From Drug Discovery to Drug Approval: A Comprehensive Review of the Pharmacogenomics Status Quo with a Special Focus on Egypt
Abstract
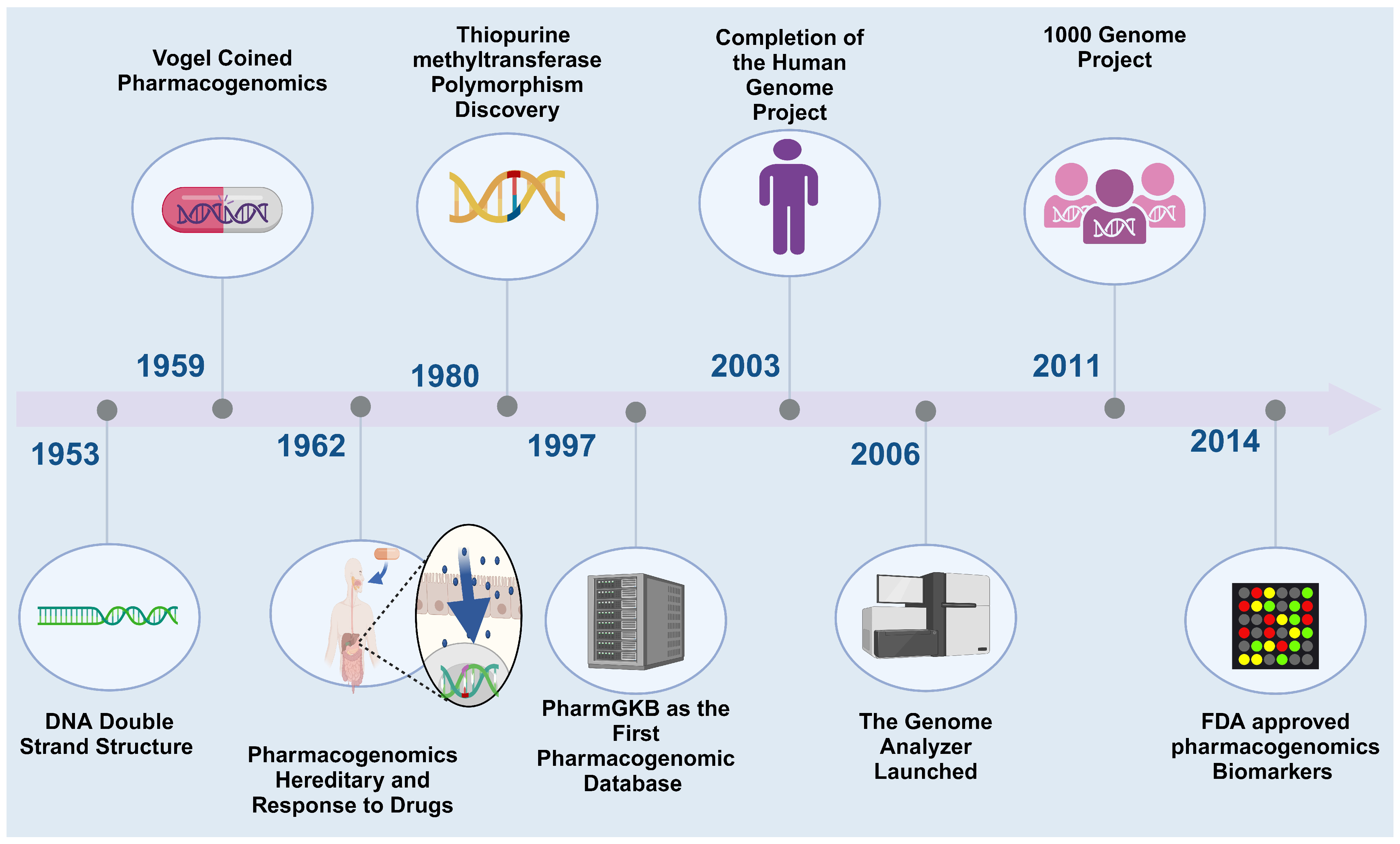
:1. Introduction
2. The Current Status of PGx
2.1. CYP2D6 and Pimozide
2.2. CYP2C9 with Warfarin and Phenytoin
2.3. CYP2C9 and VKORC1 with Warfarin
2.4. SLCO1B1 and Statins
2.5. ADRB1 and 2 Receptors with Cardiovascular Diseases (CVDs)
2.6. Other Involving Enzymes: Thiopurine S-Methyltransferase (TPMT)
2.7. The Effect of Genetic Polymorphism on Anticancer and Antiviral Treatment
3. Current Status in Egypt
4. Barriers and Challenges Hurdle the Clinical Implementation of PGx in Egypt
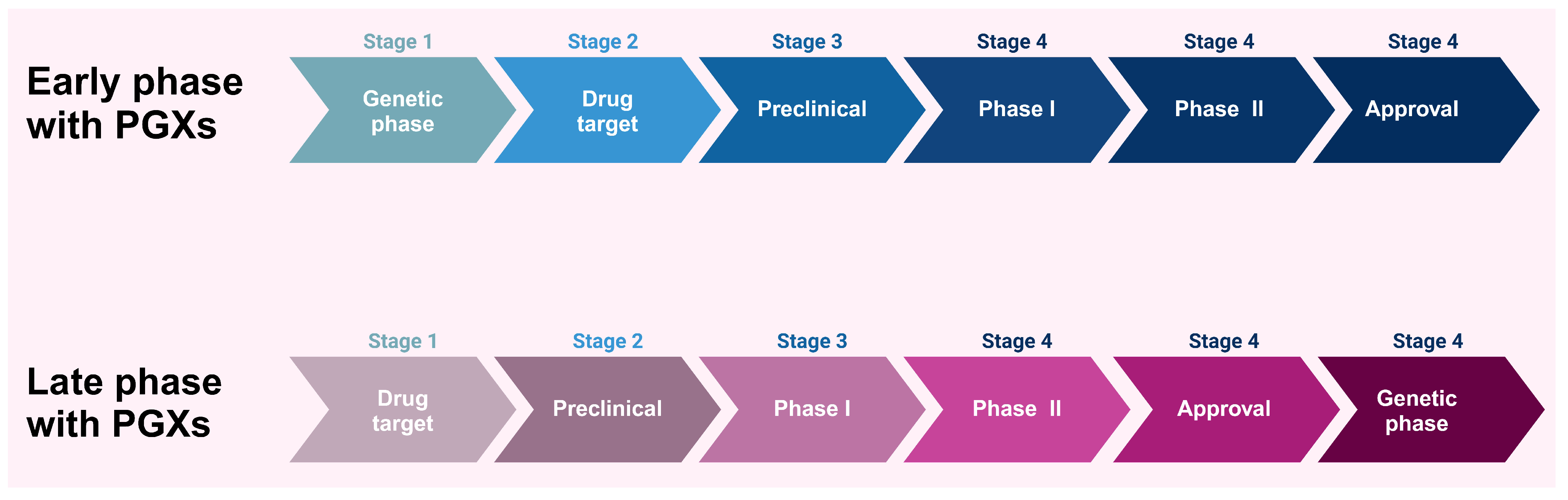
5. Impact of PGx on Drug Development and Drug Discovery
6. PGx of Repurposed Drugs
7. PGx and Pharmacoeconomics
8. Regulatory Considerations for PGx
| Aspect | FDA | EMA | Ref. |
|---|---|---|---|
| Regulatory Framework | Provides an explicit guide on data submission | Provides a detailed flexible approach on the role of PGx in drug development | [96] |
| Biomarkers | Requires validation of biomarkers during the process of approval and incorporates them in labeling | Recommends biomarkers use with flexible validation process | [97] |
| Clinical Development | Recommends the incorporation of PGx data in clinical trials | Recommends the incorporation of PGx data in clinical trials depending on case-by-case clinical situation | [98] |
| Drug Development | Warrants comprehensive PGx data for new drug applications. | Recommends the incorporation of PGx data, with emphasis on benefit–risk balance | [99] |
| Labeling | Detailed PGx data encompassed in drug labels to guide personalized prescribing | PGx data included in the drug labels | [100] |
| PGx Test | Requires PGx testing for drugs with substantial genetic polymorphism | Recommends PGx testing on the basis of evidence, considering data from post-marketing surveillance | [101] |
9. Legal and Social Implications of PGx
10. Future Perspectives and Opportunities
10.1. Integration of PGx Data in Drug Discovery and Development Pipelines
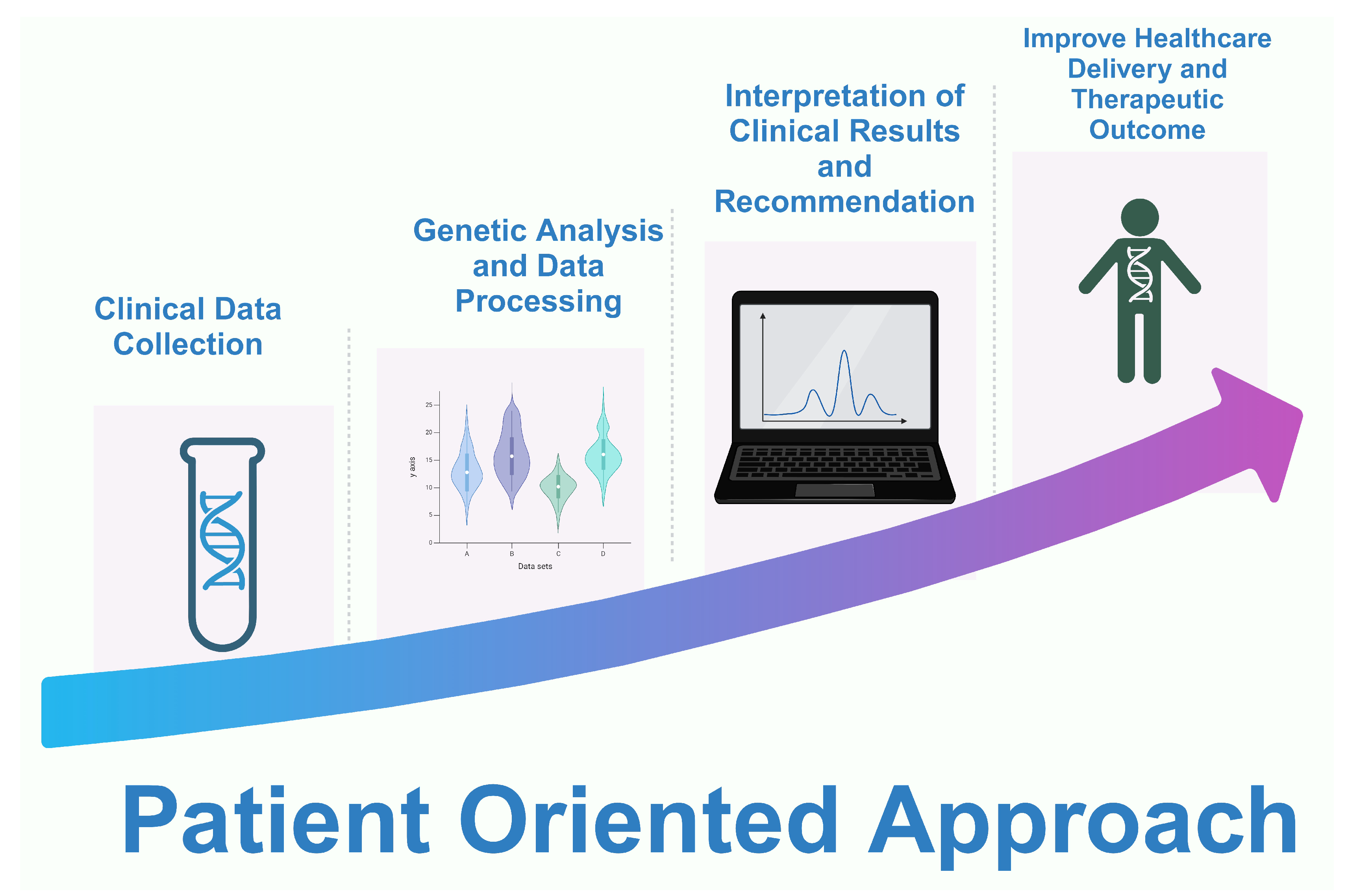
10.2. The Potential Impact of PGx on Precision Medicine and Healthcare Outcomes
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Khasawneh, L.Q.; Al-Mahayri, Z.N.; Ali, B.R. Mendelian Randomization in Pharmacogenomics: The Unforeseen Potentials. Biomed. Pharmacother. 2022, 150, 112952. [Google Scholar] [CrossRef]
- Ilyina, E.S.; Filippova, N.V.; Barylnik, Y.B. Pharmacogenetics of antidepressants (from history to the present). Vestn. Nevrol. Psihiatr. Nejrohir. Bull. Neurol. Psychiatry Neurosurg. 2021, 14, 713–729. [Google Scholar] [CrossRef]
- De Almeida Velozo, C.; De Almeida, T.B.; De Azevedo, M.C.V.M.; Espasandin, I.; Da Cunha Pinto, J.F.; López, S.; Pizzatti, L.; Tanuri, A.; Da Silva Santos, S.; Ribeiro-Alves, M.; et al. Polymorphisms at CYP Enzymes, NR1I2 and NR1I3 in Association with Virologic Response to Antiretroviral Therapy in Brazilian HIV-Positive Individuals. Pharm. J. 2022, 22, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Malsagova, K.A.; Butkova, T.V.; Kopylov, A.T.; Izotov, A.A.; Potoldykova, N.V.; Enikeev, D.V.; Grigoryan, V.; Tarasov, A.; Stepanov, A.A.; Kaysheva, A.L. Pharmacogenetic Testing: A Tool for Personalized Drug Therapy Optimization. Pharmaceutics 2020, 12, 1240. [Google Scholar] [CrossRef]
- Topić, E.; Štefanović, M.; Primorac, D.; Bach-Rojecky, L.; Höppner, W. Pharmacogenomics of Drug-Metabolizing Enzymes. In Pharmacogenomics in Clinical Practice; Primorac, D., Höppner, W., Bach-Rojecky, L., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 35–60. ISBN 978-3-031-45902-3. [Google Scholar]
- Pirmohamed, M. Pharmacogenomics: Current Status and Future Perspectives. Nat. Rev. Genet. 2023, 24, 350–362. [Google Scholar] [CrossRef]
- Shawky, A.; Sabit, H.; Nazih, M.; Baraka, K.; El-Zawahry, M. CYP2C19 Polymorphism in Ischemic Heart Disease Patients Taking Clopidogrel After Percutaneous Coronary Intervention in Egypt. J. Epidemiol. Glob. Health 2023, 13, 374–383. [Google Scholar] [CrossRef]
- Beunk, L.; Nijenhuis, M.; Soree, B.; De Boer-Veger, N.J.; Buunk, A.-M.; Guchelaar, H.J.; Houwink, E.J.F.; Risselada, A.; Rongen, G.A.P.J.M.; Van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) Guideline for the Gene-Drug Interaction between CYP2D6, CYP3A4 and CYP1A2 and Antipsychotics. Eur. J. Hum. Genet. 2024, 32, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Nofziger, C.; Turner, A.J.; Sangkuhl, K.; Whirl-Carrillo, M.; Agúndez, J.A.G.; Black, J.L.; Dunnenberger, H.M.; Ruano, G.; Kennedy, M.A.; Phillips, M.S.; et al. PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 2020, 107, 154–170. [Google Scholar] [CrossRef] [PubMed]
- Lauschke, V.M.; Milani, L.; Ingelman-Sundberg, M. Pharmacogenomic Biomarkers for Improved Drug Therapy—Recent Progress and Future Developments. AAPS J. 2018, 20, 4. [Google Scholar] [CrossRef]
- Hua Gan, S.; Ismail, R.; Adnan, W.A.W.; Zulmi, W. Impact of CYP2D6 Genetic Polymorphism on Tramadol Pharmacokinetics and Pharmacodynamics. Mol. Diagn. Ther. 2007, 11, 171–181. [Google Scholar] [CrossRef]
- Facal, F.; Portela, B.; Gil-Rodríguez, A.; Barros, F.; Maroñas, O.; Carracedo, A. Deletion of the CYP2D6 Gene as a Likely Explanation for the Serious Side Effects of the Antipsychotic Drug Pimozide: A Case Report. Front. Pharmacol. 2023, 14, 1237446. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Van Den Broek, W.W.A.; Mani, N.; Azzahhafi, J.; Ten Berg, J.M. CYP2C9 Polymorphisms and the Risk of Cardiovascular Events in Patients Treated with Clopidogrel: Combined Data from the POPular Genetics and POPular AGE Trials. Am. J. Cardiovasc. Drugs 2023, 23, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Soltani Banavandi, M.J.; Satarzadeh, N. Association between VKORC1 Gene Polymorphism and Warfarin Dose Requirement and Frequency of VKORC1 Gene Polymorphism in Patients from Kerman Province. Pharmacogenom. J. 2020, 20, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1 and Simvastatin-Induced Myopathy: 2014 Update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Charlab, R.; Zhang, L. Pharmacogenomics: Historical Perspective and Current Status. In Pharmacogenomics; Innocenti, F., Van Schaik, R.H.N., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1015, pp. 3–22. ISBN 978-1-62703-434-0. [Google Scholar]
- Leineweber, K.; Heusch, G. β1- and β2-Adrenoceptor Polymorphisms and Cardiovascular Diseases. Br. J. Pharmacol. 2009, 158, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.; Thompson, J.; Power, A. Pharmacogenomics: Integration into Drug Discovery and Development. Curr. Top. Med. Chem. 2005, 5, 1039–1046. [Google Scholar] [CrossRef]
- Gréen, H.; Skoglund, K.; Rommel, F.; Mirghani, R.A.; Lotfi, K. CYP3A activity influences imatinib response in patients with chronic myeloid leukemia: A pilot study on in vivo CYP3A activity. Eur. J. Clin. Pharmacol. 2010, 66, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Adeagbo, B.A.; Bolaji, O.O.; Olugbade, T.A.; Durosinmi, M.A.; Bolarinwa, R.A.; Masimirembwa, C. Influence of CYP3A5*3 and ABCB1 C3435T on Clinical Outcomes and Trough Plasma Concentrations of Imatinib in Nigerians with Chronic Myeloid Leukaemia. J. Clin. Pharm. Ther. 2016, 41, 546–551. [Google Scholar] [CrossRef]
- Kassogue, Y.; Quachouh, M.; Dehbi, H.; Quessar, A.; Benchekroun, S.; Nadifi, S. Functional Polymorphism of CYP2B6 G15631T Is Associated with Hematologic and Cytogenetic Response in Chronic Myeloid Leukemia Patients Treated with Imatinib. Med. Oncol. Northwood Lond. Engl. 2014, 31, 782. [Google Scholar] [CrossRef]
- Nath, A.; Wang, J.; Huang, R.S. Pharmacogenetics and pharmacogenomics of targeted therapeutics in chronic myeloid leukemia. Mol. Diagn. Ther. 2017, 21, 621–631. [Google Scholar] [CrossRef]
- Singer, J.B.; Shou, Y.; Giles, F.; Kantarjian, H.M.; Hsu, Y.; Robeva, A.S.; Rae, P.; Weitzman, A.; Meyer, J.M.; Dugan, M.; et al. UGT1A1 Promoter Polymorphism Increases Risk of Nilotinib-Induced Hyperbilirubinemia. Leukemia 2007, 21, 2311–2315. [Google Scholar] [CrossRef] [PubMed]
- Abumiya, M.; Takahashi, N.; Niioka, T.; Kameoka, Y.; Fujishima, N.; Tagawa, H.; Sawada, K.; Miura, M. Influence of UGT1A1 6, 27, and 28 Polymorphisms on Nilotinib-Induced Hyperbilirubinemia in Japanese Patients with Chronic Myeloid Leukemia. Drug Metab. Pharmacokinet. 2014, 29, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Minami, Y.; Mitsuma, A.; Morita, S.; Inada-Inoue, M.; Oguri, T.; Shimokata, T.; Sugishita, M.; Naoe, T.; Ando, Y. Association between Severe Toxicity of Nilotinib and UGT1A1 Polymorphisms in Japanese Patients with Chronic Myelogenous Leukemia. Int. J. Clin. Oncol. 2014, 19, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Di Francia, R.; De Monaco, A.; Saggese, M.; Iaccarino, G.; Crisci, S.; Frigeri, F.; De Filippi, R.; Berretta, M.; Pinto, A. Pharmacological Profile and Pharmacogenomics of Anti-Cancer Drugs Used for Targeted Therapy. Curr. Cancer Drug Targets 2018, 18, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Denard, B.; Pavia-Jimenez, A.; Chen, W.; Williams, N.S.; Naina, H.; Collins, R.; Brugarolas, J.; Ye, J. Identification of CREB3L1 as a Biomarker Predicting Doxorubicin Treatment Outcome. PLoS ONE 2015, 10, e0129233. [Google Scholar] [CrossRef] [PubMed]
- Otter, M.; Csader, S.; Keiser, M.; Oswald, S. Expression and Functional Contribution of Different Organic Cation Transporters to the Cellular Uptake of Doxorubicin into Human Breast Cancer and Cardiac Tissue. Int. J. Mol. Sci. 2021, 23, 255. [Google Scholar] [CrossRef] [PubMed]
- Kalyanaraman, B. Teaching the Basics of the Mechanism of Doxorubicin-Induced Cardiotoxicity: Have We Been Barking up the Wrong Tree? Redox Biol. 2020, 29, 101394. [Google Scholar] [CrossRef]
- Todorova, V.K.; Makhoul, I.; Dhakal, I.; Wei, J.; Stone, A.; Carter, W.; Owen, A.; Klimberg, V.S. Polymorphic Variations Associated with Doxorubicin-Induced Cardiotoxicity in Breast Cancer Patients. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 1223–1229. [Google Scholar] [CrossRef]
- Wojnowski, L.; Kulle, B.; Schirmer, M.; Schlüter, G.; Schmidt, A.; Rosenberger, A.; Vonhof, S.; Bickeböller, H.; Toliat, M.R.; Suk, E.-K.; et al. NAD(P)H Oxidase and Multidrug Resistance Protein Genetic Polymorphisms Are Associated with Doxorubicin-Induced Cardiotoxicity. Circulation 2005, 112, 3754–3762. [Google Scholar] [CrossRef]
- Ben Tanfous, M.; Sharif-Askari, B.; Ceppi, F.; Laaribi, H.; Gagné, V.; Rousseau, J.; Labuda, M.; Silverman, L.B.; Sallan, S.E.; Neuberg, D.; et al. Polymorphisms of Asparaginase Pathway and Asparaginase-Related Complications in Children with Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2015, 21, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Owen, A.; Pirmohamed, M.; Khoo, S.H.; Back, D.J. Pharmacogenetics of HIV Therapy. Pharm. Genom. 2006, 16, 693–703. [Google Scholar] [CrossRef]
- El-Attar, E.A.; Helmy Elkaffas, R.M.; Aglan, S.A.; Naga, I.S.; Nabil, A.; Abdallah, H.Y. Genomics in Egypt: Current Status and Future Aspects. Front. Genet. 2022, 13, 797465. [Google Scholar] [CrossRef]
- Aguib, Y.; Allouba, M.; Afify, A.; Halawa, S.; El-Khatib, M.; Sous, M.; Galal, A.; Abdelrahman, E.; Shehata, N.; El Sawy, A.; et al. The Egyptian Collaborative Cardiac Genomics (ECCO-GEN) Project: Defining a Healthy Volunteer Cohort. NPJ Genom. Med. 2020, 5, 46. [Google Scholar] [CrossRef]
- Saad, A.K.; Marafi, D.; Mitani, T.; Du, H.; Rafat, K.; Fatih, J.M.; Jhangiani, S.N.; Coban-Akdemir, Z.; Baylor-Hopkins Center for Mendelian Genomics; Gibbs, R.A.; et al. Neurodevelopmental Disorder in an Egyptian Family with a Biallelic ALKBH8 Variant. Am. J. Med. Genet. A 2021, 185, 1288–1293. [Google Scholar] [CrossRef]
- Elghzaly, A.A.; Sun, C.; Looger, L.L.; Hirose, M.; Salama, M.; Khalil, N.M.; Behiry, M.E.; Hegazy, M.T.; Hussein, M.A.; Salem, M.N.; et al. Genome-Wide Association Study for Systemic Lupus Erythematosus in an Egyptian Population. Front. Genet. 2022, 13, 948505. [Google Scholar] [CrossRef]
- Wohlers, I.; Künstner, A.; Munz, M.; Olbrich, M.; Fähnrich, A.; Calonga-Solís, V.; Ma, C.; Hirose, M.; El-Mosallamy, S.; Salama, M.; et al. An Integrated Personal and Population-Based Egyptian Genome Reference. Nat. Commun. 2020, 11, 4719. [Google Scholar] [CrossRef]
- Amin, N.S.; Abd El-Aziz, M.K.; Hamed, M.; Moustafa, R.R.; El Tayebi, H.M. Rs205764 and Rs547311 in Linc00513 May Influence Treatment Responses in Multiple Sclerosis Patients: A Pharmacogenomics Egyptian Study. Front. Immunol. 2023, 14, 1087595. [Google Scholar] [CrossRef]
- Donkol, H.; Ali, H.; Hamed, H.; Abdel-Rahman, M.; Abdel-Latif, M. Investigating the genetic effect of vkorc1 gene polymorphism on warfarin response in egyptian heart patients. Bull. Pharm. Sci. Assiut Univ. 2023, 46, 1047–1058. [Google Scholar] [CrossRef]
- Radouani, F.; Zass, L.; Hamdi, Y.; Rocha, J.D.; Sallam, R.; Abdelhak, S.; Ahmed, S.; Azzouzi, M.; Benamri, I.; Benkahla, A.; et al. A Review of Clinical Pharmacogenetics Studies in African Populations. Pers. Med. 2020, 17, 155–170. [Google Scholar] [CrossRef]
- Sabokbar, T.; Sharifipour, E.; Zamanlu, M.; Komeili Movahhed, T.; Aghaali, M.; Salarvand, M.; Javaherian, F.; Hejazi, S.A. Gender Differences in Response to Statin Therapy in Ischemic Stroke Patients with SLCO1B1 388A>G Polymorphism: A Clinical Study. Biomed. Res. Bull. 2023, 1, 96–104. [Google Scholar] [CrossRef]
- Center for Drug Evaluation and Research. Drug Development & Approval Process; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2022.
- Elmonem, M.A.; Soliman, N.A.; Moustafa, A.; Gad, Y.Z.; Hassan, W.A.; Taha, T.; El-Feky, G.; Sakr, M.; Amer, K. The Egypt Genome Project. Nat. Genet. 2024, 56, 1035–1037. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Zhou, B. Barriers and Solutions in Clinical Implementation of Pharmacogenomics for Personalized Medicine. In Pharmacogenomics in Precision Medicine; Cai, W., Liu, Z., Miao, L., Xiang, X., Eds.; Springer: Singapore, 2020; pp. 277–289. ISBN 9789811538940. [Google Scholar]
- Kisor, D.F.; Bright, D.R.; Smith, T.R.; Wiisanen, K. Pharmacogenomics: Foundations, Competencies, and the Pharmacists’ Patient Care Process, 2nd ed.; American Pharmacists Association: Washington, DC, USA, 2022; ISBN 978-1-58212-384-4. [Google Scholar]
- Rahma, A.T.; Elsheik, M.; Ali, B.R.; Elbarazi, I.; Patrinos, G.P.; Ahmed, L.A.; Al Maskari, F. Knowledge, Attitudes, and Perceived Barriers toward Genetic Testing and Pharmacogenomics among Healthcare Workers in the United Arab Emirates: A Cross-Sectional Study. J. Pers. Med. 2020, 10, 216. [Google Scholar] [CrossRef]
- Meli, B.A.; Fenech, A.G.; Cordina, M.; Agius, E. Ethical Aspects Pertaining to the Use of Pharmacogenetic Tests. Res. Soc. Adm. Pharm. 2021, 17, 799–804. [Google Scholar] [CrossRef]
- Moyer, A.M.; Caraballo, P.J. Translating Pharmacogenomic Research to Therapeutic Potentials (Bench to Bedside). In Comprehensive Pharmacology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 220–246. ISBN 978-0-12-820876-2. [Google Scholar]
- Nagaraj, S.H.; Toombs, M. The Gene-Drug Duality: Exploring the Pharmacogenomics of Indigenous Populations. Front. Genet. 2021, 12, 687116. [Google Scholar] [CrossRef]
- Kabbani, D.; Akika, R.; Wahid, A.; Daly, A.K.; Cascorbi, I.; Zgheib, N.K. Pharmacogenomics in Practice: A Review and Implementation Guide. Front. Pharmacol. 2023, 14, 1189976. [Google Scholar] [CrossRef]
- Schärfe, C.P.I.; Tremmel, R.; Schwab, M.; Kohlbacher, O.; Marks, D.S. Genetic Variation in Human Drug-Related Genes. Genome Med. 2017, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.C.; Kacevska, M.; Ingelman-Sundberg, M. Pharmacogenomics of Drug-Metabolizing Enzymes: A Recent Update on Clinical Implications and Endogenous Effects. Pharm. J. 2013, 13, 1–11. [Google Scholar] [CrossRef]
- Huang, R.S.; Ratain, M.J. Pharmacogenetics and Pharmacogenomics of Anticancer Agents. CA. Cancer J. Clin. 2009, 59, 42–55. [Google Scholar] [CrossRef]
- Daly, A.K. Genome-Wide Association Studies in Pharmacogenomics. Nat. Rev. Genet. 2010, 11, 241–246. [Google Scholar] [CrossRef]
- Johnson, J.A. Pharmacogenetics in Clinical Practice: How Far Have We Come and Where Are We Going? Pharmacogenomics 2013, 14, 835–843. [Google Scholar] [CrossRef]
- Roden, D.M.; Altman, R.B.; Benowitz, N.L.; Flockhart, D.A.; Giacomini, K.M.; Johnson, J.A.; Krauss, R.M.; McLeod, H.L.; Ratain, M.J.; Relling, M.V.; et al. Pharmacogenomics: Challenges and Opportunities. Ann. Intern. Med. 2006, 145, 749–757. [Google Scholar] [CrossRef]
- Van Der Wouden, C.H.; Marck, H.; Guchelaar, H.-J.; Swen, J.J.; Van Den Hout, W.B. Cost-Effectiveness of Pharmacogenomics-Guided Prescribing to Prevent Gene-Drug-Related Deaths: A Decision-Analytic Model. Front. Pharmacol. 2022, 13, 918493. [Google Scholar] [CrossRef]
- Mooij, M.G.; Nies, A.T.; Knibbe, C.A.J.; Schaeffeler, E.; Tibboel, D.; Schwab, M.; De Wildt, S.N. Development of Human Membrane Transporters: Drug Disposition and Pharmacogenetics. Clin. Pharmacokinet. 2016, 55, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, L.; Bai, L. Exploring Markers to Classify and Evaluate Ketosis Onset Diabetes: A Randomized Clinical Trails. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Takakusa, H.; Iwazaki, N.; Nishikawa, M.; Yoshida, T.; Obika, S.; Inoue, T. Drug Metabolism and Pharmacokinetics of Antisense Oligonucleotide Therapeutics: Typical Profiles, Evaluation Approaches, and Points to Consider Compared with Small Molecule Drugs. Nucleic Acid Ther. 2023, 33, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, M.; Bonifacio, M.; Breccia, M.; Castagnetti, F.; Gozzini, A.; Iurlo, A.; Pregno, P.; Stagno, F.; Specchia, G. Current Strategies and Future Directions to Achieve Deep Molecular Response and Treatment-Free Remission in Chronic Myeloid Leukemia. Front. Oncol. 2020, 10, 883. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Caldas, C. Functional Genomics Approaches to Improve Pre-clinical Drug Screening and Biomarker Discovery. EMBO Mol. Med. 2021, 13, e13189. [Google Scholar] [CrossRef]
- Wang, J.; Xu, B. Targeted Therapeutic Options and Future Perspectives for HER2-Positive Breast Cancer. Signal Transduct. Target. Ther. 2019, 4, 34. [Google Scholar] [CrossRef]
- Zaman; Wu; Bivona Targeting Oncogenic BRAF: Past, Present, and Future. Cancers 2019, 11, 1197. [CrossRef]
- Yang, S.; Sun, Z.; Sun, D.; Yu, C.; Guo, Y.; Sun, D.; Pang, Y.; Pei, P.; Yang, L.; Millwood, I.Y.; et al. Associations of Polygenic Risk Scores with Risks of Stroke and Its Subtypes in Chinese. Stroke Vasc. Neurol. 2023, svn-2023-002428. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Gruber, S.B.; Wu, Z.; Schmidt, E.M.; Zawistowski, M.; Moser, S.E.; Blanc, V.M.; Brummett, C.M.; Kheterpal, S.; Abecasis, G.R.; et al. Association of Polygenic Risk Scores for Multiple Cancers in a Phenome-Wide Study: Results from The Michigan Genomics Initiative. Am. J. Hum. Genet. 2018, 102, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Abraham, G.; Nelson, C.P.; Wood, A.M.; Sweeting, M.J.; Dudbridge, F.; Lai, F.Y.; Kaptoge, S.; Brozynska, M.; Wang, T.; et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults. J. Am. Coll. Cardiol. 2018, 72, 1883–1893. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-Wide Polygenic Scores for Common Diseases Identify Individuals with Risk Equivalent to Monogenic Mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.-H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Zhang, H.; Mehrotra, D.V.; Shen, J. Pharmacogenomics Polygenic Risk Score for Drug Response Prediction Using PRS-PGx Methods. Nat. Commun. 2022, 13, 5278. [Google Scholar] [CrossRef]
- Sperling, R.; Henley, D.; Aisen, P.S.; Raman, R.; Donohue, M.C.; Ernstrom, K.; Rafii, M.S.; Streffer, J.; Shi, Y.; Karcher, K.; et al. Findings of Efficacy, Safety, and Biomarker Outcomes of Atabecestat in Preclinical Alzheimer Disease: A Truncated Randomized Phase 2b/3 Clinical Trial. JAMA Neurol. 2021, 78, 293. [Google Scholar] [CrossRef] [PubMed]
- Babayeva, M.; Loewy, Z. Repurposing Drugs for COVID-19: Pharmacokinetics and Pharmacogenomics of Chloroquine and Hydroxychloroquine. Pharm. Pers. Med. 2020, 13, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Badary, O.A. Pharmacogenomics and COVID-19: Clinical Implications of Human Genome Interactions with Repurposed Drugs. Pharm. J. 2021, 21, 275–284. [Google Scholar] [CrossRef]
- Talevi, A.; Bellera, C.L. Challenges and Opportunities with Drug Repurposing: Finding Strategies to Find Alternative Uses of Therapeutics. Expert Opin. Drug Discov. 2020, 15, 397–401. [Google Scholar] [CrossRef]
- EUA of Chloroquine Phosphate. Fact Sheet for Healthcare Providers. FDA. 2020. Available online: https://www.fda.gov/media/136535/download (accessed on 8 June 2024).
- Antoszczak, M.; Markowska, A.; Markowska, J.; Huczyński, A. Old Wine in New Bottles: Drug Repurposing in Oncology. Eur. J. Pharmacol. 2020, 866, 172784. [Google Scholar] [CrossRef] [PubMed]
- Oates, J.T.; Lopez, D. Pharmacogenetics: An Important Part of Drug Development with A Focus on Its Application. Int. J. Biomed. Investig. 2018, 1, 111. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of Chloroquine on Viral Infections: An Old Drug against Today’s Diseases. Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Vinayagamoorthy, N.; Han, K.; Kwok, S.K.; Ju, J.H.; Park, K.S.; Jung, S.; Park, S.; Chung, Y.; Park, S. Association of Polymorphisms of Cytochrome P450 2D6 with Blood Hydroxychloroquine Levels in Patients with Systemic Lupus Erythematosus. Arthritis Rheumatol. 2016, 68, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Paludetto, M.-N.; Kurkela, M.; Kahma, H.; Backman, J.T.; Niemi, M.; Filppula, A.M. Hydroxychloroquine Is Metabolized by Cytochrome P450 2D6, 3A4, and 2C8, and Inhibits Cytochrome P450 2D6, While Its Metabolites Also Inhibit Cytochrome P450 3A In Vitro. Drug Metab. Dispos. 2023, 51, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Daglish, M.R.C.; Reilly, S.R.; Mostafa, S.; Edwards, C.; O’Gorman, T.M.; Hayllar, J.S. Cytochrome P450-2D6 Activity in People with Codeine Use Disorder. Pharm. J. 2023, 23, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Quinn, S.; Primorac, L.; Primorac, M. Economic Evaluation of Pharmacogenomic Testing. In Pharmacogenomics in Clinical Practice; Primorac, D., Höppner, W., Bach-Rojecky, L., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 373–386. ISBN 978-3-031-45902-3. [Google Scholar]
- Shah, J. Economic and Regulatory Considerations in Pharmacogenomics for Drug Licensing and Healthcare. Nat. Biotechnol. 2003, 21, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Alzarea, A.I.; Khan, Y.H.; Alanazi, A.S.; Butt, M.H.; Almalki, Z.S.; AlAhmari, A.K.; Alsahali, S.; Mallhi, T.H. Barriers and Facilitators of Pharmacoeconomic Studies: A Review of Evidence from the Middle Eastern Countries. Int. J. Environ. Res. Public Health 2022, 19, 7862. [Google Scholar] [CrossRef]
- Dinda, A. Regulatory Science: The Need for Empowering Indian Innovation. Indian J. Med. Res. 2021, 154, 770. [Google Scholar] [CrossRef]
- Alsultan, A.; Alghamdi, W.A.; Alghamdi, J.; Alharbi, A.F.; Aljutayli, A.; Albassam, A.; Almazroo, O.; Alqahtani, S. Clinical Pharmacology Applications in Clinical Drug Development and Clinical Care: A Focus on Saudi Arabia. Saudi Pharm. J. 2020, 28, 1217–1227. [Google Scholar] [CrossRef]
- Teixeira, T.; Kweder, S.L.; Saint-Raymond, A. Are the European Medicines Agency, US Food and Drug Administration, and Other International Regulators Talking to Each Other? Clin. Pharmacol. Ther. 2020, 107, 507–513. [Google Scholar] [CrossRef]
- Kandi, V.; Vadakedath, S. Clinical Trials and Clinical Research: A Comprehensive Review. Cureus 2023, 15, e35077. [Google Scholar] [CrossRef] [PubMed]
- Abdullah-Koolmees, H.; Van Keulen, A.M.; Nijenhuis, M.; Deneer, V.H.M. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front. Pharmacol. 2021, 11, 595219. [Google Scholar] [CrossRef] [PubMed]
- Reis-Pardal, J.; Rodrigues, A.; Rodrigues, E.; Fernandez-Llimos, F. Comparing Cytochrome P450 Pharmacogenetic Information.Available on United States Drug Labels and European Union Summaries of Product Characteristics. Pharm. J. 2017, 17, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Pharmacogenetics Briefing Meeting—Scientific Guideline; European Medicines Agency: Amsterdam, The Netherlands, 2006; p. 11.
- Mayzent®, a Once-Daily Pill, to Stay Ahead of Disability Progression. Treatment for Relapsing MS | MAYZENT® (Siponimod). In mayzent. 2023. Available online: https://www.mayzent.com/ (accessed on 16 April 2024).
- Omran, M.M.; Abdelfattah, R.; Moussa, H.S.; Alieldin, N.; Shouman, S.A. Association of the Trough, Peak/Trough Ratio of Imatinib, Pyridine–N-Oxide Imatinib and ABCG2 SNPs 34 G>A and SLCO1B3 334 T>G with Imatinib Response in Egyptian Chronic Myeloid Leukemia Patients. Front. Oncol. 2020, 10, 1348. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.A.; Van Bebber, S.L. Regulatory Perspectives on Pharmacogenomics: A Review of the Literature on Key Issues Faced by the United States Food and Drug Administration. Med. Care Res. Rev. 2006, 63, 301–326. [Google Scholar] [CrossRef] [PubMed]
- Vivot, A.; Boutron, I.; Ravaud, P.; Porcher, R. Guidance for Pharmacogenomic Biomarker Testing in Labels of FDA-Approved Drugs. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Joppi, R.; Bertele, V.; Vannini, T.; Garattini, S.; Banzi, R. Food and Drug Administration vs European Medicines Agency: Review Times and Clinical Evidence on Novel Drugs at the Time of Approval. Br. J. Clin. Pharmacol. 2020, 86, 170–174. [Google Scholar] [CrossRef]
- Kashoki, M.; Hanaizi, Z.; Yordanova, S.; Veselý, R.; Bouygues, C.; Llinares, J.; Kweder, S.L. A Comparison of EMA and FDA Decisions for New Drug Marketing Applications 2014-2016: Concordance, Discordance, and Why. Clin. Pharmacol. Ther. 2020, 107, 195–202. [Google Scholar] [CrossRef]
- Gnanasakthy, A.; Doward, L.; Clark, M.; Mordin, M.; DeMuro, C. Concordance of Pro Labeling Claims between the FDA and EMA. Value Health 2012, 15, A321. [Google Scholar] [CrossRef]
- Kordou, Z.; Skokou, M.; Tsermpini, E.E.; Chantratita, W.; Fukunaga, K.; Mushiroda, T.; Patrinos, G.P.; Koromina, M. Discrepancies and similarities in the genome-informed guidance for psychiatric disorders amongst different regulatory bodies and research consortia using next generation sequencing-based clinical pharmacogenomics data. Pharmacol. Res. 2021, 167, 105538. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Liu, Z.; Miao, L.; Xiang, X. (Eds.) Pharmacogenomics in Precision Medicine: From a Perspective of Ethnic Differences; Springer: Singapore, 2020; ISBN 9789811538940. [Google Scholar]
- Cheng, C.M.; So, T.W.; Bubp, J.L. Characterization of Pharmacogenetic Information in Food and Drug Administration Drug Labeling and the Table of Pharmacogenetic Associations. Ann. Pharmacother. 2021, 55, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, S.; Mitra Ghosh, T.; Mukherjee, U.K.; Chakravarti, S.; Amiri, F.; Waliagha, R.S.; Hemmati, F.; Mistriotis, P.; Ahmed, S.; Elhussin, I.; et al. Integrating Pharmacogenomics Data-Driven Computational Drug Prediction with Single-Cell RNAseq to Demonstrate the Efficacy of a NAMPT Inhibitor against Aggressive, Taxane-Resistant, and Stem-like Cells in Lethal Prostate Cancer. Cancers 2022, 14, 6009. [Google Scholar] [CrossRef] [PubMed]
- Primorac, D.; Bach-Rojecky, L.; Vađunec, D.; Juginović, A.; Žunić, K.; Matišić, V.; Skelin, A.; Arsov, B.; Boban, L.; Erceg, D.; et al. Pharmacogenomics at the Center of Precision Medicine: Challenges and Perspective in an Era of Big Data. Pharmacogenomics 2020, 21, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.B. The Impact of Pharmacogenomics in Personalized Medicine. In Current Applications of Pharmaceutical Biotechnology; Silva, A.C., Moreira, J.N., Lobo, J.M.S., Almeida, H., Eds.; Advances in Biochemical Engineering/Biotechnology, Springer International Publishing: Cham, Switzerland, 2019; Volume 171, pp. 369–394. ISBN 978-3-030-40463-5. [Google Scholar]
- Bousman, C.A.; Maruf, A.A.; Marques, D.F.; Brown, L.C.; Müller, D.J. The Emergence, Implementation, and Future Growth of Pharmacogenomics in Psychiatry: A Narrative Review. Psychol. Med. 2023, 53, 7983–7993. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.D.; Cavallari, L.H. Pharmacogenetics to Guide Cardiovascular Drug Therapy. Nat. Rev. Cardiol. 2021, 18, 649–665. [Google Scholar] [CrossRef]
- Scionti, F.; Di Martino, M.T.; Caracciolo, D.; Pensabene, L.; Tagliaferri, P.; Arbitrio, M. Tools in Pharmacogenomics Biomarker Identification for Cancer Patients. In Microarray Data Analysis; Agapito, G., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2401, pp. 1–12. ISBN 978-1-07-161838-7. [Google Scholar]
- Chaudhary, R.; Singh, B.; Kumar, M.; Gakhar, S.K.; Saini, A.K.; Parmar, V.S.; Chhillar, A.K. Role of Single Nucleotide Polymorphisms in Pharmacogenomics and Their Association with Human Diseases. Drug Metab. Rev. 2015, 47, 281–290. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elgarhy, F.M.; Borham, A.; Alziny, N.; AbdElaal, K.R.; Shuaib, M.; Musaibah, A.S.; Hussein, M.A.; Abdelnaser, A. From Drug Discovery to Drug Approval: A Comprehensive Review of the Pharmacogenomics Status Quo with a Special Focus on Egypt. Pharmaceuticals 2024, 17, 881. https://doi.org/10.3390/ph17070881
Elgarhy FM, Borham A, Alziny N, AbdElaal KR, Shuaib M, Musaibah AS, Hussein MA, Abdelnaser A. From Drug Discovery to Drug Approval: A Comprehensive Review of the Pharmacogenomics Status Quo with a Special Focus on Egypt. Pharmaceuticals. 2024; 17(7):881. https://doi.org/10.3390/ph17070881
Chicago/Turabian StyleElgarhy, Fadya M., Abdallah Borham, Noha Alziny, Khlood R. AbdElaal, Mahmoud Shuaib, Abobaker Salem Musaibah, Mohamed Ali Hussein, and Anwar Abdelnaser. 2024. "From Drug Discovery to Drug Approval: A Comprehensive Review of the Pharmacogenomics Status Quo with a Special Focus on Egypt" Pharmaceuticals 17, no. 7: 881. https://doi.org/10.3390/ph17070881
APA StyleElgarhy, F. M., Borham, A., Alziny, N., AbdElaal, K. R., Shuaib, M., Musaibah, A. S., Hussein, M. A., & Abdelnaser, A. (2024). From Drug Discovery to Drug Approval: A Comprehensive Review of the Pharmacogenomics Status Quo with a Special Focus on Egypt. Pharmaceuticals, 17(7), 881. https://doi.org/10.3390/ph17070881








