Novel Protein Biomarkers and Therapeutic Targets for Type 1 Diabetes and Its Complications: Insights from Summary-Data-Based Mendelian Randomization and Colocalization Analysis
Abstract
:1. Introduction
2. Results
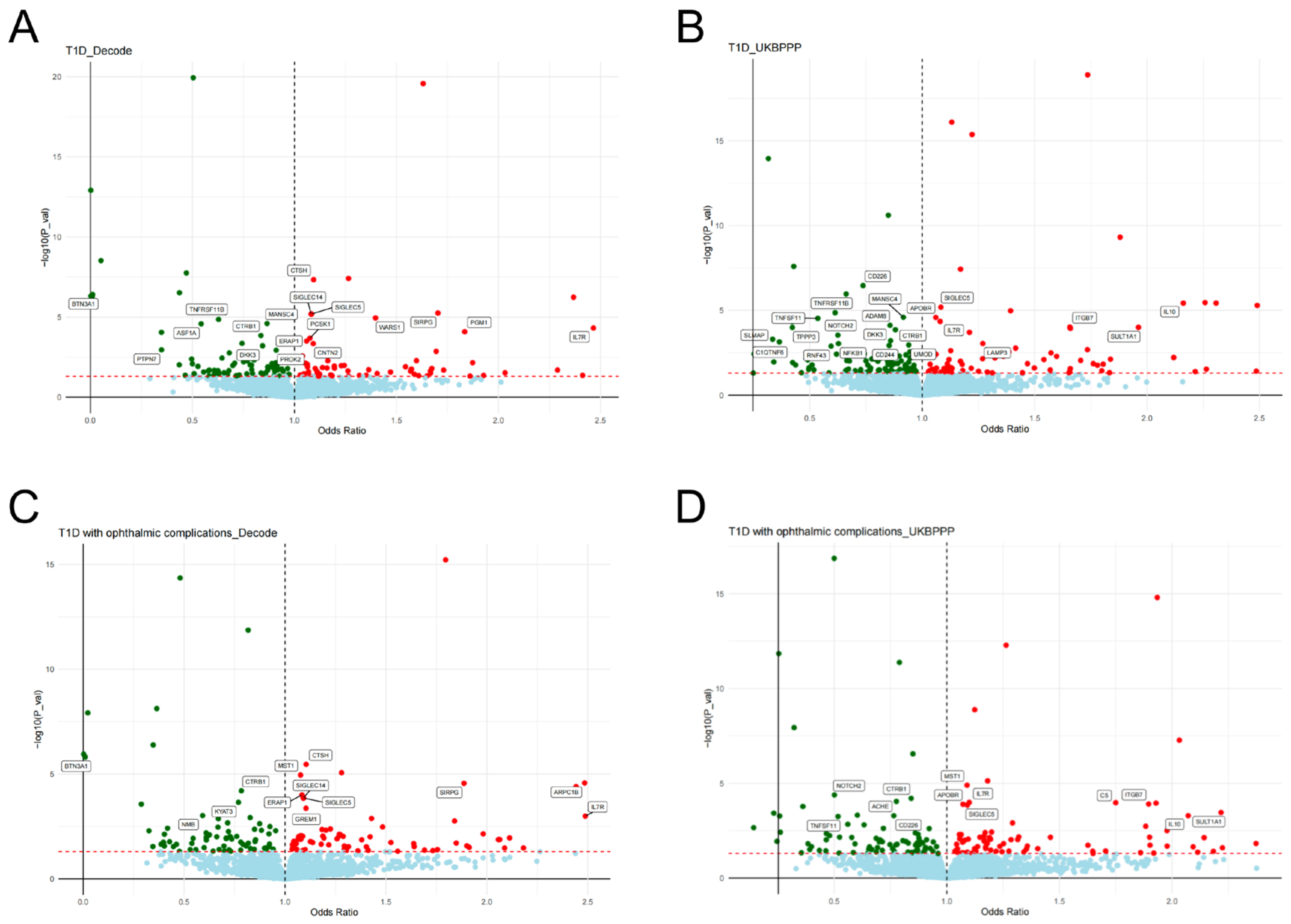
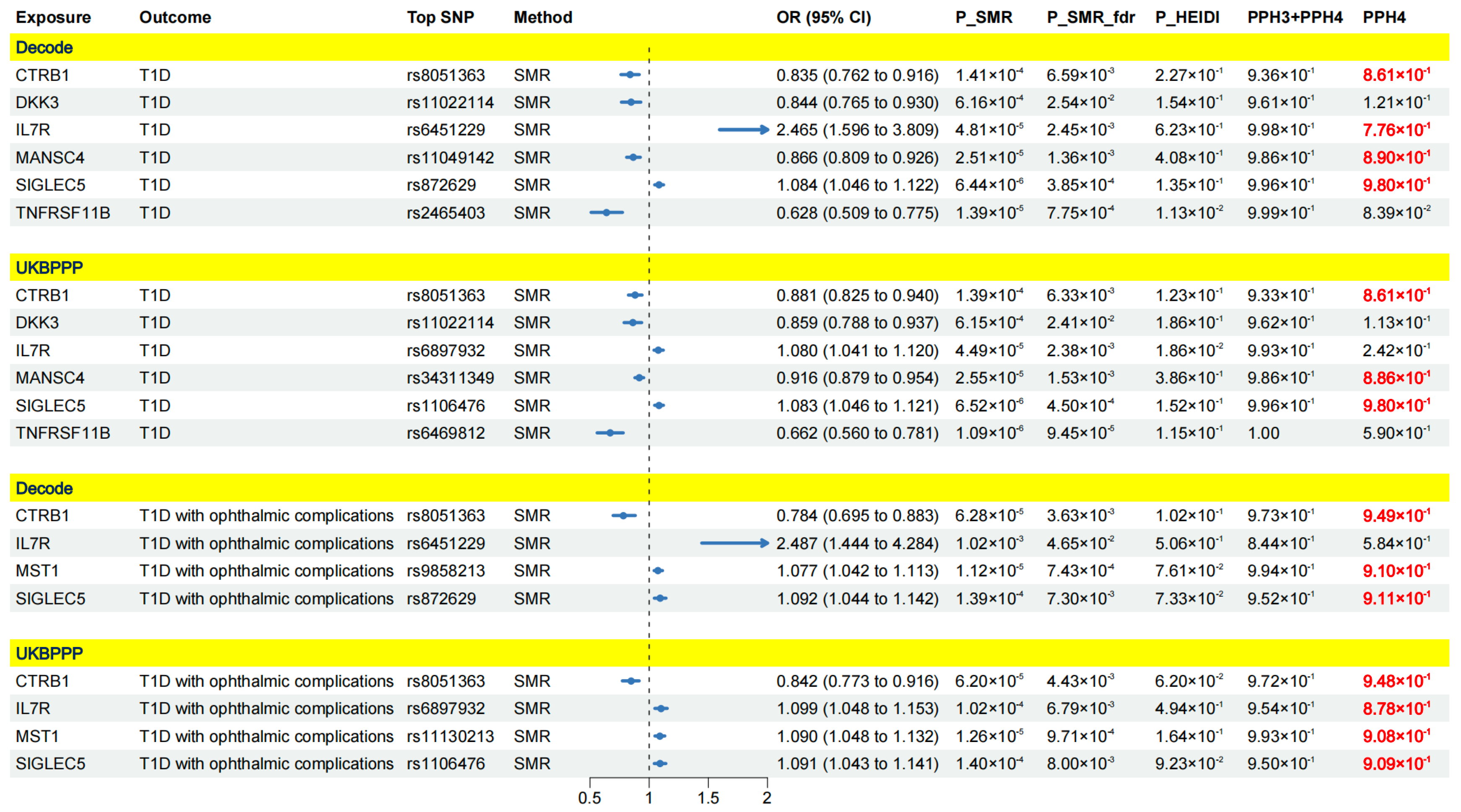
2.1. Results of SMR Analysis and HEIDI Test
2.2. Colocalization Analysis
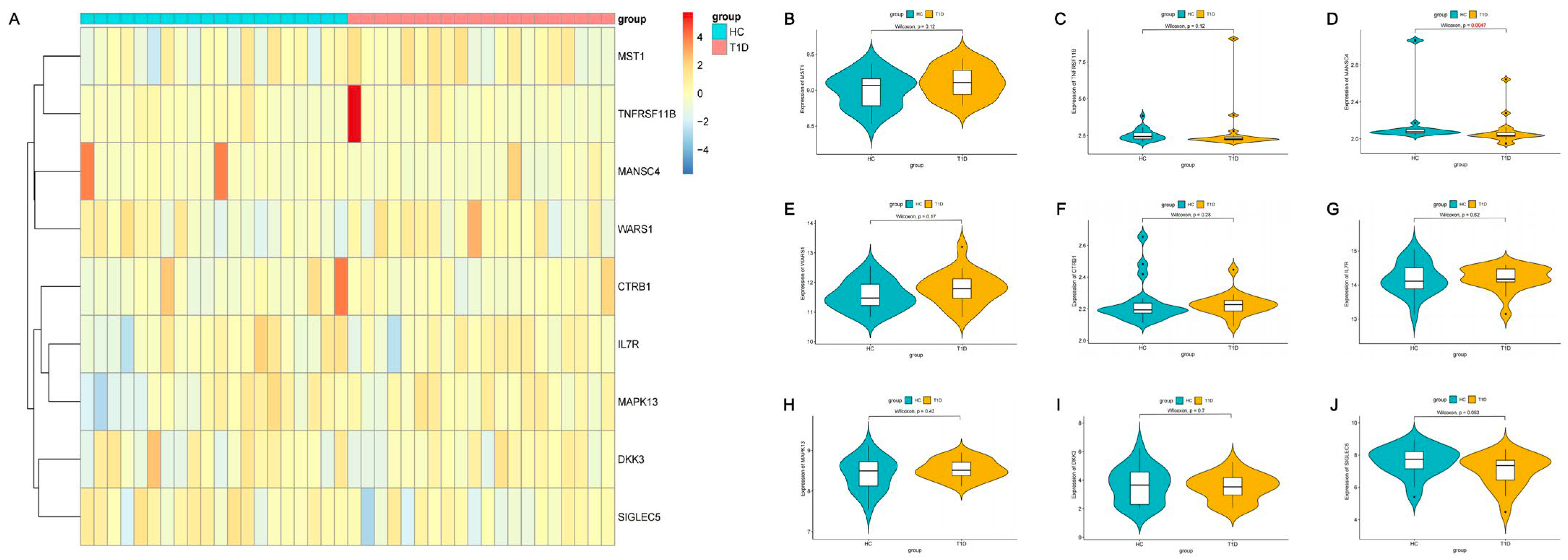
2.3. Expression of Identified Proteins in T1D Patients and Healthy Individuals
2.4. Associations of Identified Protein Targets with Current T1D Medications
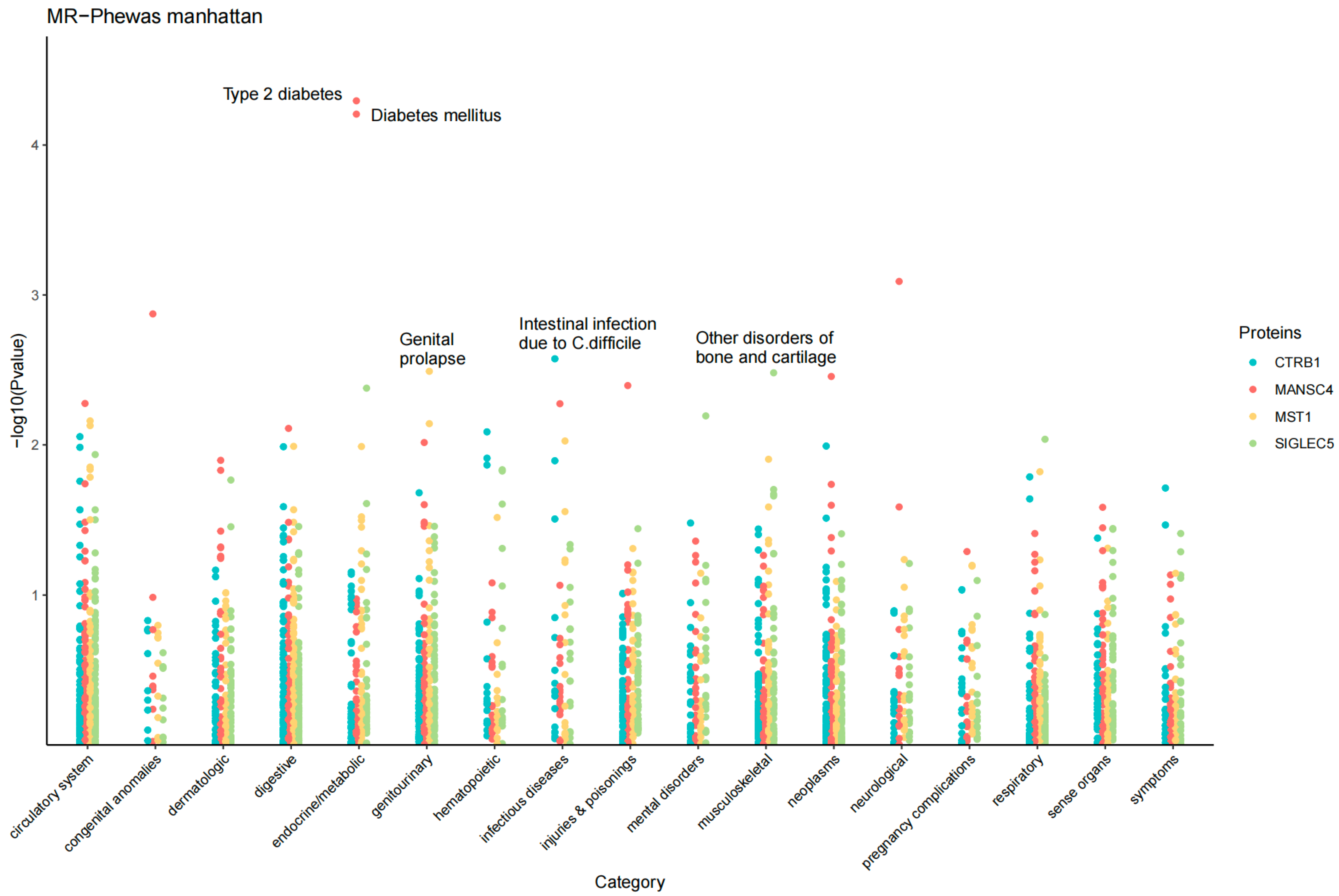
2.5. MR Phenome-Wide Association Studies of Core Therapeutic Targets of T1D
3. Discussion
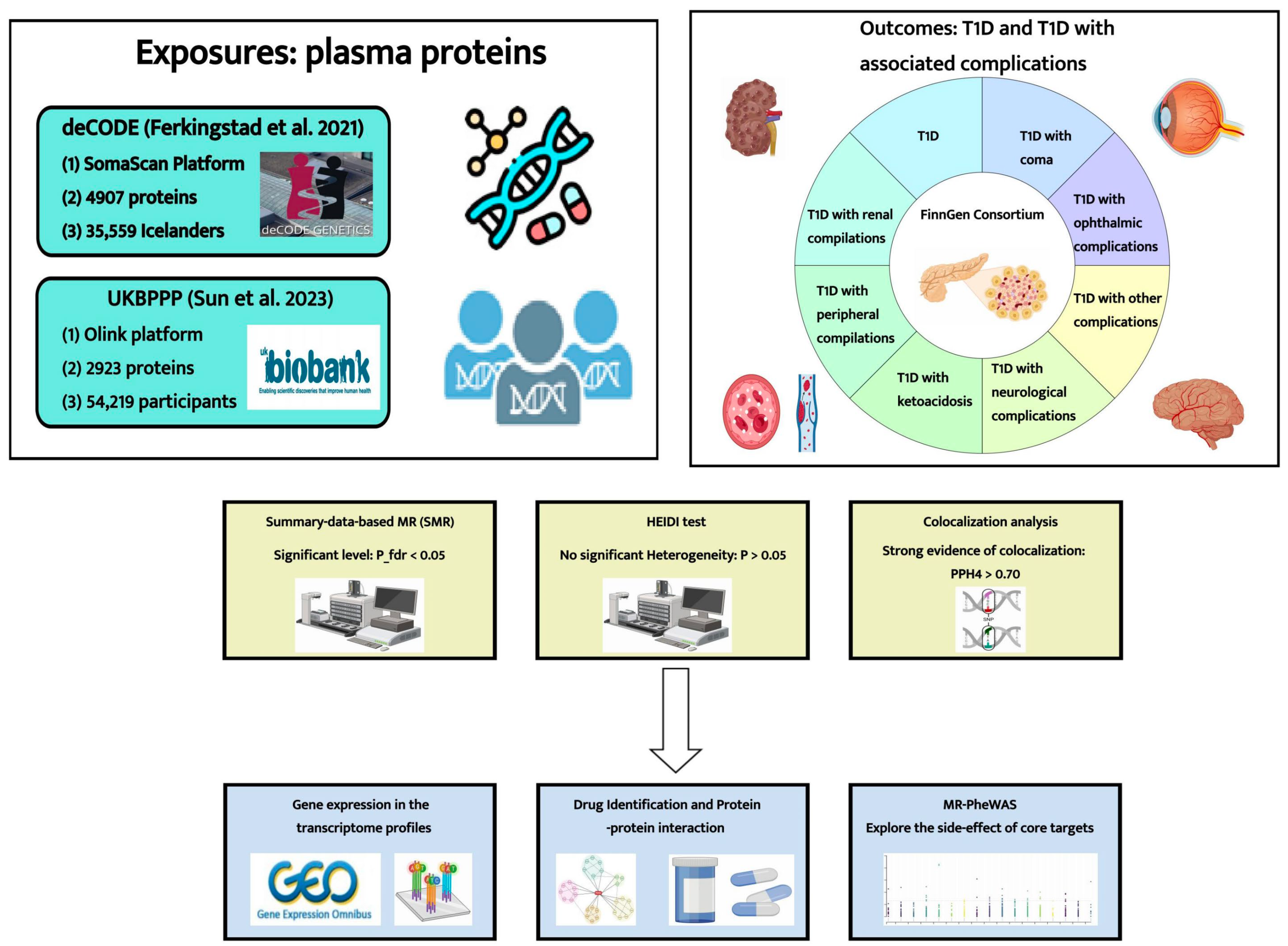
4. Materials and Methods
4.1. Data Sources
4.2. Summary-Data-Based MR (SMR) Analysis
4.3. Bayesian Colocalization Analysis
4.4. Integrating Results from Promote-Wide SMR Analysis (Classification Hierarchy)
4.5. Analysis of Expression Patterns of Identified Proteins in T1D Patients and Healthy Individuals
4.6. Drug Identification and Protein–Protein Interaction (PPI) Network
4.7. MR Phenome-Wide Association Study
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katsarou, A.; Gudbjörnsdottir, S.; Rawshani, A.; Dabelea, D.; Bonifacio, E.; Anderson, B.J.; Jacobsen, L.M.; Schatz, D.A.; Lernmark, Å. Type 1 diabetes mellitus. Nat. Rev. Dis. Primers 2017, 3, 17016. [Google Scholar] [CrossRef] [PubMed]
- Patterson, C.C.; Dahlquist, G.G.; Gyürüs, E.; Green, A.; Soltész, G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: A multicentre prospective registration study. Lancet 2009, 373, 2027–2033. [Google Scholar] [CrossRef] [PubMed]
- Green, A.; Hede, S.M.; Patterson, C.C.; Wild, S.H.; Imperatore, G.; Roglic, G.; Beran, D. Type 1 diabetes in 2017: Global estimates of incident and prevalent cases in children and adults. Diabetologia 2021, 64, 2741–2750. [Google Scholar] [CrossRef] [PubMed]
- Ogle, G.D.; James, S.; Dabelea, D.; Pihoker, C.; Svennson, J.; Maniam, J.; Klatman, E.L.; Patterson, C.C. Global estimates of incidence of type 1 diabetes in children and adolescents: Results from the International Diabetes Federation Atlas, 10th edition. Diabetes Res. Clin. Pract. 2022, 183, 109083. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, S.J.; Levin, D.; Looker, H.C.; Lindsay, R.S.; Wild, S.H.; Joss, N.; Leese, G.; Leslie, P.; McCrimmon, R.J.; Metcalfe, W.; et al. Estimated life expectancy in a Scottish cohort with type 1 diabetes, 2008–2010. JAMA 2015, 313, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, E.S.; Bramer, L.M.; Ansong, C.; Schepmoes, A.A.; Fillmore, T.L.; Gritsenko, M.A.; Clauss, T.R.; Gao, Y.; Piehowski, P.D.; Stanfill, B.A.; et al. Plasma protein biomarkers predict the development of persistent autoantibodies and type 1 diabetes 6 months prior to the onset of autoimmunity. Cell Rep. Med. 2023, 4, 101093. [Google Scholar] [CrossRef] [PubMed]
- Quattrin, T.; Mastrandrea, L.D.; Walker, L.S.K. Type 1 diabetes. Lancet 2023, 401, 2149–2162. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Arnold, M.; Bhagwat, A.M.; Cotton, R.J.; Engelke, R.; Raffler, J.; Sarwath, H.; Thareja, G.; Wahl, A.; DeLisle, R.K.; et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017, 8, 14357. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef]
- Rudman, N.; Kaur, S.; Simunović, V.; Kifer, D.; Šoić, D.; Keser, T.; Štambuk, T.; Klarić, L.; Pociot, F.; Morahan, G.; et al. Integrated glycomics and genetics analyses reveal a potential role for N-glycosylation of plasma proteins and IgGs, as well as the complement system, in the development of type 1 diabetes. Diabetologia 2023, 66, 1071–1083. [Google Scholar] [CrossRef]
- Jia, X.; Wang, S.; Lu, J. Plasma protein biomarkers trailblaze as early predictors of type 1 diabetes. Cell Rep. Med. 2023, 4, 101116. [Google Scholar] [CrossRef] [PubMed]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef] [PubMed]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef]
- Sun, B.B.; Chiou, J.; Traylor, M.; Benner, C.; Hsu, Y.H.; Richardson, T.G.; Surendran, P.; Mahajan, A.; Robins, C.; Vasquez-Grinnell, S.G.; et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 2023, 622, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ruan, X.; Sun, Y.; Lu, S.; Hu, S.; Yuan, S.; Li, X. Multi-omic insight into the molecular networks of mitochondrial dysfunction in the pathogenesis of inflammatory bowel disease. EBioMedicine 2024, 99, 104934. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Shao, Z. Proteome-Wide Mendelian Randomization and Colocalization Analysis Identify Therapeutic Targets for Knee and Hip Osteoarthritis. Biomolecules 2024, 14, 355. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhao, J.; Jiang, F.; Wang, L.; Xiao, Q.; Han, F.; Chen, J.; Yuan, S.; Wei, J.; Larsson, S.C.; et al. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med. 2023, 15, 75. [Google Scholar] [CrossRef]
- Yuan, S.; Jiang, F.; Chen, J.; Lebwohl, B.; Green, P.H.R.; Leffler, D.; Larsson, S.C.; Li, X.; Ludvigsson, J.F. Phenome-wide Mendelian randomization analysis reveals multiple health comorbidities of coeliac disease. EBioMedicine 2024, 101, 105033. [Google Scholar] [CrossRef]
- Yuan, S.; Xu, F.; Li, X.; Chen, J.; Zheng, J.; Mantzoros, C.S.; Larsson, S.C. Plasma proteins and onset of type 2 diabetes and diabetic complications: Proteome-wide Mendelian randomization and colocalization analyses. Cell Rep. Med. 2023, 4, 101174. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, S.; Huang, C.; Chen, L.; Studholme, D.J.; Zhao, S.; Yu, L. MANSC: A seven-cysteine-containing domain present in animal membrane and extracellular proteins. Trends Biochem. Sci. 2004, 29, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Lafleur, S.; Bodein, A.; Mbuya Malaïka Mutombo, J.; Mathieu, A.; Joly Beauparlant, C.; Minne, X.; Chandad, F.; Droit, A.; Houde, V.P. Multi-Omics Data Integration Reveals Key Variables Contributing to Subgingival Microbiome Dysbiosis-Induced Inflammatory Response in a Hyperglycemic Microenvironment. Int. J. Mol. Sci. 2023, 24, 8832. [Google Scholar] [CrossRef] [PubMed]
- Rosendahl, J.; Kirsten, H.; Hegyi, E.; Kovacs, P.; Weiss, F.U.; Laumen, H.; Lichtner, P.; Ruffert, C.; Chen, J.M.; Masson, E.; et al. Genome-wide association study identifies inversion in the CTRB1-CTRB2 locus to modify risk for alcoholic and non-alcoholic chronic pancreatitis. Gut 2018, 67, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.Y.; Zou, W.B.; Masson, E.; Hu, L.H.; Ferec, C.; Chen, J.M.; Li, Z.S.; Liao, Z. The CTRB1-CTRB2 risk allele for chronic pancreatitis discovered in European populations does not contribute to disease risk variation in the Chinese population due to near allele fixation. Gut 2018, 67, 1368–1369. [Google Scholar] [CrossRef] [PubMed]
- Inshaw, J.R.J.; Sidore, C.; Cucca, F.; Stefana, M.I.; Crouch, D.J.M.; McCarthy, M.I.; Mahajan, A.; Todd, J.A. Analysis of overlapping genetic association in type 1 and type 2 diabetes. Diabetologia 2021, 64, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- t Hart, L.M.; Fritsche, A.; Nijpels, G.; van Leeuwen, N.; Donnelly, L.A.; Dekker, J.M.; Alssema, M.; Fadista, J.; Carlotti, F.; Gjesing, A.P.; et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes 2013, 62, 3275–3281. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Rodríguez, R.; Avendaño-Ortíz, J.; Montalbán-Hernández, K.; Ruiz-Rodríguez, J.C.; Ferrer, R.; Martín-Quirós, A.; Maroun-Eid, C.; González-López, J.J.; Fàbrega, A.; Terrón-Arcos, V.; et al. The prognostic impact of SIGLEC5-induced impairment of CD8(+) T cell activation in sepsis. EBioMedicine 2023, 97, 104841. [Google Scholar] [CrossRef] [PubMed]
- Munz, M.; Willenborg, C.; Richter, G.M.; Jockel-Schneider, Y.; Graetz, C.; Staufenbiel, I.; Wellmann, J.; Berger, K.; Krone, B.; Hoffmann, P.; et al. A genome-wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum. Mol. Genet. 2017, 26, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.; Freitag-Wolf, S.; Weiner, J., 3rd; Chopra, A.; Top, T.; Dommisch, H.; Schaefer, A.S. Case-only design identifies interactions of genetic risk variants at SIGLEC5 and PLG with the lncRNA CTD-2353F22.1 implying the importance of periodontal wound healing for disease aetiology. J. Clin. Periodontol. 2023, 50, 90–101. [Google Scholar] [CrossRef]
- Mueller, R.; Chopra, A.; Dommisch, H.; Schaefer, A.S. Periodontitis Risk Variants at SIGLEC5 Impair ERG and MAFB Binding. J. Dent. Res. 2022, 101, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Wei, Z.; Yin, J.; Zhang, B.; Zhang, T.; Deng, C.; Huang, Y.; Zhang, N. Genetic susceptibility of common polymorphisms in NIN and SIGLEC5 to chronic periodontitis. Sci. Rep. 2019, 9, 2088. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, L.; Zhang, Y.; Li, W.; Li, J.; Wang, Y.; Meng, C.; Qin, J.; Zheng, Z.H.; Lan, H.Y.; et al. Tubule-Specific Mst1/2 Deficiency Induces CKD via YAP and Non-YAP Mechanisms. J. Am. Soc. Nephrol. 2020, 31, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Ardestani, A.; Maedler, K. MST1: A promising therapeutic target to restore functional beta cell mass in diabetes. Diabetologia 2016, 59, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wang, Y.; Sun, L.; Zhou, S.; Xu, J.; Xing, D. MST1: A future novel target for cardiac diseases. Int. J. Biol. Macromol. 2023, 239, 124296. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ni, H.; Rui, Q.; Gao, R.; Chen, G. Mst1: Function and Mechanism in Brain and Myocardial Ischemia Reperfusion Injury. Curr. Neuropharmacol. 2018, 16, 1358–1364. [Google Scholar] [CrossRef] [PubMed]
- Mathis, D.; Vence, L.; Benoist, C. beta-Cell death during progression to diabetes. Nature 2001, 414, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Ardestani, A.; Paroni, F.; Azizi, Z.; Kaur, S.; Khobragade, V.; Yuan, T.; Frogne, T.; Tao, W.; Oberholzer, J.; Pattou, F.; et al. MST1 is a key regulator of beta cell apoptosis and dysfunction in diabetes. Nat. Med. 2014, 20, 385–397. [Google Scholar] [CrossRef]
- McKinnon, C.M.; Docherty, K. Pancreatic duodenal homeobox-1, PDX-1, a major regulator of beta cell identity and function. Diabetologia 2001, 44, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Brissova, M.; Shiota, M.; Nicholson, W.E.; Gannon, M.; Knobel, S.M.; Piston, D.W.; Wright, C.V.; Powers, A.C. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J. Biol. Chem. 2002, 277, 11225–11232. [Google Scholar] [CrossRef]
- Zheng, J.; Haberland, V.; Baird, D.; Walker, V.; Haycock, P.C.; Hurle, M.R.; Gutteridge, A.; Erola, P.; Liu, Y.; Luo, S.; et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020, 52, 1122–1131. [Google Scholar] [CrossRef]
- Deng, Y.T.; Ou, Y.N.; Wu, B.S.; Yang, Y.X.; Jiang, Y.; Huang, Y.Y.; Liu, Y.; Tan, L.; Dong, Q.; Suckling, J.; et al. Identifying causal genes for depression via integration of the proteome and transcriptome from brain and blood. Mol. Psychiatry 2022, 27, 2849–2857. [Google Scholar] [CrossRef]
- Glickman, M.E.; Rao, S.R.; Schultz, M.R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J. Clin. Epidemiol. 2014, 67, 850–857. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Su, W.M.; Gu, X.J.; Dou, M.; Duan, Q.Q.; Jiang, Z.; Yin, K.F.; Cai, W.C.; Cao, B.; Wang, Y.; Chen, Y.P. Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 2023, 94, 954–961. [Google Scholar] [CrossRef]
- Liu, B.; Gloudemans, M.J.; Rao, A.S.; Ingelsson, E.; Montgomery, S.B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019, 51, 768–769. [Google Scholar] [CrossRef]
- Santos, A.S.; Cunha-Neto, E.; Gonfinetti, N.V.; Bertonha, F.B.; Brochet, P.; Bergon, A.; Moreira-Filho, C.A.; Chevillard, C.; da Silva, M.E.R. Prevalence of Inflammatory Pathways Over Immuno-Tolerance in Peripheral Blood Mononuclear Cells of Recent-Onset Type 1 Diabetes. Front. Immunol. 2021, 12, 765264. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef]
- Zhou, W.; Nielsen, J.B.; Fritsche, L.G.; Dey, R.; Gabrielsen, M.E.; Wolford, B.N.; LeFaive, J.; VandeHaar, P.; Gagliano, S.A.; Gifford, A.; et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018, 50, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Scott, R.A.; Timpson, N.J.; Davey Smith, G.; Thompson, S.G. Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 2015, 30, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qin, F.; Li, X.; Du, X.; Li, T. Identification of novel proteins for lacunar stroke by integrating genome-wide association data and human brain proteomes. BMC Med. 2022, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, M.; Yang, J. Novel Protein Biomarkers and Therapeutic Targets for Type 1 Diabetes and Its Complications: Insights from Summary-Data-Based Mendelian Randomization and Colocalization Analysis. Pharmaceuticals 2024, 17, 766. https://doi.org/10.3390/ph17060766
Zou M, Yang J. Novel Protein Biomarkers and Therapeutic Targets for Type 1 Diabetes and Its Complications: Insights from Summary-Data-Based Mendelian Randomization and Colocalization Analysis. Pharmaceuticals. 2024; 17(6):766. https://doi.org/10.3390/ph17060766
Chicago/Turabian StyleZou, Mingrui, and Jichun Yang. 2024. "Novel Protein Biomarkers and Therapeutic Targets for Type 1 Diabetes and Its Complications: Insights from Summary-Data-Based Mendelian Randomization and Colocalization Analysis" Pharmaceuticals 17, no. 6: 766. https://doi.org/10.3390/ph17060766
APA StyleZou, M., & Yang, J. (2024). Novel Protein Biomarkers and Therapeutic Targets for Type 1 Diabetes and Its Complications: Insights from Summary-Data-Based Mendelian Randomization and Colocalization Analysis. Pharmaceuticals, 17(6), 766. https://doi.org/10.3390/ph17060766





