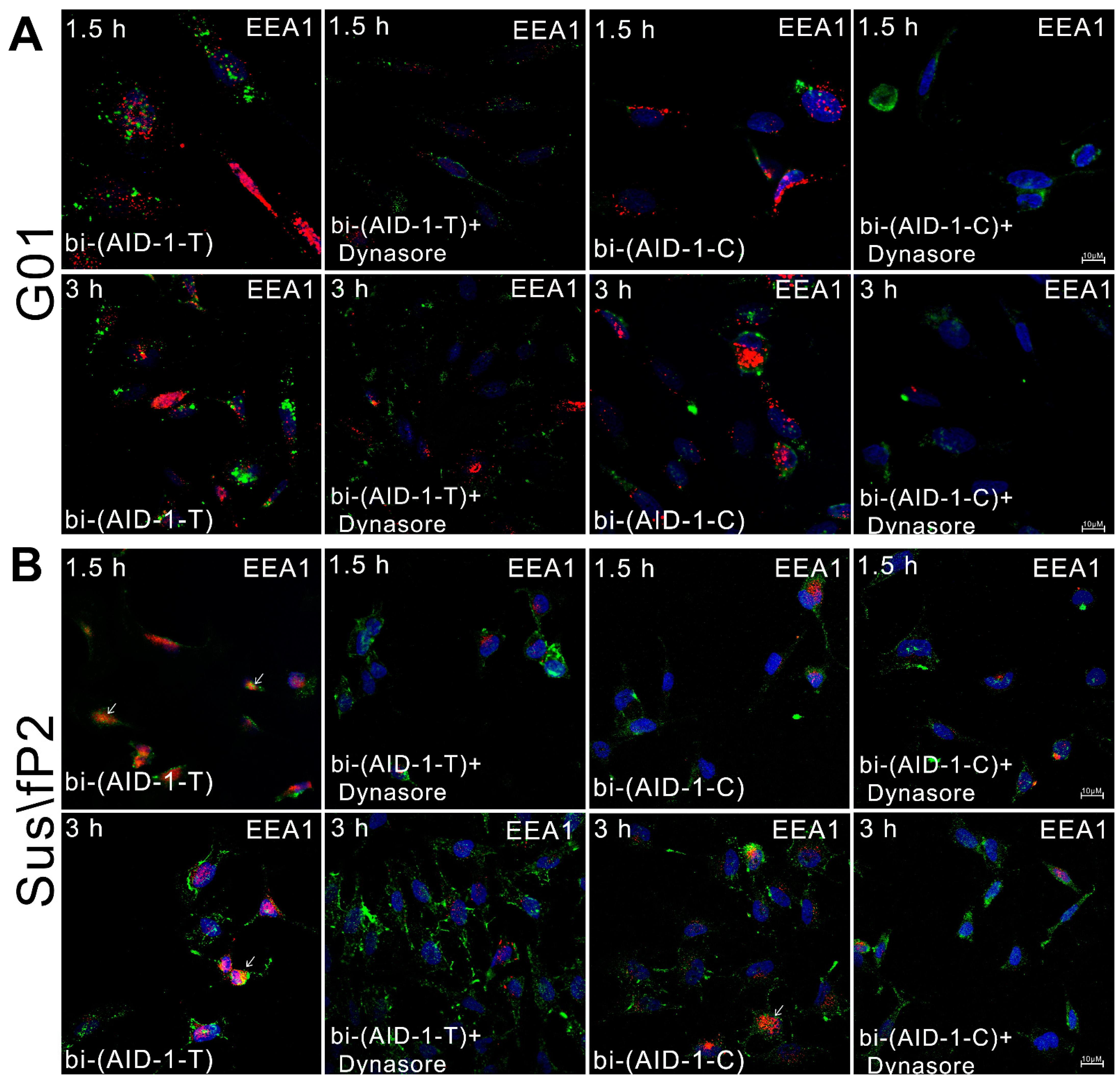
4.5. Endocytosis in Cellular Uptake of Aptamers
4.6. Aptamer Localization in Early Endosomes and Lysosomes
4.7. Treatment with Aptamers
4.8. Cell Viability Assessment by xCELLigence RTCA
4.9. Cell Proliferation Assessment by MTS Assay
4.10. Cell Migration Assessment by Transwell Inserts
4.11. Immunocytochemistry (ICC)
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.F.; Ottaviani, D.; Tazare, J.; Gregson, J.; Kitchen, N.; Brandner, S.; Fersht, N.; Mulholland, P. Survival Outcomes and Prognostic Factors in Glioblastoma. Cancers 2022, 14, 3161. [Google Scholar] [CrossRef] [PubMed]
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme–Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.J.; Kessler, A.F.; Feldheim, J.J.; Schulz, E.; Wend, D.; Lazaridis, L.; Kleinschnitz, C.; Glas, M.; Ernestus, R.-I.; Brandner, S.; et al. Effects of Long-Term Temozolomide Treatment on Glioblastoma and Astrocytoma WHO Grade 4 Stem-like Cells. Int. J. Mol. Sci. 2022, 23, 5238. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ishida, J.; Kurozumi, K.; Ichikawa, T.; Otani, Y.; Oka, T.; Tomita, Y.; Hattori, Y.; Uneda, A.; Matsumoto, Y.; et al. δ-Catenin Promotes Bevacizumab-Induced Glioma Invasion. Mol. Cancer Ther. 2019, 18, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Merrick, M.; Mimlitz, M.J.; Weeder, C.; Akhter, H.; Bray, A.; Walther, A.; Nwakama, C.; Bamesberger, J.; Djam, H.; Abid, K.; et al. In Vitro Radiotherapy and Chemotherapy Alter Migration of Brain Cancer Cells before Cell Death. Biochem. Biophys. Rep. 2021, 27, 101071. [Google Scholar] [CrossRef]
- Kochanowski, P.; Catapano, J.; Pudełek, M.; Wróbel, T.; Madeja, Z.; Ryszawy, D.; Czyż, J. Temozolomide Induces the Acquisition of Invasive Phenotype by O6-Methylguanine-DNA Methyltransferase (MGMT)+ Glioblastoma Cells in a Snail-1/Cx43-Dependent Manner. Int. J. Mol. Sci. 2021, 22, 4150. [Google Scholar] [CrossRef]
- Kang, M.-K.; Kang, S.-K. Tumorigenesis of Chemotherapeutic Drug-Resistant Cancer Stem-Like Cells in Brain Glioma. Stem Cells Dev. 2007, 16, 837–848. [Google Scholar] [CrossRef]
- Jovčevska, I. Genetic Secrets of Long-Term Glioblastoma Survivors. Bosn. J. Basic Med. Sci. 2018, 19, 116. [Google Scholar] [CrossRef]
- Osuka, S.; Van Meir, E.G. Overcoming Therapeutic Resistance in Glioblastoma: The Way Forward. J. Clin. Invest. 2017, 127, 415–426. [Google Scholar] [CrossRef]
- Tsuji, Y.; Nonoguchi, N.; Okuzaki, D.; Wada, Y.; Motooka, D.; Hirota, Y.; Toho, T.; Yoshikawa, N.; Furuse, M.; Kawabata, S.; et al. Chronic Pathophysiological Changes in the Normal Brain Parenchyma Caused by Radiotherapy Accelerate Glioma Progression. Sci. Rep. 2021, 11, 22110. [Google Scholar] [CrossRef] [PubMed]
- Shankar, A.; Kumar, S.; Iskander, A.; Varma, N.R.; Janic, B.; DeCarvalho, A.; Mikkelsen, T.; Frank, J.A.; Ali, M.M.; Knight, R.A.; et al. Subcurative Radiation Significantly Increases Cell Proliferation, Invasion, and Migration of Primary Glioblastoma Multiforme in Vivo. Chin. J. Cancer 2014, 33, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Falk, A.T.; Moncharmont, C.; Guilbert, M.; Guy, J.-B.; Alphonse, G.; Trone, J.-C.; Rivoirard, R.; Gilormini, M.; Toillon, R.-A.; Rodriguez-Lafrasse, C.; et al. La Radiothérapie Induit-Elle Une Agressivité Accrue Des Cellules Tumorales Du Glioblastome? Bull. Cancer 2014, 101, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shi, S.; Wang, H.; Zhao, T.; Wang, Y.; Huang, S.; Su, Y.; Zhao, C.; Yang, M. Advances in Antibody-Based Drugs and Their Delivery through the Blood-Brain Barrier for Targeted Therapy and Immunotherapy of Gliomas. Int. Immunopharmacol. 2023, 117, 109990. [Google Scholar] [CrossRef]
- Okada, H.; Kohanbash, G.; Zhu, X.; Kastenhuber, E.R.; Hoji, A.; Ueda, R.; Fujita, M. Immunotherapeutic Approaches for Glioma. Crit. Rev. Immunol. 2009, 29, 1–42. [Google Scholar] [CrossRef]
- Boskovitz, A.; Wikstrand, C.J.; Kuan, C.-T.; Zalutsky, M.R.; Reardon, D.A.; Bigner, D.D. Monoclonal Antibodies for Brain Tumour Treatment. Expert Opin. Biol. Ther. 2004, 4, 1453–1471. [Google Scholar] [CrossRef]
- Fu, Z.; Xiang, J. Aptamers, the Nucleic Acid Antibodies, in Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 2793. [Google Scholar] [CrossRef]
- Agnello, L.; Camorani, S.; Fedele, M.; Cerchia, L. Aptamers and Antibodies: Rivals or Allies in Cancer Targeted Therapy? Explor. Target. Anti-Tumor Ther. 2021, 2, 107. [Google Scholar] [CrossRef]
- Amundarain, A.; Pastor, F.; Prósper, F.; Agirre, X. Aptamers, a New Therapeutic Opportunity for the Treatment of Multiple Myeloma. Cancers 2022, 14, 5471. [Google Scholar] [CrossRef]
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Monaco, I.; Camorani, S.; Colecchia, D.; Locatelli, E.; Calandro, P.; Oudin, A.; Niclou, S.; Arra, C.; Chiariello, M.; Cerchia, L.; et al. Aptamer Functionalization of Nanosystems for Glioblastoma Targeting through the Blood–Brain Barrier. J. Med. Chem. 2017, 60, 4510–4516. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, S.; Yan, H.; Li, X.X.; Yazd, H.S.; Li, X.X.; Huang, T.; Cui, C.; Jiang, J.; Tan, W. Nucleic Acid Aptamers for Molecular Diagnostics and Therapeutics: Advances and Perspectives. Angew. Chem. Int. Ed. 2021, 60, 2221–2231. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, S.; Fab, L.; Savchenko, E.; Ryabova, A.; Ryzhova, M.; Revishchin, A.; Pronin, I.; Usachev, D.; Kopylov, A.; Pavlova, G. The Bi-(AID-1-T) G-Quadruplex Has a Janus Effect on Primary and Recurrent Gliomas: Anti-Proliferation and Pro-Migration. Pharmaceuticals 2024, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-H.; Nie, X.; Liu, H.-Y.; Fang, Y.-M.; Zhao, Y.; Xia, L.-X. TMPyP4 Promotes Cancer Cell Migration at Low Doses, but Induces Cell Death at High Doses. Sci. Rep. 2016, 6, 26592. [Google Scholar] [CrossRef] [PubMed]
- Thanh, H.D.; Lee, S.; Nguyen, T.T.; Huu, T.N.; Ahn, E.-J.; Cho, S.-H.; Kim, M.S.; Moon, K.-S.; Jung, C. Temozolomide Promotes Matrix Metalloproteinase 9 Expression through P38 MAPK and JNK Pathways in Glioblastoma Cells. Sci. Rep. 2024, 14, 14341. [Google Scholar] [CrossRef]
- Cesarini, V.; Scopa, C.; Silvestris, D.A.; Scafidi, A.; Petrera, V.; Del Baldo, G.; Gallo, A. Aptamer-Based In Vivo Therapeutic Targeting of Glioblastoma. Molecules 2020, 25, 4267. [Google Scholar] [CrossRef]
- Yang, J.; Shi, Z.; Liu, R.; Wu, Y.; Zhang, X. Combined-Therapeutic Strategies Synergistically Potentiate Glioblastoma Multiforme Treatment via Nanotechnology. Theranostics 2020, 10, 3223–3239. [Google Scholar] [CrossRef]
- Li, D.; Zhao, J.; Ma, J.; Yang, H.; Zhang, X.; Cao, Y.; Liu, P. GMT8 Aptamer Conjugated PEGylated Ag@Au Core-Shell Nanoparticles as a Novel Radiosensitizer for Targeted Radiotherapy of Glioma. Colloids Surf. B Biointerfaces 2022, 211, 112330. [Google Scholar] [CrossRef]
- Chan, H.Y.; Choi, J.; Jackson, C.; Lim, M. Combination Immunotherapy Strategies for Glioblastoma. J. Neurooncol. 2021, 151, 375–391. [Google Scholar] [CrossRef]
- Guo, G.; Zhang, Z.; Zhang, J.; Wang, D.; Xu, S.; Liu, G.; Gao, Y.; Mei, J.; Yan, Z.; Zhao, R.; et al. Predicting Recurrent Glioblastoma Clinical Outcome to Immune Checkpoint Inhibition and Low-Dose Bevacizumab with Tumor in Situ Fluid Circulating Tumor DNA Analysis. Cancer Immunol. Immunother. 2024, 73, 193. [Google Scholar] [CrossRef]
- Chen, H.-C.; Lin, H.-Y.; Chiang, Y.-H.; Yang, W.-B.; Wang, C.-H.; Yang, P.-Y.; Hu, S.-L.; Hsu, T.-I. Progesterone Boosts Abiraterone-Driven Target and NK Cell Therapies against Glioblastoma. J. Exp. Clin. Cancer Res. 2024, 43, 218. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liao, L.; Du, K.; Mo, J.; Zou, X.; Liang, J.; Chen, J.; Tang, W.; Su, L.; Wu, J.; et al. Clinical Activity and Safety of Sintilimab, Bevacizumab, and TMZ in Patients with Recurrent Glioblastoma. BMC Cancer 2024, 24, 133. [Google Scholar] [CrossRef]
- Legatova, V.; Samoylenkova, N.; Arutyunyan, A.; Tashlitsky, V.; Zavyalova, E.; Usachev, D.; Pavlova, G.; Kopylov, A. Covalent Bi-Modular Parallel and Antiparallel G-Quadruplex DNA Nanocostructs Reduce Viability of Patient Glioma Primary Cell Cultures. Int. J. Mol. Sci. 2021, 22, 3372. [Google Scholar] [CrossRef] [PubMed]
- Kopylov, A.M.; Samoylenkova, N.; Bizayeva, A.; Arutyunyan, A.; Tashlitsky, V.; Golbin, D.; Usachev, D.; Pavlova, G. P13.19 Bi-Modular G-Quadruplex DNA-Crypto-Aptamers Diminish Viability of Glioma Primary Cell Cultures of Patients. Neuro-Oncology 2021, 23 (Suppl. S2), ii36–ii37. [Google Scholar] [CrossRef]
- Nile, D.L.; Rae, C.; Hyndman, I.J.; Gaze, M.N.; Mairs, R.J. An Evaluation in Vitro of PARP-1 Inhibitors, Rucaparib and Olaparib, as Radiosensitisers for the Treatment of Neuroblastoma. BMC Cancer 2016, 16, 621. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Murphy, C.G.; Doubrovina, E.; Jasin, M.; Moynahan, M.E. PARP Inhibitors in Clinical Use Induce Genomic Instability in Normal Human Cells. PLoS ONE 2016, 11, e0159341. [Google Scholar] [CrossRef]
- Murnyák, B.; Kouhsari, M.C.; Hershkovitch, R.; Kálmán, B.; Marko-Varga, G.; Klekner, Á.; Hortobágyi, T. PARP1 Expression and Its Correlation with Survival Is Tumour Molecular Subtype Dependent in Glioblastoma. Oncotarget 2017, 8, 46348–46362. [Google Scholar] [CrossRef]
- Malyuchenko, N.V.; Kotova, E.Y.; Kulaeva, O.I.; Kirpichnikov, M.P.; Studitskiy, V.M. PARP1 Inhibitors: Antitumor Drug Design. Acta Naturae 2015, 7, 27–37. [Google Scholar] [CrossRef]
- Musumeci, D.; Montesarchio, D. G-quadruplex-based aptamers in therapeutic applications. In Handbook of Chemical Biology of Nucleic Acids; Springer Nature Singapore: Singapore, 2023; pp. 1–26. [Google Scholar] [CrossRef]
- Weerasinghe, P.; Li, Y.; Guan, Y.; Zhang, R.; Tweardy, D.J.; Jing, N. T40214/PEI Complex: A Potent Therapeutics for Prostate Cancer That Targets STAT3 Signaling. Prostate 2008, 68, 1430–1442. [Google Scholar] [CrossRef]
- Esposito, V.; Benigno, D.; Bello, I.; Panza, E.; Bucci, M.; Virgilio, A.; Galeone, A. Structural and Biological Features of G-Quadruplex Aptamers as Promising Inhibitors of the STAT3 Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 9524. [Google Scholar] [CrossRef]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a Cell-Permeable Inhibitor of Dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef]
- Preta, G.; Cronin, J.G.; Sheldon, I.M. Dynasore—Not Just a Dynamin Inhibitor. Cell Commun. Signal. 2015, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 SnRNP Regulates Chromatin Retention of Noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Lié, O.; Virolle, T.; Gabut, M.; Pasquier, C.; Zemmoura, I.; Augé-Gouillou, C. SETMAR Shorter Isoform: A New Prognostic Factor in Glioblastoma. Front. Oncol. 2022, 11, 638397. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Zhu, X.-L.; An, Y.; Li, Y. Clinical Significance of Small Nuclear Ribonucleoprotein U1 Subunit 70 in Patients with Hepatocellular Carcinoma. PeerJ 2024, 12, e16876. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, A.; Stieber, D.; Enger, P.Ø.; Golebiewska, A.; Molven, A.; Svendsen, A.; Westermark, B.; Niclou, S.P.; Olsen, T.K.; Chekenya Enger, M.; et al. U-251 Revisited: Genetic Drift and Phenotypic Consequences of Long-term Cultures of Glioblastoma Cells. Cancer Med. 2014, 3, 812–824. [Google Scholar] [CrossRef]
- Allen, M.; Bjerke, M.; Edlund, H.; Nelander, S.; Westermark, B. Origin of the U87MG Glioma Cell Line: Good News and Bad News. Sci. Transl. Med. 2016, 8, 354re3. [Google Scholar] [CrossRef]
- Miserocchi, G.; Mercatali, L.; Liverani, C.; De Vita, A.; Spadazzi, C.; Pieri, F.; Bongiovanni, A.; Recine, F.; Amadori, D.; Ibrahim, T. Management and Potentialities of Primary Cancer Cultures in Preclinical and Translational Studies. J. Transl. Med. 2017, 15, 229. [Google Scholar] [CrossRef]
- Mitra, A.; Mishra, L.; Li, S. Technologies for Deriving Primary Tumor Cells for Use in Personalized Cancer Therapy. Trends Biotechnol. 2013, 31, 347–354. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The Multifaceted Roles of PARP1 in DNA Repair and Chromatin Remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Schiewer, M.J.; Knudsen, K.E. Transcriptional Roles of PARP1 in Cancer. Mol. Cancer Res. 2014, 12, 1069–1080. [Google Scholar] [CrossRef]
- Ji, Y.; Tulin, A.V. The Roles of PARP1 in Gene Control and Cell Differentiation. Curr. Opin. Genet. Dev. 2010, 20, 512–518. [Google Scholar] [CrossRef]
- Domagala, P.; Huzarski, T.; Lubinski, J.; Gugala, K.; Domagala, W. PARP-1 Expression in Breast Cancer Including BRCA1-Associated, Triple Negative and Basal-like Tumors: Possible Implications for PARP-1 Inhibitor Therapy. Breast Cancer Res. Treat. 2011, 127, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Tentori, L.; Ricci-Vitiani, L.; Muzi, A.; Ciccarone, F.; Pelacchi, F.; Calabrese, R.; Runci, D.; Pallini, R.; Caiafa, P.; Graziani, G. Pharmacological Inhibition of Poly(ADP-Ribose) Polymerase-1 Modulates Resistance of Human Glioblastoma Stem Cells to Temozolomide. BMC Cancer 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Karpel-Massler, G.; Pareja, F.; Aimé, P.; Shu, C.; Chau, L.; Westhoff, M.-A.; Halatsch, M.-E.; Crary, J.F.; Canoll, P.; Siegelin, M.D. PARP Inhibition Restores Extrinsic Apoptotic Sensitivity in Glioblastoma. PLoS ONE 2014, 9, e114583. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Robert, I.; Reina-San-Martin, B.; Schreiber, V.; Dantzer, F. Poly(ADP-Ribose) Polymerases in Double-Strand Break Repair: Focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014, 329, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Livraghi, L.; Garber, J.E. PARP Inhibitors in the Management of Breast Cancer: Current Data and Future Prospects. BMC Med. 2015, 13, 188. [Google Scholar] [CrossRef]
- Csonka, T.; Murnyák, B.; Szepesi, R.; Kurucz, A.; Klekner, Á.; Hortobágyi, T. Original Article Poly(ADP-Ribose) Polymerase-1 (PARP1) and P53 Labelling Index Correlates with Tumour Grade in Meningiomas. Folia Neuropathol. 2014, 2, 111–120. [Google Scholar] [CrossRef]
- Lacza, Z.; Horváth, E.; Komjáti, K.; Hortobágyi, T.; Szabó, C.; Busija, D. PARP Inhibition Improves the Effectiveness of Neural Stem Cell Transplantation in Experimental Brain Trauma. Int. J. Mol. Med. 2003, 13, 153–159. [Google Scholar] [CrossRef]
- Hortobágyi, T.; Görlach, C.; Benyó, Z.; Lacza, Z.; Hortobágyi, S.; Wahl, M.; Harkany, T. Inhibition of Neuronal Nitric Oxide Synthase-Mediated Activation of Poly(ADP-Ribose) Polymerase in Traumatic Brain Injury: Neuroprotection by 3-Aminobenzamide. Neuroscience 2003, 121, 983–990. [Google Scholar] [CrossRef]
- Galia, A.; Calogero, A.E.; Condorelli, R.; Fraggetta, F.; La Corte, A.; Ridolfo, F.; Bosco, P.; Castiglione, R.; Salemi, M. PARP-1 Protein Expression in Glioblastoma Multiforme. Eur. J. Histochem. 2012, 56, 9. [Google Scholar] [CrossRef]
- Vuurden, D.G.; Hulleman, E.; Meijer, O.L.M.; Wedekind, L.E.; Kool, M.; Witt, H.; Vandertop, W.P.; Würdinger, T.; Noske, D.P.; Kaspers, G.J.L.; et al. PARP Inhibition Sensitizes Childhood High Grade Glioma, Medulloblastoma and Ependymoma to Radiation. Oncotarget 2011, 2, 984–996. [Google Scholar] [CrossRef] [PubMed]
- Barton, V.N.; Donson, A.M.; Kleinschmidt-DeMasters, B.K.; Gore, L.; Liu, A.K.; Foreman, N.K. PARP1 Expression in Pediatric Central Nervous System Tumors. Pediatr. Blood Cancer 2009, 53, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Makise, N.; Shinozaki-Ushiku, A.; Zhang, L.; Ishibashi, Y.; Ikegami, M.; Tsuda, Y.; Kohsaka, S.; Ushiku, T.; Oda, K.; et al. Dramatic Response to Entrectinib in a Patient with Malignant Peripheral Nerve Sheath Tumor Harboring Novel SNRNP70-NTRK3 Fusion Gene. Genes, Chromosom. Cancer 2023, 62, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Spritz, R.A.; Strunk, K.; Surowy, C.S.; Mohrenweiser, H.W. Human U1-70K Ribonucleoprotein Antigen Gene: Organization, Nucleotide Sequence, and Mapping to Locus 19q13.3. Genomics 1990, 8, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-M.; Venters, C.C.; Di, C.; Pinto, A.M.; Wan, L.; Younis, I.; Cai, Z.; Arai, C.; So, B.R.; Duan, J.; et al. U1 SnRNP Regulates Cancer Cell Migration and Invasion in Vitro. Nat. Commun. 2020, 11, 1. [Google Scholar] [CrossRef]
- Kucinska, M.; Pospieszna, J.; Tang, J.; Lisiak, N.; Toton, E.; Rubis, B.; Murias, M. The Combination Therapy Using Tyrosine Kinase Receptors Inhibitors and Repurposed Drugs to Target Patient-Derived Glioblastoma Stem Cells. Biomed. Pharmacother. 2024, 176, 116892. [Google Scholar] [CrossRef]
- Kim, Y.S.; Chung, J.; Song, M.Y.; Jurng, J.; Kim, B.C. Aptamer Cocktails: Enhancement of Sensing Signals Compared to Single Use of Aptamers for Detection of Bacteria. Biosens. Bioelectron. 2014, 54, 195–198. [Google Scholar] [CrossRef]
- Cao, X.; Li, S.; Chen, L.; Ding, H.; Xu, H.; Huang, Y.; Li, J.; Liu, N.; Cao, W.; Zhu, Y.; et al. Combining Use of a Panel of SsDNA Aptamers in the Detection of Staphylococcus Aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [Google Scholar] [CrossRef]
- Guan, B.; Zhang, X. Aptamers as Versatile Ligands for Biomedical and Pharmaceutical Applications. Int. J. Nanomed. 2020, 15, 1059–1071. [Google Scholar] [CrossRef]
- An, S.; Song, I.H.; Woo, C.G. Diagnostic Value of Nestin Expression in Adult Gliomas. Int. J. Surg. Pathol. 2023, 31, 1014–1020. [Google Scholar] [CrossRef]
- Naito, Z. Neuroepithelial Stem Cell Marker Nestin Regulates the Migration, Invasion and Growth of Human Gliomas. Oncol. Rep. 2011, 26, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Arai, H.; Ikota, H.; Sugawara, K.; Nobusawa, S.; Hirato, J.; Nakazato, Y. Nestin Expression in Brain Tumors: Its Utility for Pathological Diagnosis and Correlation with the Prognosis of High-Grade Gliomas. Brain Tumor Pathol. 2012, 29, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Strojnik, T.; Røsland, G.V.; Sakariassen, P.O.; Kavalar, R.; Lah, T. Neural Stem Cell Markers, Nestin and Musashi Proteins, in the Progression of Human Glioma: Correlation of Nestin with Prognosis of Patient Survival. Surg. Neurol. 2007, 68, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.J.; Lan, F.; He, Q.; Lee, A.; Tang, C.Z.; Dong, L.; Lan, B.; Ma, X.; Wu, J.C.; Shen, L. Inducible Expression of Stem Cell Associated Intermediate Filament Nestin Reveals an Important Role in Glioblastoma Carcinogenesis. Int. J. Cancer 2011, 128, 343–351. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, H.; Hu, J.; Fu, H.; Qu, Y.; Yang, Y.; Cai, K.Q.; Efimov, A.; Wu, M.; Yen, T.; et al. Nestin Is Required for Spindle Assembly and Cell-Cycle Progression in Glioblastoma Cells. Mol. Cancer Res. 2021, 19, 1651–1665. [Google Scholar] [CrossRef]
- Murphy, M.J.; Wilson, A.; Trumpp, A. More than Just Proliferation: Myc Function in Stem Cells. Trends Cell Biol. 2005, 15, 128–137. [Google Scholar] [CrossRef]
- Wong, D.J.; Liu, H.; Ridky, T.W.; Cassarino, D.; Segal, E.; Chang, H.Y. Module Map of Stem Cell Genes Guides Creation of Epithelial Cancer Stem Cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef]
- Vita, M.; Henriksson, M. The Myc Oncoprotein as a Therapeutic Target for Human Cancer. Semin. Cancer Biol. 2006, 16, 318–330. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Li, Z.; Wu, Q.; Lathia, J.D.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. C-Myc Is Required for Maintenance of Glioma Cancer Stem Cells. PLoS ONE 2008, 3, e3769. [Google Scholar] [CrossRef]
- Annibali, D.; Whitfield, J.R.; Favuzzi, E.; Jauset, T.; Serrano, E.; Cuartas, I.; Redondo-Campos, S.; Folch, G.; Gonzàlez-Juncà, A.; Sodir, N.M.; et al. Myc Inhibition Is Effective against Glioma and Reveals a Role for Myc in Proficient Mitosis. Nat. Commun. 2014, 5, 4632. [Google Scholar] [CrossRef]
- Bidwell, G.L.; Perkins, E.; Hughes, J.; Khan, M.; James, J.R.; Raucher, D. Thermally Targeted Delivery of a C-Myc Inhibitory Polypeptide Inhibits Tumor Progression and Extends Survival in a Rat Glioma Model. PLoS ONE 2013, 8, e55104. [Google Scholar] [CrossRef] [PubMed]
- Bidwell, G.L.; Perkins, E.; Raucher, D. A Thermally Targeted C-Myc Inhibitory Polypeptide Inhibits Breast Tumor Growth. Cancer Lett. 2012, 319, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xiao, W.; Song, T.; Feng, G.; Dai, Z. Expression and Prognostic Significance of P53 in Glioma Patients: A Meta-Analysis. Neurochem. Res. 2016, 41, 1723–1731. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, W.; Chen, B.; Jiang, T.; Wang, Z. Prognostic and Predictive Value of P53 in Low MGMT Expressing Glioblastoma Treated with Surgery, Radiation and Adjuvant Temozolomide Chemotherapy. Neurol. Res. 2010, 32, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.; Hao, S.; Su, Z.; Zhang, P.; Li, Y.; Song, G.; Yu, L.; Wang, J.; Ji, N.; et al. Analysis of Treatment Tolerance and Factors Associated with Overall Survival in Elderly Patients with Glioblastoma. World Neurosurg. 2016, 95, 77–84. [Google Scholar] [CrossRef]
- Wachowiak, R.; Krause, M.; Mayer, S.; Peukert, N.; Suttkus, A.; Müller, W.C.; Lacher, M.; Meixensberger, J.; Nestler, U. Increased L1CAM (CD171) Levels Are Associated with Glioblastoma and Metastatic Brain Tumors. Medicine 2018, 97, e12396. [Google Scholar] [CrossRef]
- Mohanan, V.; Temburni, M.K.; Kappes, J.C.; Galileo, D.S. L1CAM Stimulates Glioma Cell Motility and Proliferation through the Fibroblast Growth Factor Receptor. Clin. Exp. Metastasis 2013, 30, 507–520. [Google Scholar] [CrossRef]
- Held-Feindt, J.; Schmelz, S.; Hattermann, K.; Mentlein, R.; Mehdorn, H.M.; Sebens, S. The Neural Adhesion Molecule L1CAM Confers Chemoresistance in Human Glioblastomas. Neurochem. Int. 2012, 61, 1183–1191. [Google Scholar] [CrossRef]
- Giordano, M.; Cavallaro, U. Different Shades of L1CAM in the Pathophysiology of Cancer Stem Cells. J. Clin. Med. 2020, 9, 1502. [Google Scholar] [CrossRef]
- Parat, M.-O.; Riggins, G.J. Caveolin-1, Caveolae, and Glioblastoma. Neuro-Oncology 2012, 14, 679–688. [Google Scholar] [CrossRef]
- Barresi, V.; Buttarelli, F.R.; Vitarelli E, E.; Arcella, A.; Antonelli, M.; Giangaspero, F. Caveolin-1 Expression in Diffuse Gliomas: Correlation with the Proliferation Index, Epidermal Growth Factor Receptor, P53, and 1p/19q Status. Hum. Pathol. 2009, 40, 1738–1746. [Google Scholar] [CrossRef] [PubMed]
- Moriconi, C.; Civita, P.; Neto, C.; Pilkington, G.J.; Gumbleton, M. Caveolin-1, a Key Mediator Across Multiple Pathways in Glioblastoma and an Independent Negative Biomarker of Patient Survival. Front. Oncol. 2021, 11, 701933. [Google Scholar] [CrossRef] [PubMed]
- Senetta, R.; Trevisan, E.; Rudà, R.; Maldi, E.; Molinaro, L.; Lefranc, F.; Chiusa, L.; Lanotte, M.; Soffietti, R.; Cassoni, P. Caveolin 1 Expression Independently Predicts Shorter Survival in Oligodendrogliomas. J. Neuropathol. Exp. Neurol. 2009, 68, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Cassoni, P.; Senetta, R.; Castellano, I.; Ortolan, E.; Bosco, M.; Magnani, I.; Ducati, A. Caveolin-1 Expression Is Variably Displayed in Astroglial-Derived Tumors and Absent in Oligodendrogliomas: Concrete Premises for a New Reliable Diagnostic Marker in Gliomas. Am. J. Surg. Pathol. 2007, 31, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Eser Ocak, P.; Ocak, U.; Tang, J.; Zhang, J.H. The Role of Caveolin-1 in Tumors of the Brain—Functional and Clinical Implications. Cell. Oncol. 2019, 42, 423–447. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Wang, F.; Lin, J.; Tan, X.; Chen, C.; Wu, L.; Zhang, X.; Wang, Y.; Shi, Y.; et al. Caveolin-1 Promotes Glioma Progression and Maintains Its Mitochondrial Inhibition Resistance. Discov. Oncol. 2023, 14, 161. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
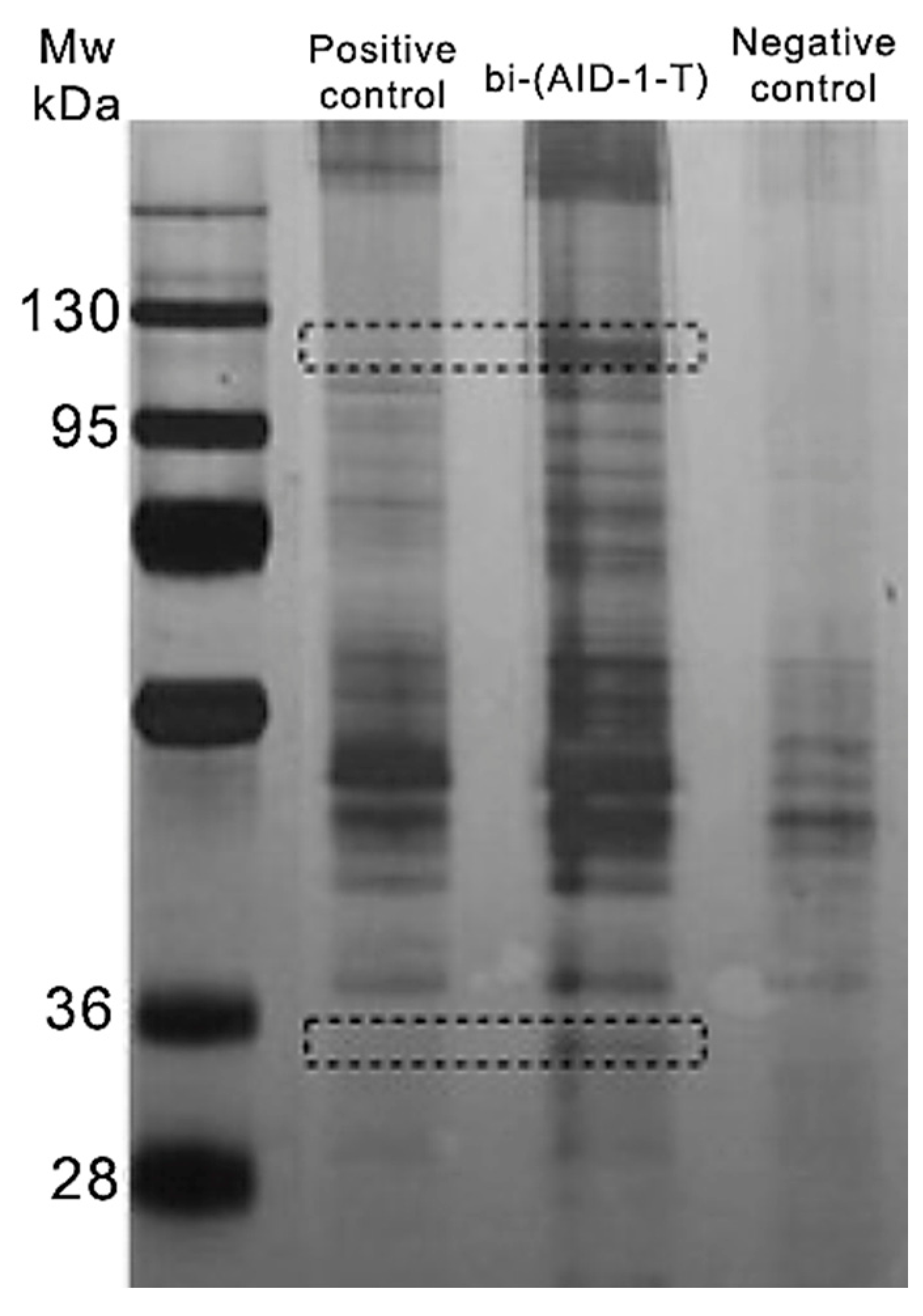
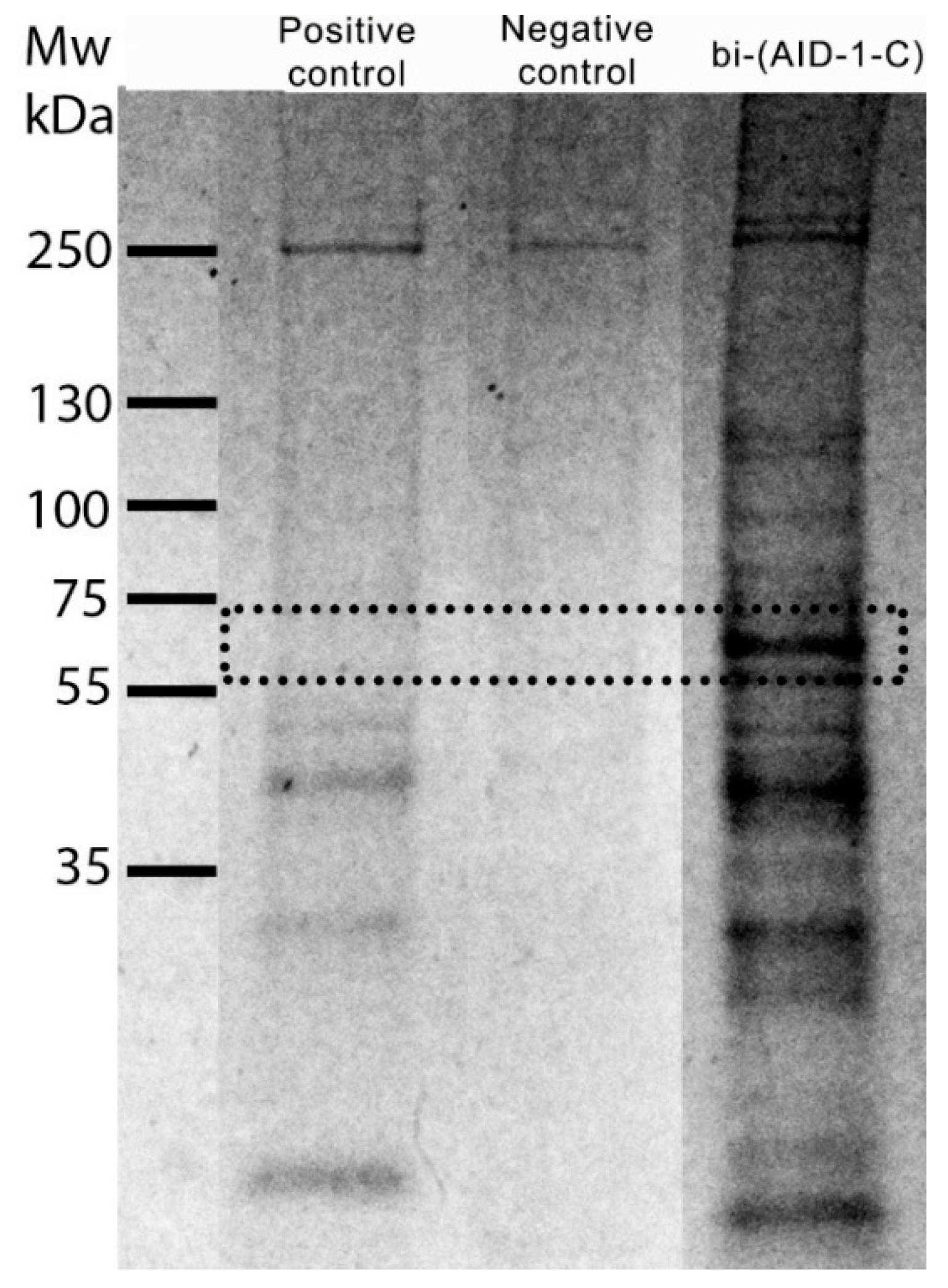

| Protein ID | Accession | −10lgP | Coverage (%) | Area Sample | Peptides | Unique | Spec Sample | Avg. Mass | Description |
|---|---|---|---|---|---|---|---|---|---|
| P09874 | PARP1_HUMAN | 219.24 | 36 | 1.60 × 108 | 60 | 60 | 114 | 113,084 | Poly [ADP-ribose] polymerase 1 OS = Homo sapiens OX = 9606 GN = PARP1 PE = 1 SV = 4 |
| Q9H2U1 | DHX36_HUMAN | 211.11 | 28 | 1.44 × 108 | 47 | 47 | 101 | 114,760 | ATP-dependent DNA/RNA helicase DHX36 OS = Homo sapiens OX = 9606 GN = DHX36 PE = 1 SV = 2 |
| Q1KMD3 | HNRL2_HUMAN | 166.63 | 27 | 7.70 × 107 | 29 | 29 | 92 | 85,105 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 OS = Homo sapiens OX = 9606 GN = HNRNPUL2 PE = 1 SV = 1 |
| P68371 | TBB4B_HUMAN | 125.29 | 16 | 0 | 7 | 1 | 24 | 49,831 | Tubulin beta-4B chain OS = Homo sapiens OX = 9606 GN = TUBB4B PE = 1 SV = 1 |
| Q07157 | ZO1_HUMAN | 119.68 | 7 | 2.47 × 106 | 15 | 13 | 18 | 195,457 | Tight junction protein ZO-1 OS = Homo sapiens OX = 9606 GN = TJP1 PE = 1 SV = 3 |
| P16615 | AT2A2_HUMAN | 114.27 | 12 | 2.83 × 106 | 14 | 14 | 16 | 114,757 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 OS = Homo sapiens OX = 9606 GN = ATP2A2 PE = 1 SV = 1 |
| Q9UDY2 | ZO2_HUMAN | 112.37 | 14 | 2.50 × 106 | 21 | 19 | 24 | 133,958 | Tight junction protein ZO-2 OS = Homo sapiens OX = 9606 GN = TJP2 PE = 1 SV = 2 |
| P08621 | RU17_HUMAN | 108.03 | 19 | 1.86 × 106 | 9 | 9 | 13 | 51,557 | U1 small nuclear ribonucleoprotein 70 kDa OS = Homo sapiens OX = 9606 GN = SNRNP70 PE = 1 SV = 2 |
| Q9NR30 | DDX21_HUMAN | 104.2 | 18 | 2.01 × 106 | 15 | 12 | 17 | 87,344 | Nucleolar RNA helicase 2 OS = Homo sapiens OX = 9606 GN = DDX21 PE = 1 SV = 5 |
| Q12797 | ASPH_HUMAN | 103.01 | 17 | 3.38 × 106 | 14 | 14 | 18 | 85,863 | Aspartyl/asparaginyl beta-hydroxylase OS = Homo sapiens OX = 9606 GN = ASPH PE = 1 SV = 3 |
| Affinity Parameter | Kinetic Association Constant, kon (×105 M−1⋅s−1) | Kinetic Dissociation Constant, koff (×102 s−1) | Apparent Dissociation Constant, aKD nM |
|---|---|---|---|
| bi-(AID-1-T) | 6.6 ± 0.34 | 1.28 ± 0.04 | 19.4 ± 0.7 |
| Name | Sequence |
|---|---|
| bi-(AID-1-T) | GGG TGG GTG GGT GGG TTT GGG TGG GTG GGT GGG |
| bi-(AID-1-C) | GGG CGG GCG GGC GGG TTT GGG CGG GCG GGC GGG |
| Protein ID | Accession | −10lgP | Coverage (%) | Area Sample | Peptides | Unique | Spec Sample | Avg. Mass | Description |
|---|---|---|---|---|---|---|---|---|---|
| P08621-2 | RU17_HUMAN | 146.32 | 25 | 6.95 × 108 | 9 | 9 | 27 | 50,618 | Isoform 2 of U1 small nuclear ribonucleoprotein 70 kDa OS = Homo sapiens OX = 9606 GN = SNRNP70 |
| P08621 | RU17_HUMAN | 146.32 | 24 | 6.95 × 108 | 9 | 9 | 27 | 51,557 | U1 small nuclear ribonucleoprotein 70 kDa OS = Homo sapiens OX = 9606 GN = SNRNP70 PE = 1 SV = 2 |
| O76021 | RL1D1_HUMAN | 92.3 | 11 | 6.86 × 107 | 5 | 5 | 6 | 54,973 | Ribosomal L1 domain-containing protein 1 OS = Homo sapiens OX = 9606 GN = RSL1D1 PE = 1 SV = 3 |
| O60506-2 | HNRPQ_HUMAN | 66.87 | 4 | 1.15 × 107 | 3 | 3 | 3 | 65,682 | Isoform 2 of Heterogeneous nuclear ribonucleoprotein Q OS = Homo sapiens OX = 9606 GN = SYNCRIP |
| O60506 | HNRPQ_HUMAN | 66.87 | 4 | 1.15 × 107 | 3 | 3 | 3 | 69,603 | Heterogeneous nuclear ribonucleoprotein Q OS = Homo sapiens OX = 9606 GN = SYNCRIP PE = 1 SV = 2 |
| O43390-4 | HNRPR_HUMAN | 66.87 | 4 | 1.15 × 107 | 3 | 3 | 3 | 59,953 | Isoform 4 of Heterogeneous nuclear ribonucleoprotein R OS = Homo sapiens OX = 9606 GN = HNRNPR |
| O43390 | HNRPR_HUMAN | 66.87 | 4 | 1.15 × 107 | 3 | 3 | 3 | 70,943 | Heterogeneous nuclear ribonucleoprotein R OS = Homo sapiens OX = 9606 GN = HNRNPR PE = 1 SV = 1 |
| O43390-2 | HNRPR_HUMAN | 66.87 | 4 | 1.15 × 107 | 3 | 3 | 3 | 71,214 | Isoform 2 of Heterogeneous nuclear ribonucleoprotein R OS = Homo sapiens OX = 9606 GN = HNRNPR |
| P27694 | RFA1_HUMAN | 62.93 | 6 | 5.18 × 106 | 3 | 3 | 5 | 68,138 | Replication protein A 70 kDa DNA-binding subunit OS = Homo sapiens OX = 9606 GN = RPA1 PE = 1 SV = 2 |
| Q8IYB3-2 | SRRM1_HUMAN | 62.27 | 4 | 6.64 × 106 | 3 | 3 | 5 | 102,126 | Isoform 2 of Serine/arginine repetitive matrix protein 1 OS = Homo sapiens OX = 9606 GN = SRRM1 |
| Q8IYB3 | SRRM1_HUMAN | 62.27 | 4 | 6.64 × 106 | 3 | 3 | 5 | 102,335 | Serine/arginine repetitive matrix protein 1 OS = Homo sapiens OX = 9606 GN = SRRM1 PE = 1 SV = 2 |
| Combination Name | Added aptamers | |||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | |
| T | bi-(AID-1-T) | — | — | Start of observation |
| TT | bi-(AID-1-T) | bi-(AID-1-T) | — | |
| TC | bi-(AID-1-T) | bi-(AID-1-C) | — | |
| TTT | bi-(AID-1-T) | bi-(AID-1-T) | bi-(AID-1-T) | |
| TCT | bi-(AID-1-T) | bi-(AID-1-C) | bi-(AID-1-T) | |
| T-T | bi-(AID-1-T) | — | bi-(AID-1-T) | |
| T-C | bi-(AID-1-T) | — | bi-(AID-1-C) | |
| T + C | bi-(AID-1-T) bi-(AID-1-C) | — | — | |
| C | bi-(AID-1-C) | — | — | |
| CC | bi-(AID-1-C) | bi-(AID-1-C) | — | |
| CT | bi-(AID-1-C) | bi-(AID-1-T) | — | |
| CCC | bi-(AID-1-C) | bi-(AID-1-C) | bi-(AID-1-C) | |
| CTC | bi-(AID-1-C) | bi-(AID-1-T) | bi-(AID-1-C) | |
| C-C | bi-(AID-1-C) | — | bi-(AID-1-C) | |
| C-T | bi-(AID-1-C) | — | bi-(AID-1-T) | |
| Culture Name | Grade | IDH1 | Location |
|---|---|---|---|
| Sus\fP2 | IV | IDH1 wild type | Left temporal lobe |
| BU307 | IV | IDH1 wild type | Left temporal lobe |
| G-11 | III–IV | IDH1 wild type | Right frontal–parietal–insular region. |
| G-01 | IV | IDH1 wild type | Left frontal lobe |
| G-22 | IV | IDH1 wild type | Left frontal lobe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlova, S.; Fab, L.; Dzarieva, F.; Ryabova, A.; Revishchin, A.; Panteleev, D.; Antipova, O.; Usachev, D.; Kopylov, A.; Pavlova, G. Unite and Conquer: Association of Two G-Quadruplex Aptamers Provides Antiproliferative and Antimigration Activity for Cells from High-Grade Glioma Patients. Pharmaceuticals 2024, 17, 1435. https://doi.org/10.3390/ph17111435
Pavlova S, Fab L, Dzarieva F, Ryabova A, Revishchin A, Panteleev D, Antipova O, Usachev D, Kopylov A, Pavlova G. Unite and Conquer: Association of Two G-Quadruplex Aptamers Provides Antiproliferative and Antimigration Activity for Cells from High-Grade Glioma Patients. Pharmaceuticals. 2024; 17(11):1435. https://doi.org/10.3390/ph17111435
Chicago/Turabian StylePavlova, Svetlana, Lika Fab, Fatima Dzarieva, Anastasia Ryabova, Alexander Revishchin, Dmitriy Panteleev, Olga Antipova, Dmitry Usachev, Alexey Kopylov, and Galina Pavlova. 2024. "Unite and Conquer: Association of Two G-Quadruplex Aptamers Provides Antiproliferative and Antimigration Activity for Cells from High-Grade Glioma Patients" Pharmaceuticals 17, no. 11: 1435. https://doi.org/10.3390/ph17111435
APA StylePavlova, S., Fab, L., Dzarieva, F., Ryabova, A., Revishchin, A., Panteleev, D., Antipova, O., Usachev, D., Kopylov, A., & Pavlova, G. (2024). Unite and Conquer: Association of Two G-Quadruplex Aptamers Provides Antiproliferative and Antimigration Activity for Cells from High-Grade Glioma Patients. Pharmaceuticals, 17(11), 1435. https://doi.org/10.3390/ph17111435









