Network Pharmacology and Experimental Validation to Explore the Potential Mechanism of Nigella sativa for the Treatment of Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. Network Pharmacology Study
2.1.1. Active Constituents of Nigella sativa
2.1.2. Target Prediction for Nigella sativa
2.1.3. Screening of Breast Cancer-Related Targets
2.1.4. Network Construction of Phytochemicals and Their Targets
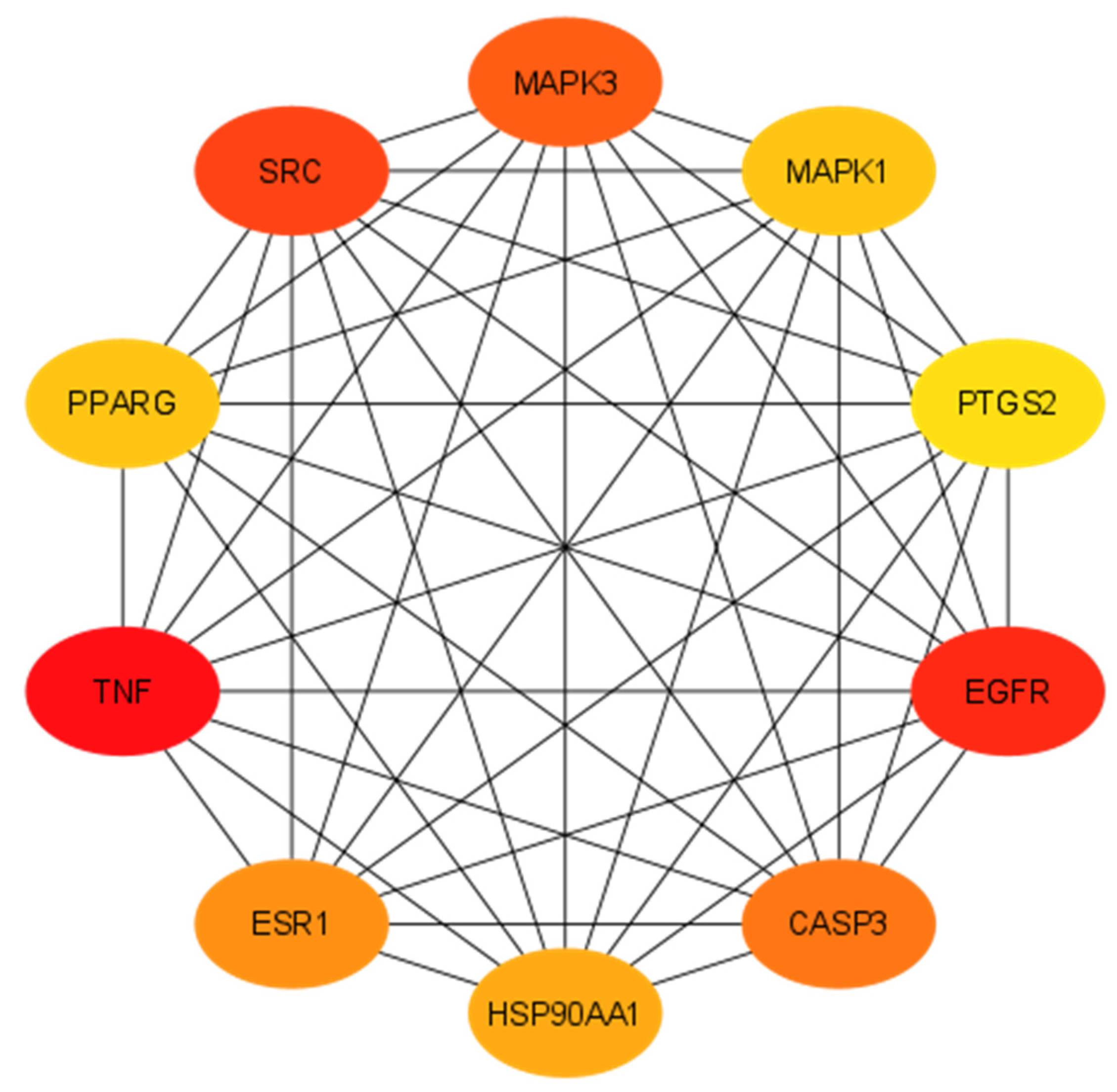
2.1.5. Protein–Protein Interactions
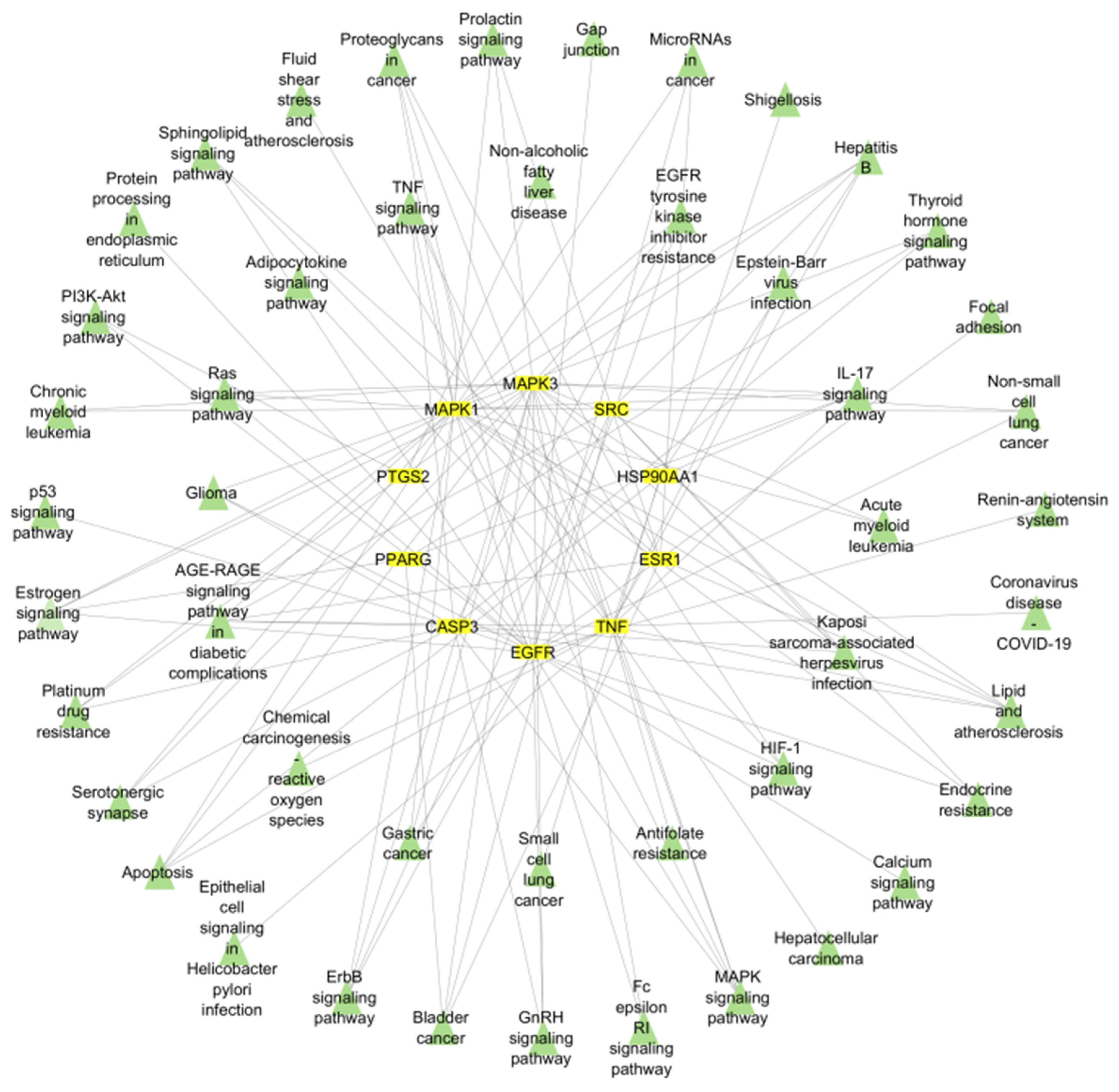
2.1.6. Functional Enrichment Analysis
2.1.7. Construction of the Merged Compound–Target–Pathway Network
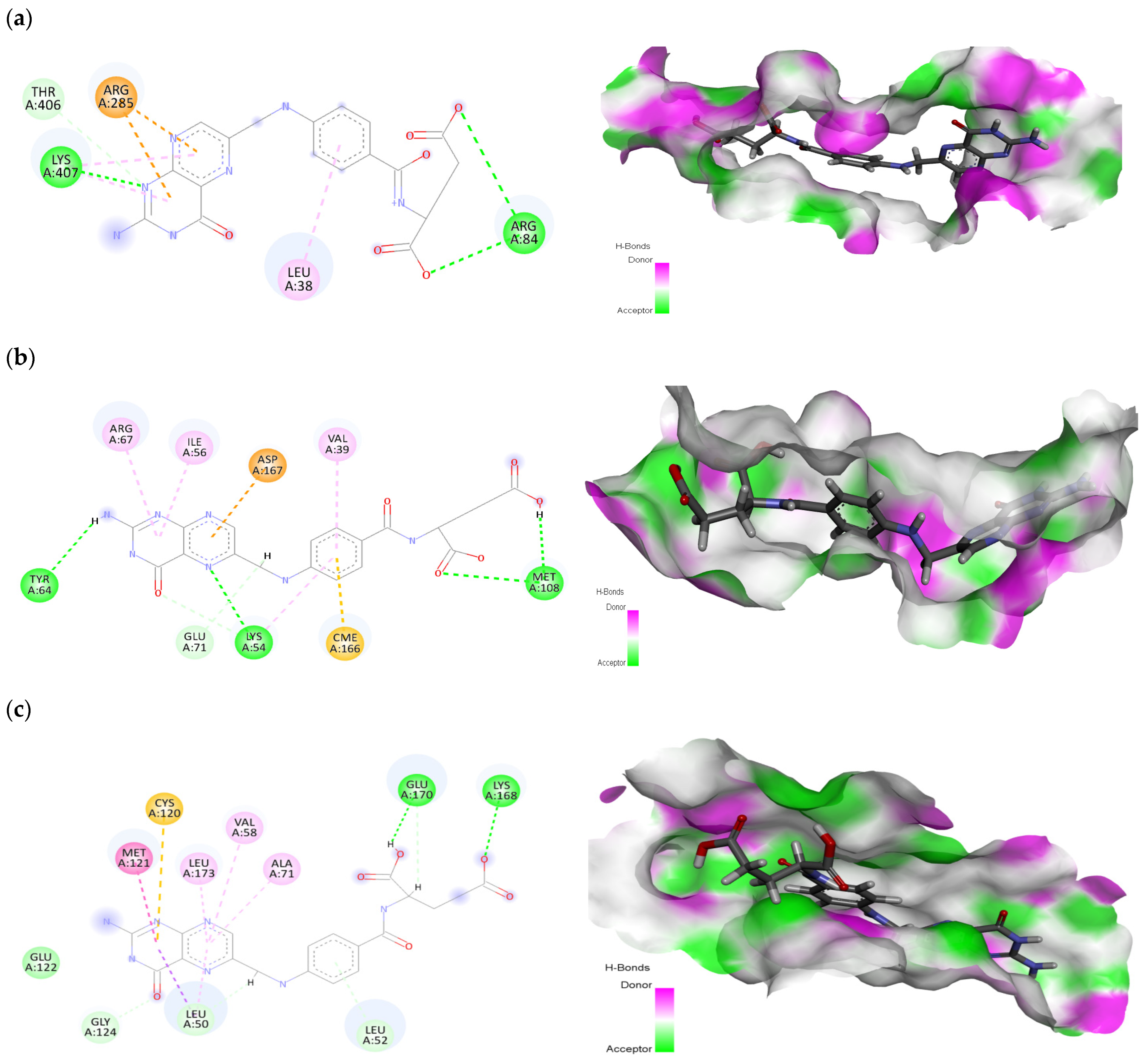
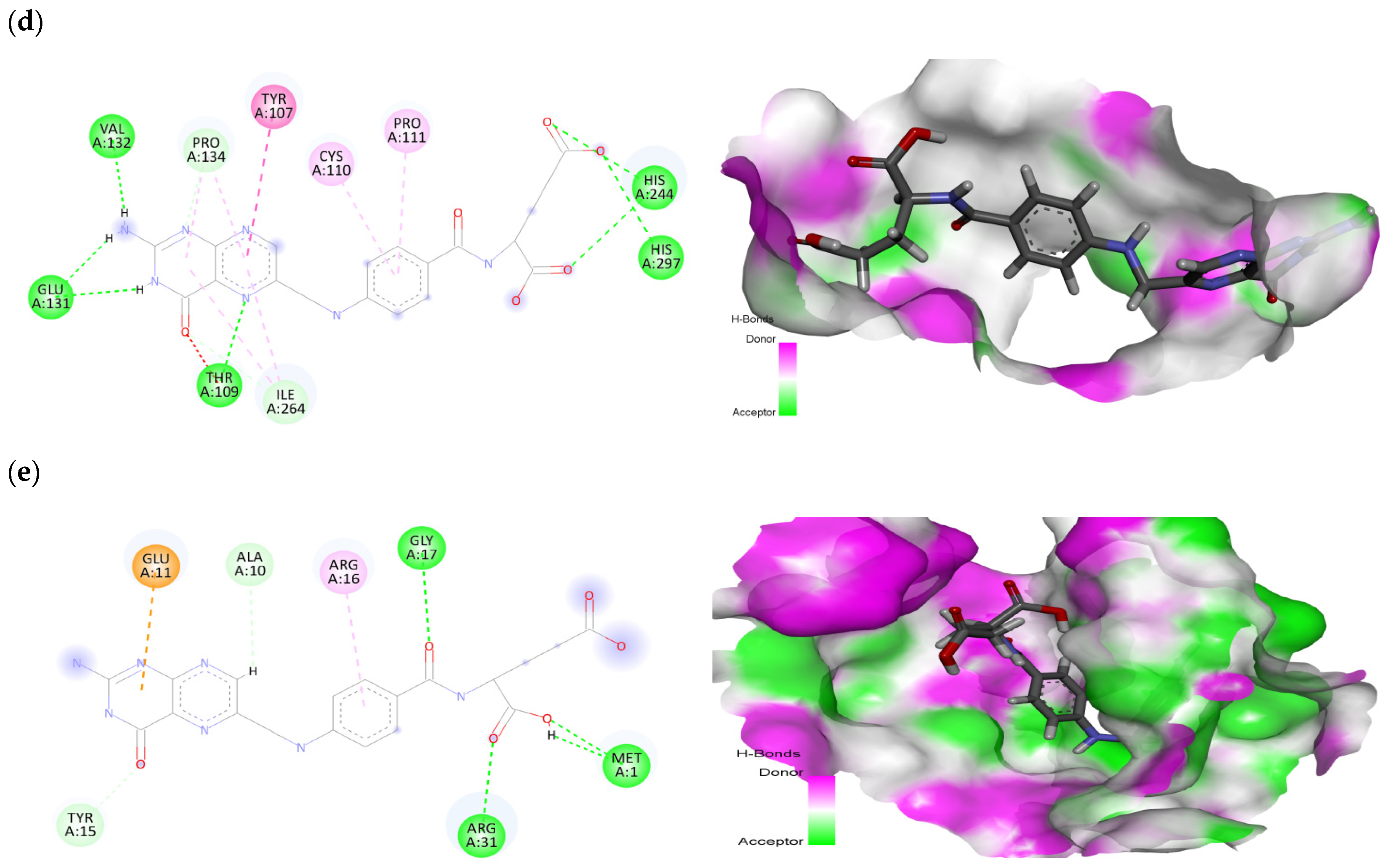
2.2. Ligand-Protein Interactions
2.3. Druggability Analyses
2.4. In Vitro Evaluation of the Best Selected Phytochemicals in MDA-MB-231 Cells
2.5. In Vivo Evaluation
2.5.1. Effect of Stigmasterol and Betulinic Acid on Serum Parameters
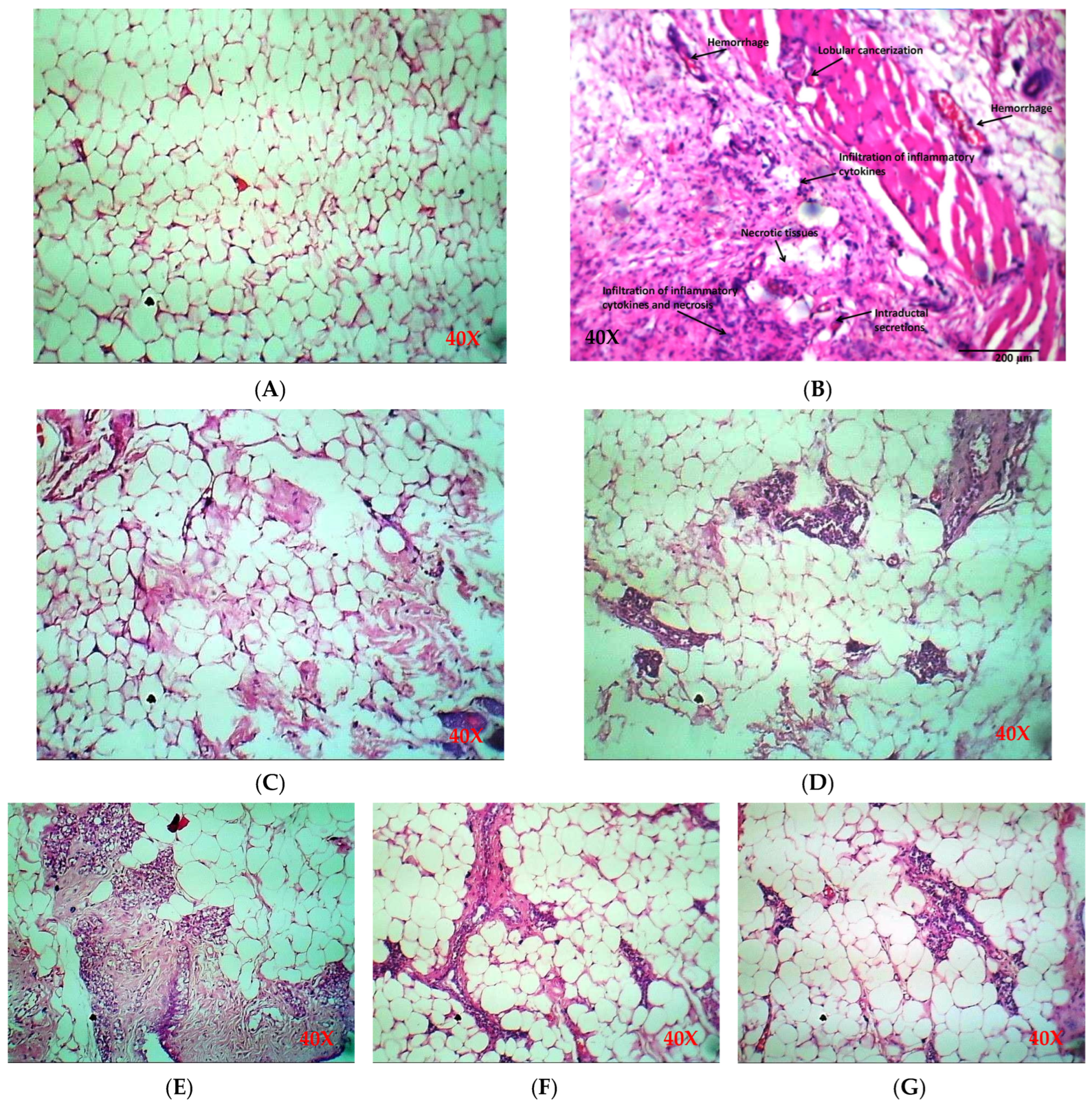
2.5.2. Histology of Breast Tissue
3. Discussion
4. Materials and Methods
4.1. Network Pharmacology Study
4.1.1. Screening of Active Constituents of Nigella sativa
4.1.2. Therapeutic Targets of Nigella sativa
4.1.3. Screening of Targets Related to Breast Cancer
4.1.4. Network Construction of Phytochemicals and Targets
4.1.5. Construction of the Protein–Protein Interaction Network and Identification of Main Targets
4.1.6. GO and KEGG Pathway Enrichment Analyses
4.1.7. Active Compound–Target–Pathway Network (C-T-P)
4.2. Ligand–Protein Docking Study
4.3. Assessment of Pharmacokinetic Parameters
4.4. In Vitro Evaluation
MDA-MB-231 Cell Culture and Cell Viability Determination
4.5. In Vivo Evaluation
4.5.1. Grouping of Animals
4.5.2. Blood Collection
4.5.3. Determination of Serum Parameters
4.5.4. Histopathology of Breast Tissues
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yedjou, C.G.; Sims, J.N.; Miele, L.; Noubissi, F.; Lowe, L.; Fonseca, D.D.; Alo, R.A.; Payton, M.; Tchounwou, P.B. Health and racial disparity in breast cancer. Breast Cancer Metastasis Drug Resist. Chall. Prog. 2019, 1152, 31–49. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-c. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Sharif, S.; Atta, A.; Huma, T.; Shah, A.A.; Afzal, G.; Rashid, S.; Shahid, M.; Mustafa, G. Anticancer, antithrombotic, antityrosinase, and anti-α-glucosidase activities of selected wild and commercial mushrooms from Pakistan. Food Sci. Nutr. 2018, 6, 2170–2176. [Google Scholar] [CrossRef]
- Ai, Y.; Zhao, Z.; Wang, H.; Zhang, X.; Qin, W.; Guo, Y.; Zhao, M.; Tang, J.; Ma, X.; Zeng, J. Pull the plug: Anti-angiogenesis potential of natural products in gastrointestinal cancer therapy. Phytother. Res. 2022, 36, 3371–3393. [Google Scholar] [CrossRef]
- Huang, M.; Lu, J.-J.; Ding, J. Natural products in cancer therapy: Past, present and future. Nat. Prod. Bioprospecting 2021, 11, 5–13. [Google Scholar] [CrossRef]
- Dong, S.; Guo, X.; Han, F.; He, Z.; Wang, Y. Emerging role of natural products in cancer immunotherapy. Acta Pharm. Sin. B 2022, 12, 1163–1185. [Google Scholar] [CrossRef]
- Naeem, M.; Iqbal, M.O.; Khan, H.; Ahmed, M.M.; Farooq, M.; Aadil, M.M.; Jamaludin, M.I.; Hazafa, A.; Tsai, W.-C. A review of twenty years of research on the regulation of signaling pathways by natural products in breast cancer. Molecules 2022, 27, 3412. [Google Scholar] [CrossRef]
- Kordestani, Z.; Shahrokhi-Farjah, M.; Rouholamini, S.E.Y.; Saberi, A. Reduced IKK/NF-kB Expression by Nigella Sativa Extract in Breast Cancer. Middle East J. Cancer 2020, 11, 150–158. [Google Scholar]
- Korak, T.; Ergül, E.; Sazci, A. Nigella sativa and cancer: A review focusing on breast cancer, inhibition of metastasis and enhancement of natural killer cell cytotoxicity. Curr. Pharm. Biotechnol. 2020, 21, 1176–1185. [Google Scholar] [CrossRef]
- Li, T.; Guo, R.; Zong, Q.; Ling, G. Application of molecular docking in elaborating molecular mechanisms and interactions of supramolecular cyclodextrin. Carbohydr. Polym. 2022, 276, 118644. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Younas, S.; Mahrosh, H.S.; Albeshr, M.F.; Bhat, E.A. Molecular Docking and Simulation-Binding Analysis of Plant Phytochemicals with the Hepatocellular Carcinoma Targets Epidermal Growth Factor Receptor and Caspase-9. Molecules 2023, 28, 3583. [Google Scholar] [CrossRef]
- Mustafa, G.; Mahrosh, H.S.; Attique, S.A.; Arif, R.; Farah, M.A.; Al-Anazi, K.M.; Ali, S. Identification of Plant Peptides as Novel Inhibitors of Orthohepevirus A (HEV) Capsid Protein by Virtual Screening. Molecules 2023, 28, 2675. [Google Scholar] [CrossRef]
- Brown, K.A. Metabolic pathways in obesity-related breast cancer. Nat. Rev. Endocrinol. 2021, 17, 350–363. [Google Scholar] [CrossRef]
- Mustafa, G.; Majid, M.; Ghaffar, A.; Yameen, M.; Samad, H.A.; Mahrosh, H.S. Screening and molecular docking of selected phytochemicals against NS5B polymerase of hepatitis C virus. Pak. J. Pharm. Sci. 2020, 33, 2317–2322. [Google Scholar] [PubMed]
- Noor, F.; Tahir ul Qamar, M.; Ashfaq, U.A.; Albutti, A.; Alwashmi, A.S.; Aljasir, M.A. Network pharmacology approach for medicinal plants: Review and assessment. Pharmaceuticals 2022, 15, 572. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, L.; Chen, C.; Nie, J.; Jiao, B. Can EGFR be a therapeutic target in breast cancer? Biochim. Biophys. Acta (BBA)-Rev. Cancer 2022, 1877, 188789. [Google Scholar] [CrossRef]
- Huang, S.; Chen, Y.; Pan, L.; Fei, C.; Wang, N.; Chu, F.; Peng, D.; Duan, X.; Wang, Y. Exploration of the potential mechanism of Tao Hong Si Wu decoction for the treatment of breast cancer based on network pharmacology and in vitro experimental verification. Front. Oncol. 2021, 11, 731522. [Google Scholar] [CrossRef]
- Wang, F.; Wu, K.; Li, Y.; Song, R.; Wu, Y.; Zhang, X.; Song, M.; Liang, L.; Smith-Warner, S.A.; Giovannucci, E.L. Association of folate intake and colorectal cancer risk in the postfortification era in US women. Am. J. Clin. Nutr. 2021, 114, 49–58. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Liu, B.; Liu, Y.; Shi, X.; Li, W.; Xin, H.; Xin, J.; Hao, C. Anticancer Effects of Herbal Medicines in Pancreatic Cancer Through Modulation of Steroid Hormone Response Proteins. Sci. Rep. 2022, 12, 9910. [Google Scholar] [CrossRef]
- Dustin, D.; Gu, G.; Fuqua, S.A. ESR1 mutations in breast cancer. Cancer 2019, 125, 3714–3728. [Google Scholar] [CrossRef]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef] [PubMed]
- Ríos, J.L.; Máñez, S. New pharmacological opportunities for betulinic acid. Planta Medica 2018, 84, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zheng, Y.; Gu, J.; Wang, S.; Wang, N.; Yang, B.; Zhang, F.; Wang, D.; Fu, W.; Wang, Z. Betulinic acid chemosensitizes breast cancer by triggering ER stress-mediated apoptosis by directly targeting GRP78. Cell Death Dis. 2018, 9, 636. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.-Q.; Yu, Y.; Yao, Y.-Q.; Yang, F.-F.; Liao, M.; Song, L.-J.; Li, Y.-L.; Yu, Y.; Li, Y.-J.; Deng, Y.-L. Betulinic acid impairs metastasis and reduces immunosuppressive cells in breast cancer models. Oncotarget 2018, 9, 3794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, J.; Dong, Y.; Zhai, D.; Lai, L.; Dai, F.; Deng, H.; Chen, Y.; Liu, M.; Yi, Z. Cucurbitacin E inhibits breast tumor metastasis by suppressing cell migration and invasion. Breast Cancer Res. Treat. 2012, 135, 445–458. [Google Scholar] [CrossRef]
- Gao, Y.; Ma, Q.; Ma, Y.-B.; Ding, L.; Xu, X.-L.; Wei, D.-F.; Wei, L.; Zhang, J.-W. Betulinic acid induces apoptosis and ultrastructural changes in MDA-MB-231 breast cancer cells. Ultrastruct. Pathol. 2018, 42, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, R.; Bhatti, H.N. Stigmasterol. In A Centum of Valuable Plant Bioactives; Elsevier: Amsterdam, The Netherlands, 2021; pp. 213–232. [Google Scholar]
- Dube, N.P.; Tembu, V.J.; Nyemba, G.R.; Davison, C.; Rakodi, G.H.; Kemboi, D.; de la Mare, J.-A.; Siwe-Noundou, X.; Manicum, A.-L.E. In vitro cytotoxic effect of stigmasterol derivatives against breast cancer cells. BMC Complement. Med. Ther. 2023, 23, 316. [Google Scholar] [CrossRef]
- Muhammad, S.N.; Yaacob, N.S.; Safuwan, N.A.; Fauzi, A.N. Antiglycolytic activities of Strobilanthes crispus active fraction and its bioactive components on triple-negative breast cancer cells in vitro. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2022, 22, 1363–1369. [Google Scholar] [CrossRef]
- Omran, G.; Abd-Alhaseeb, M.; HOUSSEN, M. Chemotherapeutic effect of stigmasterol in sorafenib treated breast cancer cell lines via modulation of NF-κB and ERK signaling pathways. Egypt. J. Chem. 2024, 67, 615–622. [Google Scholar]
- Vundru, S.S.; Kale, R.K.; Singh, R.P. β-sitosterol induces G1 arrest and causes depolarization of mitochondrial membrane potential in breast carcinoma MDA-MB-231 cells. BMC Complement. Altern. Med. 2013, 13, 280. [Google Scholar] [CrossRef] [PubMed]
- Field, M.S.; Stover, P.J. Safety of folic acid. Ann. N. Y. Acad. Sci. 2018, 1414, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Xu, P.; Zhang, D.; Liu, K.; Song, D.; Zheng, Y.; Yang, S.; Li, N.; Hao, Q.; Wu, Y. Association of folate intake and plasma folate level with the risk of breast cancer: A dose-response meta-analysis of observational studies. Aging 2020, 12, 21355. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Gu, Y.; Fu, H.; Liu, C.; Zou, Y.; Chang, H. Association between one-carbon metabolism-related vitamins and risk of breast cancer: A systematic review and meta-analysis of prospective studies. Clin. Breast Cancer 2020, 20, e469–e480. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.; Guddati, A.K. Carcinoembryonic antigen, carbohydrate antigen 19-9, cancer antigen 125, prostate-specific antigen and other cancer markers: A primer on commonly used cancer markers. World J. Oncol. 2023, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Mizejewski, G.J. An Alpha-fetoprotein derived peptide suppresses growth in Breast Cancer and other Malignancies: A Review and Prospectus. Med. Res. Arch. 2023, 11, 1–15. [Google Scholar] [CrossRef]
- Li, J.; Liu, L.; Feng, Z.; Wang, X.; Huang, Y.; Dai, H.; Zhang, L.; Song, F.; Wang, D.; Zhang, P. Tumor markers CA15-3, CA125, CEA and breast cancer survival by molecular subtype: A cohort study. Breast Cancer 2020, 27, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Vivek-Ananth, R.; Mohanraj, K.; Sahoo, A.K.; Samal, A. IMPPAT 2.0: An enhanced and expanded phytochemical atlas of Indian medicinal plants. ACS Omega 2023, 8, 8827–8845. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminformatics 2014, 6, 13. [Google Scholar] [CrossRef]
- Guo, W.; Huang, J.; Wang, N.; Tan, H.-Y.; Cheung, F.; Chen, F.; Feng, Y. Integrating network pharmacology and pharmacological evaluation for deciphering the action mechanism of herbal formula zuojin pill in suppressing hepatocellular carcinoma. Front. Pharmacol. 2019, 10, 1185. [Google Scholar] [CrossRef]
- Tao, W.; Xu, X.; Wang, X.; Li, B.; Wang, Y.; Li, Y.; Yang, L. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J. Ethnopharmacol. 2013, 145, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]
- UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The genecards suite. In Practical Guide to Life Science Databases; Springer: Singapore, 2021; pp. 27–56. [Google Scholar]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Arif, R.; Zia, M.A.; Mustafa, G. Structural and functional annotation of napin-like protein from momordica charantia to explore its medicinal importance. Biochem. Genet. 2022, 60, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Chem. Biol. Methods Protoc. 2015, 1263, 243–250. [Google Scholar]
- Bittrich, S.; Rose, Y.; Segura, J.; Lowe, R.; Westbrook, J.D.; Duarte, J.M.; Burley, S.K. RCSB Protein Data Bank: Improved annotation, search and visualization of membrane protein structures archived in the PDB. Bioinformatics 2022, 38, 1452–1454. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharma, A.; Gupta, U. Molecular Docking studies on the Anti-fungal activity of Allium sativum (Garlic) against Mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1.0.0. Res. Sq. 2021, preprint. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Azeem, M.; Mustafa, G.; Mahrosh, H.S. Virtual screening of phytochemicals by targeting multiple proteins of severe acute respiratory syndrome coronavirus 2: Molecular docking and molecular dynamics simulation studies. Int. J. Immunopathol. Pharmacol. 2022, 36, 03946320221142793. [Google Scholar] [CrossRef]
- Rasul, A.; Khan, M.; Yu, B.; Ali, M.; Bo, Y.J.; Yang, H.; Ma, T. Isoalantolactone, a sesquiterpene lactone, induces apoptosis in SGC-7901 cells via mitochondrial and phosphatidylinositol 3-kinase/Akt signaling pathways. Arch. Pharmacal Res. 2013, 36, 1262–1269. [Google Scholar] [CrossRef]
- Ubhenin, A.E.; Adefolalu, A.A.; Oriakhi, K.; Adamude, F.A.; Dingwoke, E.J.; Ikebuiro, J.O.; Chiwendu, B.C.; Muhammad, M.L.; Omage, K. Caesalpinia pulcherrima lowered serum carcinoembryonic antigen and antigen 125 in 7, 12-Dimethylbenz [a] anthracene-induced Mammary Carcinogenesis in Female Albino Rats. Heliyon 2024, 10, e23401. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Loginova, A.I.; Vyushkov, D.M.; Choi, E.D. Potential Diagnostic Value of Salivary Tumor Markers in Breast, Lung and Ovarian Cancer: A Preliminary Study. Curr. Issues Mol. Biol. 2023, 45, 5084–5098. [Google Scholar] [CrossRef]
- Karnam, K.C.; Ellutla, M.; Bodduluru, L.N.; Kasala, E.R.; Uppulapu, S.K.; Kalyankumarraju, M.; Lahkar, M. Preventive effect of berberine against DMBA-induced breast cancer in female Sprague Dawley rats. Biomed. Pharmacother. 2017, 92, 207–214. [Google Scholar] [CrossRef]
- Abdel-Hamid, N.M.; EL-Gharieb, M.S.; El-Senduny, F.F.; Alnakib, N.A.-A. Effect of Doxorubicin and Cisplatin on Alpha-fetoprotein levels in Hepatocellular Carcinoma Cell lines. Alfarama J. Basic Appl. Sci. 2022, 3, 35–44. [Google Scholar]




| Sr. No. | Genes | Full Name | Degrees |
|---|---|---|---|
| 1 | TNF | Tumor Necrosis Factor | 117 |
| 2 | EGFR | Epidermal Growth Factor Receptor | 92 |
| 3 | SRC | Proto-Oncogene Tyrosine-Protein Kinase Src | 89 |
| 4 | MAPK3 | Mitogen-Activated Protein Kinase 3 | 88 |
| 5 | CASP3 | Caspase 3 | 83 |
| 6 | ESR1 | Estrogen Receptor 1 | 80 |
| 7 | HSP90AA1 | Heat Shock Protein 90 Alpha Family Class A Member 1 | 79 |
| 8 | MAPK1 | Mitogen-Activated Protein Kinase 1 | 76 |
| 9 | PPARG | Peroxisome Proliferator-Activated Receptor Gamma | 76 |
| 10 | PTGS2 | Prostaglandin-Endoperoxide Synthase 2 | 72 |
| Sr. No. | Ligand | Receptor | Docking Score (kcal/mol) | Interacting Residues |
|---|---|---|---|---|
| 1 | Folic acid | EGFR | −9.28 | Leu38, Arg84, Arg285, Thr406, Lys407 |
| 2 | Betulinic acid | −7.27 | Val6, Leu38, Phe263, Tyr275 | |
| 3 | Folic acid | MAPK1 | −8.96 | Val39, Lys54, Ile56, Tyr64, Arg67, Glu71, Met108, Asp167 |
| 4 | Stigmasterol | −6.33 | Ile31, Val39, Lys54, Glu71, Leu156 | |
| 5 | Folic acid | MAPK3 | −8.98 | Leu50, Leu52, Val58, Ala71, Cys120, Met121, Glu122, Gly124, Lys168, Glu170, Leu173 |
| 6 | Stigmasterol | −8.57 | Leu50, Val58, Ala71, Met118, Glu119, Cys120, Met121, Leu173 | |
| 7 | Folic acid | PTGS2 | −8.14 | Tyr107, Thr109, Cys110, Pro111, Glu131, Val132, Pro134, His244, Ile264, His297 |
| 8 | Betulinic acid | −6.95 | Pro111, His244, Ile246, Tyr251, Ile264, Arg296, His297, Val343 | |
| 9 | Folic acid | ESR1 | −7.33 | Met1, Ala10, Glu11, Tyr15, Arg16, Gly17, Arg31 |
| 10 | Betulinic acid | −6.01 | Val8, Lys14, Val18, Arg39 |
| Ligands | Molecular Properties | Violations | |||||
|---|---|---|---|---|---|---|---|
| MW | HBD | HBA | Nrotb | LogP | A | ||
| Stigmasterol | 412.69 | 1 | 1 | 5 | 5.08 | 132.75 | 1 |
| Folic acid | 441.4 | 6 | 3 | 10 | −0.25 | 111.92 | 1 |
| Betulinic acid | 456.7 | 2 | 9 | 2 | 3.83 | 136.91 | 1 |
| Parameter | Groups | ||||||
|---|---|---|---|---|---|---|---|
| Healthy | DMBA | Standard Drug | Stigmasterol (LD) | Stigmasterol (HD) | Betulinic Acid (LD) | Betulinic Acid (HD) | |
| AFP (ng/mL) | 2.1 ± 0.25 | 33.6 ± 6.1 | 14 ± 3 *** | 17 ± 4 ** | 11.3 ± 2.5 *** | 21 ± 4 * | 16 ± 3 *** |
| CA125 (ng/mL) | 3.83 ± 1.45 | 41.3 ± 4.5 | 14.6 ± 2.5 *** | 21.3 ± 2.5 *** | 12 ± 3.6 *** | 16.6 ± 1.5 *** | 11 ± 2 *** |
| Breast Project | ||||||||
|---|---|---|---|---|---|---|---|---|
| Specimen ID | Lobular Cancerization | Lobular Carcinoma Glands | Ductal Hyperplasia | Acute Inflammation | Chronic Inflammation | Necrosis | Hemorrhage | Intraductal Secretions |
| G1 | Intact | Intact | Intact | Not Seen | Not Seen | Not Seen | Not Seen | Seen |
| G2 | Lobular cancerization | Invasive Lobular carcinoma + | Ductal hyperplasia ++ | +++ | +++ | Fat necrosis +++ | +++ | +++ |
| G3 | Intact | Intact | Intact | Not Seen | Not Seen | Not Seen | Not Seen | Seen |
| G4 | Slightly distorted | Intact | Intact | Not Seen | Mild Lymphocyte | Not Seen | Not Seen | Seen |
| G5 | Slightly distorted | Intact | Intact | Not Seen | Mild Lymphocyte | Not Seen | Not Seen | Seen |
| G6 | Intact | Intact | Intact | Not Seen | Not Seen | Not Seen | Not Seen | Seen |
| G7 | Intact | Intact | Intact | Not Seen | Not Seen | Not Seen | Not Seen | Seen |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arif, R.; Bukhari, S.A.; Mustafa, G.; Ahmed, S.; Albeshr, M.F. Network Pharmacology and Experimental Validation to Explore the Potential Mechanism of Nigella sativa for the Treatment of Breast Cancer. Pharmaceuticals 2024, 17, 617. https://doi.org/10.3390/ph17050617
Arif R, Bukhari SA, Mustafa G, Ahmed S, Albeshr MF. Network Pharmacology and Experimental Validation to Explore the Potential Mechanism of Nigella sativa for the Treatment of Breast Cancer. Pharmaceuticals. 2024; 17(5):617. https://doi.org/10.3390/ph17050617
Chicago/Turabian StyleArif, Rawaba, Shazia Anwer Bukhari, Ghulam Mustafa, Sibtain Ahmed, and Mohammed Fahad Albeshr. 2024. "Network Pharmacology and Experimental Validation to Explore the Potential Mechanism of Nigella sativa for the Treatment of Breast Cancer" Pharmaceuticals 17, no. 5: 617. https://doi.org/10.3390/ph17050617
APA StyleArif, R., Bukhari, S. A., Mustafa, G., Ahmed, S., & Albeshr, M. F. (2024). Network Pharmacology and Experimental Validation to Explore the Potential Mechanism of Nigella sativa for the Treatment of Breast Cancer. Pharmaceuticals, 17(5), 617. https://doi.org/10.3390/ph17050617








