Abstract
According to the World Health Organization, over 422 million people worldwide have diabetes, with the majority residing in low- and middle-income countries. Diabetes causes 1.5 million fatalities a year. The number of diabetes cases and its prevalence have progressively increased over the last few decades. This study aims to determine the phytochemicals in the edible part of Perkia javanica, predict their α-glucosidase inhibitory potential, one of the promising targets for diabetes, and then carry out in vitro and in vivo studies. The phytochemicals present in the n-butanol fraction of the methanol extract of P. javanica pods were analyzed using UHPLC-QTOF-MS/MS (Ultra-High-Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry). The UHPLC-QTOF analysis revealed the presence of 79 different compounds in the n-butanol fraction. Among these, six compounds demonstrated excellent binding affinities with α-glucosidase, surpassing the performance of two standard inhibitors, Miglitol and Voglibose. In vitro α-glucosidase inhibitory activities were assessed by the n-butanol fraction, followed by in vivo studies. According to the in vitro study, the inhibitory efficiency against α-glucosidase was determined to have an IC50 value of 261.9 µg/mL. The in vivo findings revealed a significant reduction in blood glucose levels in Swiss albino mice treated with the same extract, decreasing from 462.66 mg/dL to 228.66 mg/dL. Additionally, the extract significantly increased the activity of the enzymes catalase and superoxide dismutase (SOD) and decreased the amount of malondialdehyde (MDA) in the liver and kidney tissue. The predicted physicochemical parameters indicated that most of the compounds would be excreted from the body after inhibition in the small intestine without being absorbed. Considering the low cost and wide availability of raw materials, P. javanica pods can serve as a good food supplement that may help prevent type 2 diabetes management.
1. Introduction
Just about 500 million people worldwide have diabetes at the moment, but by 2030 and 2045, that number is predicted to increase to 51% and 25%, respectively [1]. In 2003, the anticipated yearly cost of diabetes treatment in India was between Rs. 10,000 and Rs. 12,000 crore. By 2025, this amount is expected to reach up to Rs. 126,000 crore [2]. Diabetes has numerous financial repercussions, both directly and indirectly, around the globe, and these effects impose a substantial burden on society. The two main types of diabetes mellitus are type 1 and type 2. Type 2 diabetes comprises more than 95% of people with diabetes. Type 2 diabetes is referred to as adult-onset or non-insulin-dependent diabetes. Major classes of drugs used in the treatment of DM are insulins, sulfonylureas (SUs), thiazolidinediones (TZDs), biguanides, meglitinides, α-glucosidase inhibitors (AGIs), dipeptidyl peptidase-4 (DPP-4) inhibitors, glucagon-like peptide-1 (GLP-1) agonists, sodium-glucose cotransporter 2 (SGLT2) inhibitors, and dopamine agonists [3]. One effective treatment approach for lowering blood glucose associated with type 2 diabetes is to inhibit α-amylase, α-glucosidase, or both, the enzymes that catalyze the intestinal hydrolysis of starch. In the last stage of glucose digestion, α-glucosidase catalyzes the hydrolysis of the α-(1,4)-glycosidic bond of sugar, releasing free monosaccharide (α-D-glucose) [4]. α-Glucosidase enzymes, in particular, are inhibited by α-glucosidase inhibitors in the small intestine. The recognized α-glucosidase inhibitors at the moment are Voglibose, Miglitol, and Acarbose [5,6]. Current long-term anti-diabetes medications have several side effects, which emphasizes the significance of adopting natural products. Medicinal plants commonly used for the treatment and management of diabetes in different systems of medicine like Traditional Chinese Medicine (TCM), Traditional Japanese Medicine (Kampo), Traditional Indian Medicine (Ayurveda), European Herbal Medicine, and Russian Traditional Medicine [7,8,9,10]. Recent research has identified anti-diabetes compounds sourced from plants using computational approaches [11,12,13,14]. This study focuses on finding α-glucosidase inhibitors or a mixture of compounds as inhibitors from natural sources. Thus, efforts to find new inhibitors with better efficacy and reduced side effects are still ongoing. It has been suggested that more than 1200 blooming plants have anti-diabetic potential. Among them, one-third were scientifically validated and published in around 460 articles [15]. Some plants exhibit hypoglycemic, anti-hyperglycemic, and anti-diabetic properties with α-amylase and α-glucosidase inhibitory potential [16,17]. The indigenous inhabitants of the eastern Himalayan region of India are known to use herbal remedies either on their own or in conjunction with other types of treatment to cure diabetes [18]. Therefore, natural products may be safer than synthetic medications in treating diabetes [19]. α-Glucosidase inhibitors, such as Acarbose, Miglitol, and Voglibose, have been used clinically since 1990 to manage type 2 diabetes by slowing down the digestion and absorption of carbohydrates. Despite being useful in regulating blood glucose levels, they are often associated with gastrointestinal side effects [20,21]. Therefore, finding new α-glucosidase inhibitors or edible herbal products with fewer side effects is urgently needed. Target-based in silico analysis, combined with modern chemical profiling methods, saves both money and time [12,22,23]. Numerous research teams have identified several plant-based anti-diabetic agents [24,25,26,27]. The aforementioned results motivate the current researcher to use cutting-edge methods to determine the potential of edible pods of P. javanica to prevent diabetes. Parkia javanica (local name: Kuki Tetai, Youngchak) is a medium-sized tree of the Mimosaceae family that grows abundantly in the northeastern states of India. Tender pods are mostly used as vegetables [28]. Various parts of this plant are used in traditional medicine [29]. It was stated that 49 Parkia timoriana compounds were identified by GC-MS, revealing their anti-inflammatory and anti-cancer properties [30]. An investigation using pod extract revealed its anti-biofilm activity against Pseudomonas aeruginosa. The pods have been used traditionally to treat diabetes, kidney pain, and cholera [31]. The P. javanica pods have the ability to induce apoptosis in cancer cells, and the methanolic extract of bark exhibits cytotoxic potential against a colon cancer cell line [32,33,34]. Studies showed that P. javanica pods have high antioxidant properties [35] and in vitro activity against the Leishmania donovani parasite [36,37]. P. javanica pod extract also possesses potent anti-angiogenic and antiproliferative properties [38]. The major compounds reported so far from P. javanica are 2,4-Di-tert-butylphenol, baicalein, quercetin, chrysin, cinnamic acid, tannic acid, resorcinol, 8-O-p-hydroxybenzoyl-6′-O-p-coumaroyl-mussaenosidic acid, and 7-O-E-3,4-dimethoxycinnamoyl-6′-O-b-d-glucopyranosylloganic acid [31,39,40,41]. The total number of compounds reported so far is inadequate. Exploring more compounds from this important medicinal plant is urgently needed, which may open up new avenues in the field of anti-cancer and anti-diabetes drug discovery. As of now, no study has been conducted to investigate the UHPLC-QTOF-MS/MS-based chemical profiling of compounds of P. javanica and their anti-diabetic potential. In the present study, UHPLC-QTOF-MS/MS-based chemical profiling, in vitro α-glucosidase inhibitory potential, in vivo anti-diabetic activity, and oxidative stress of the n-butanol fraction of the methanol extract of P. javanica pods were investigated.
2. Results
2.1. Chemical Profiling
The n-butanol fraction of the methanol extract of P. javanica was used for UHPLC-QTOF-MS/MS-based analysis. The positive and negative ionization modes were employed, and a series of 56 (PJ_01–PJ_56) and 23 (PJ_57–PJ_79) compounds were identified, respectively. The spectra of both positive and negative ionization modes are shown in Figures S1 and S2, respectively. The separated compounds were analyzed within the m/z 100–1500 mass range with increasing retention time and identified by comparing the METLIN Database [42]. All the compounds detected by UPLC-MS through the positive and negative modes are shown in Table 1 and Table 2, respectively. The structures of all the compounds are shown in Figure S3.

Table 1.
UHPLC-QTOF-MS/MS-based positive mode identifications of compounds; RT, retention time; [M+H]+, [M+Na]+, and [M+NH4]+ ions; experimental exact mass; and score.

Table 2.
UHPLC-QTOF-MS/MS-based negative mode identifications of compounds; RT, retention time; [M+H]−, [M+CH3COO]+, and [2M+Cl]−; experimental exact mass; and score.
2.2. In Silico Studies of Phytochemicals
Molecular Docking
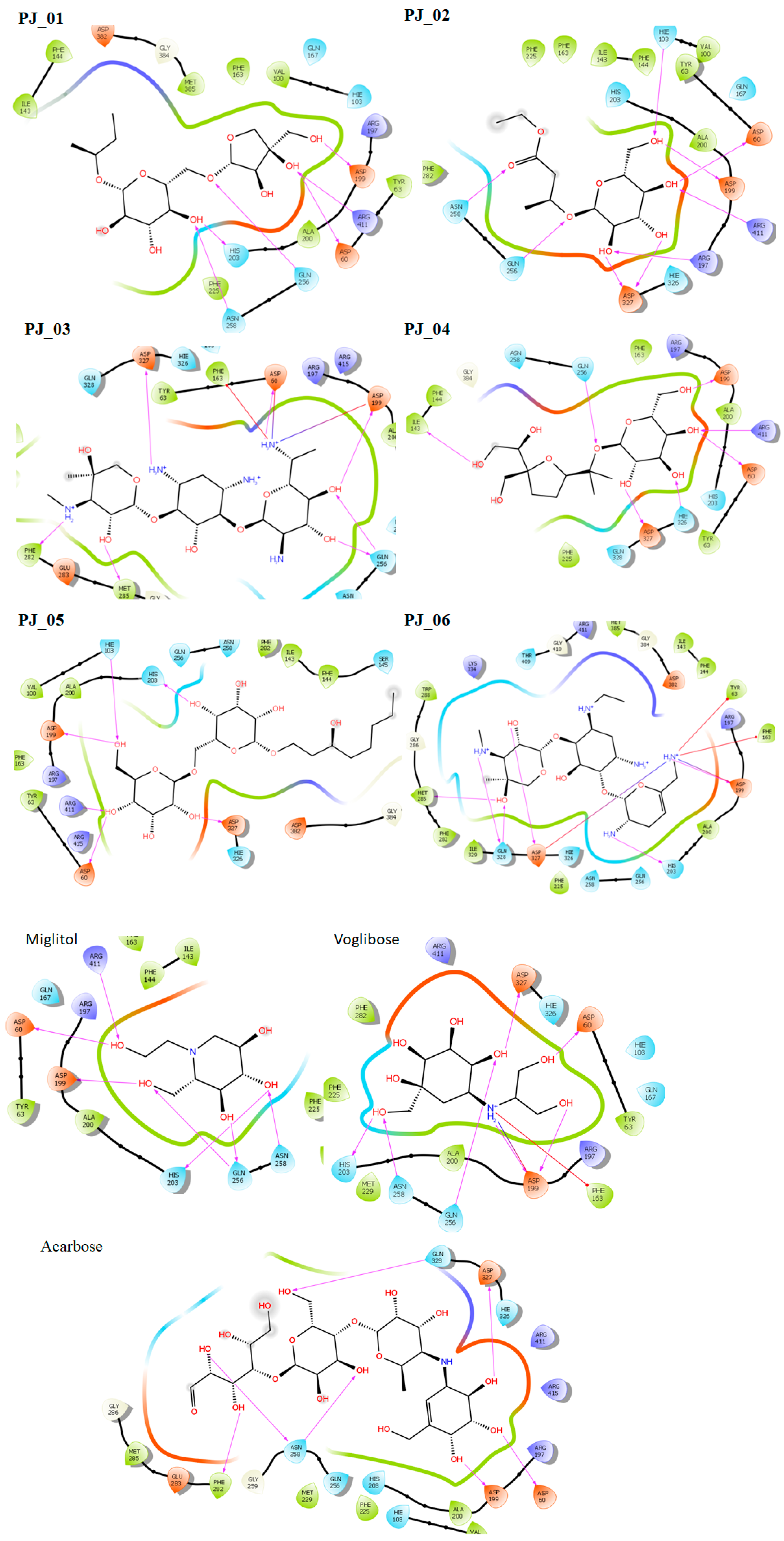
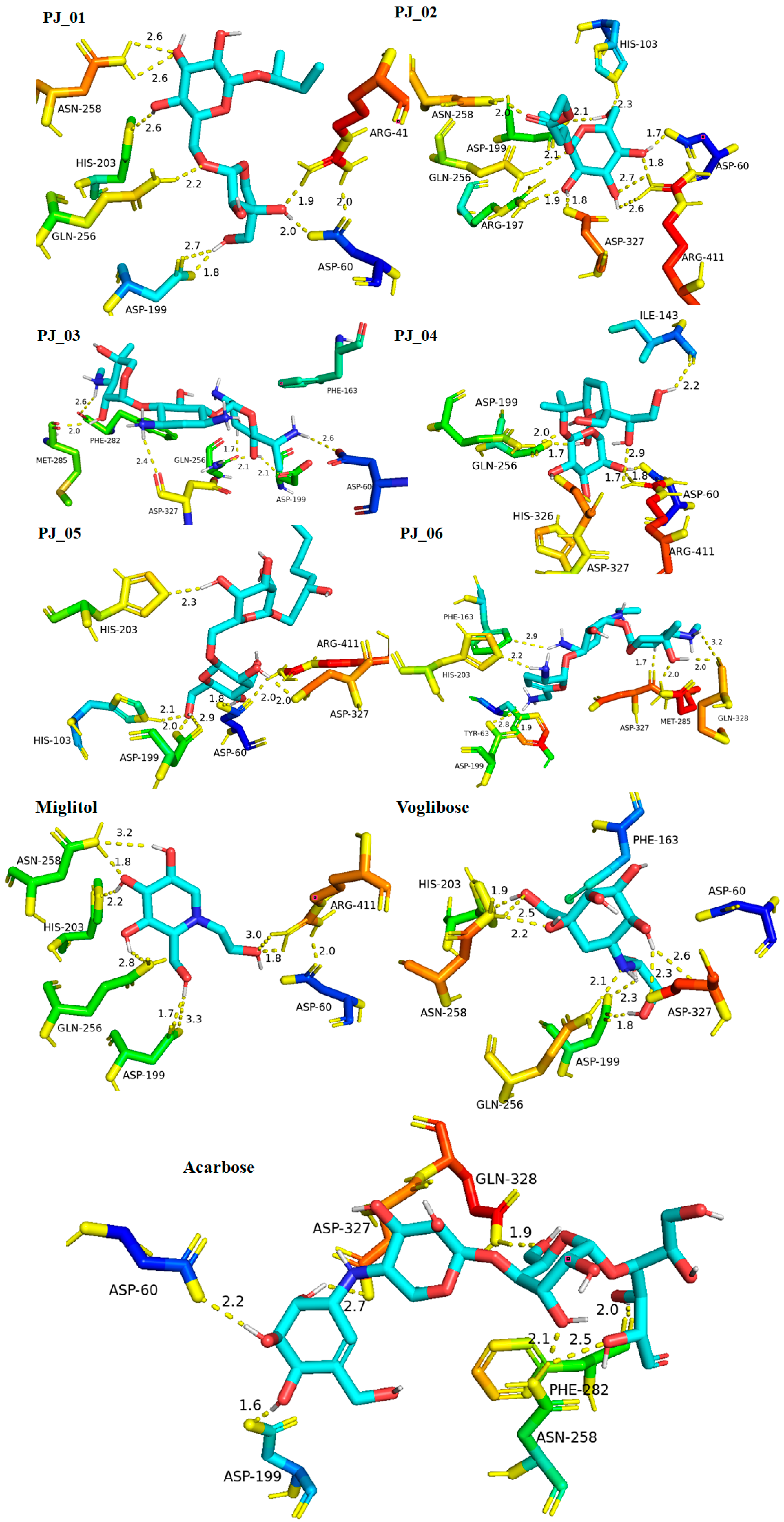
Six compounds were identified from the virtual screening of the seventy-nine compounds of P. javanica that exhibited better binding affinity than known α-glucosidase inhibitors. The XP Glide score of these six top hits exceeds those of the approved known α-glucosidase inhibitors, Miglitol and Voglibose. The XP Glide scores of the known α-glucosidase inhibitors Miglitol, Voglibose, and Acarbose were −7.327, −8.662, and −11.975 kcal/mol, respectively. The binding affinities of the top six hits, PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06, were −9.092, −8.579, −13.632, −10.506, −11.013, and −13.818 kcal/mol, respectively. One common method to determine the binding free energy of small organic molecules to receptors is to combine molecular mechanics energies with generalized Born and surface area continuum solvation (MM-GBSA) techniques. The XP glide post-process MM-GBSA dG binding energies predicted by the Prime of known inhibitors Miglitol, Voglibose, and Acarbose were −22.722, −32.823, and −50.035 kcal/mol, respectively. Similarly, glide post-process MM-GBSA dG binding energies of top hits PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 were −26.259, −36.846, −57.542, −27.704, −22.205, and −80.784 kcal/mol, respectively. The 2D ligand interaction diagram showed that the compound PJ_01 interacts with active site amino acid residues through H-bonding with ASP-60, ASP-199, HIS-203, GLN-256, ASN-258, and ARG-411. The compounds PJ_02 that interact with active site amino acid residues through H-bonding are ASP-60, HIE-103, ARG-197, ASP-199, GLN-256, ASN-258, ASP-327 (2), and ARG-411. The active site-interacting amino acid residues with compound PJ_03 were ASP-60, ASP-199, GLN-256 (2), PHE-282, MET-285, ASP-327 through hydrogen bonding; PHE-163 through π-cation; and ASP-60, ASP-199 through salt bridge. The compound PJ_04 interacts through H-bonding with ASP-60, ILE-143, ASP-196, GLN-256, ASP-326, ASP-327, and ARG-411. The amino acid residues interacting with PJ_05 through H-bonding were ASP-60, HIE-103, ASP-199, HIS-203, ASP-327, and ARG-411. The compound with the highest binding affinity was PJ_06, and the interacting residues were ASP-199, HIS-203, MET-285, ASP-327, and GLN-328, which interact through H-bonding; TYR-63 and PHE-163 interact through π-cation; and ASP-327 interacts via a salt bridge. The 2D interactions of six selected top hits and known inhibitors with α-glucosidase are depicted in Figure 1. Table 3 represents an overview of the types of interactions, the residues associated with each interaction, XP Glide scores, and MM-GBSA dG binding free energies of the top six hits and known inhibitors. Most selected top six hits interact with active site amino acid residues from close distances ≤ 3.0 Å. A visual 3D interaction of selected top hits PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 and known inhibitors can be seen in Figure 2. All the selected hits are highly soluble in water and violate Lipinski’s rule of five, the Ghose rule, and the Veber rule. Except for compound PJ_02, it does not violate Lipinski’s rule of five. The other important parameter, gastrointestinal absorption of all compounds, was low. The physiochemical parameters of all the molecules are listed in Supplementary Table S1.
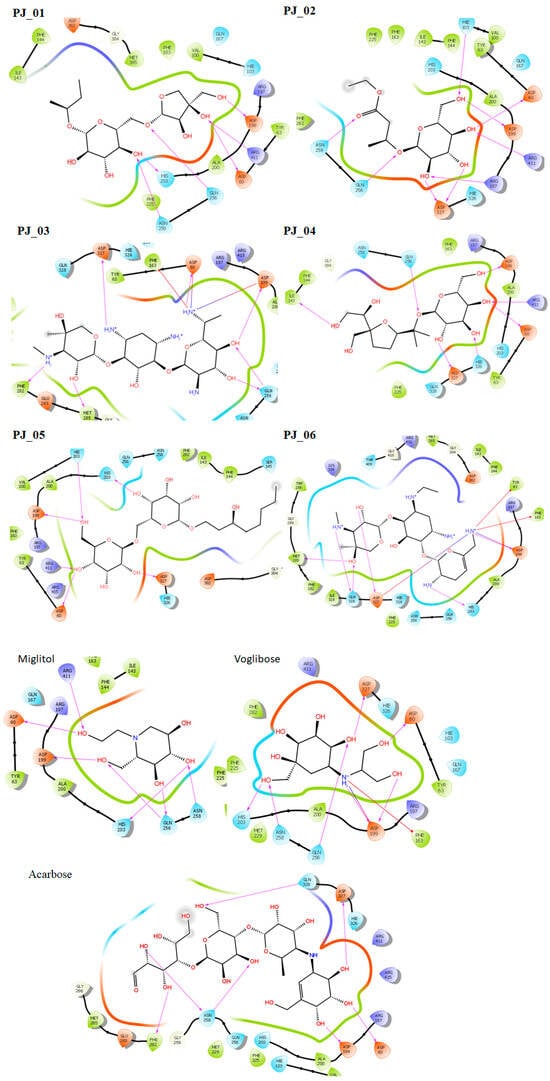
Figure 1.
Post-docking 2D interactions of α-glucosidase (PDB ID: 5ZCC)-ligand (PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, PJ_06, and control ligands).

Table 3.
Extra precision Glide score (XPGS), MM-GBSA dG-binding free energies, and interacting amino acid residues of α-glucosidase with the top six selected hits.
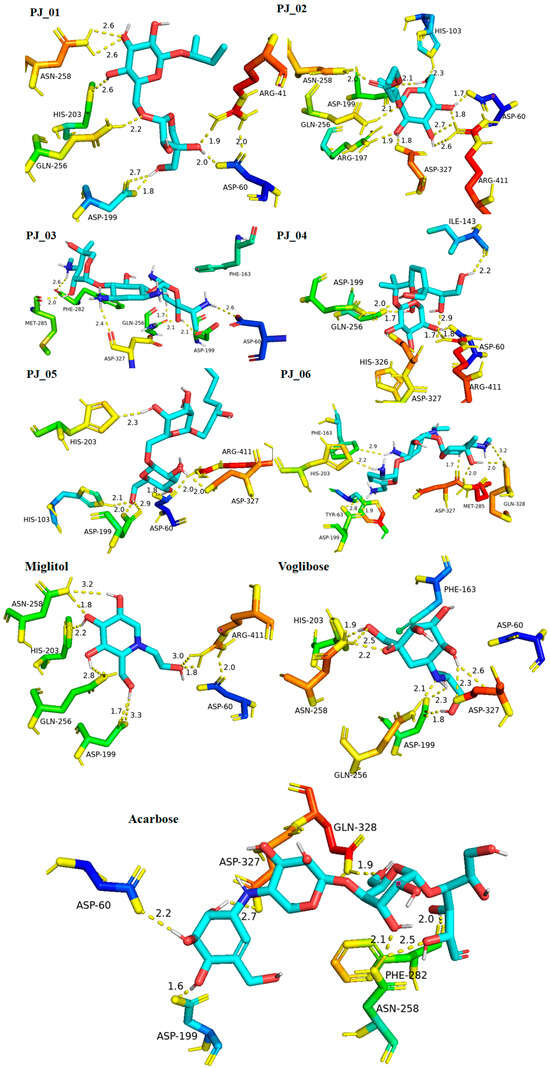
Figure 2.
Post-docking protein–ligand 3D interactions of selected compounds (PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06) and control ligands with their interacting distances. The hydrogen-bonding interactions were depicted in a yellow dotted line.
2.3. Biological Evaluation
2.3.1. In Vitro α-Glucosidase Inhibitory Assay
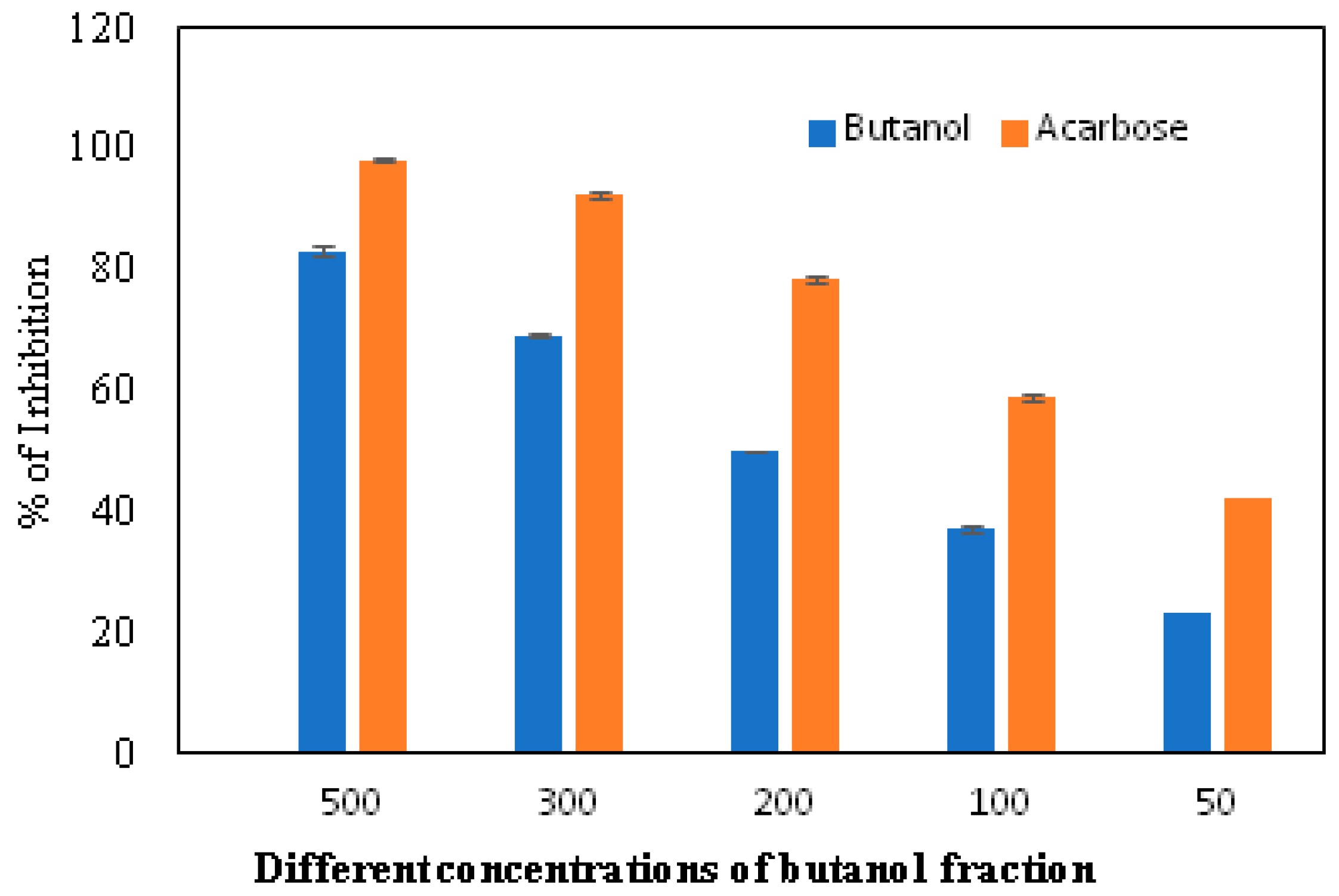
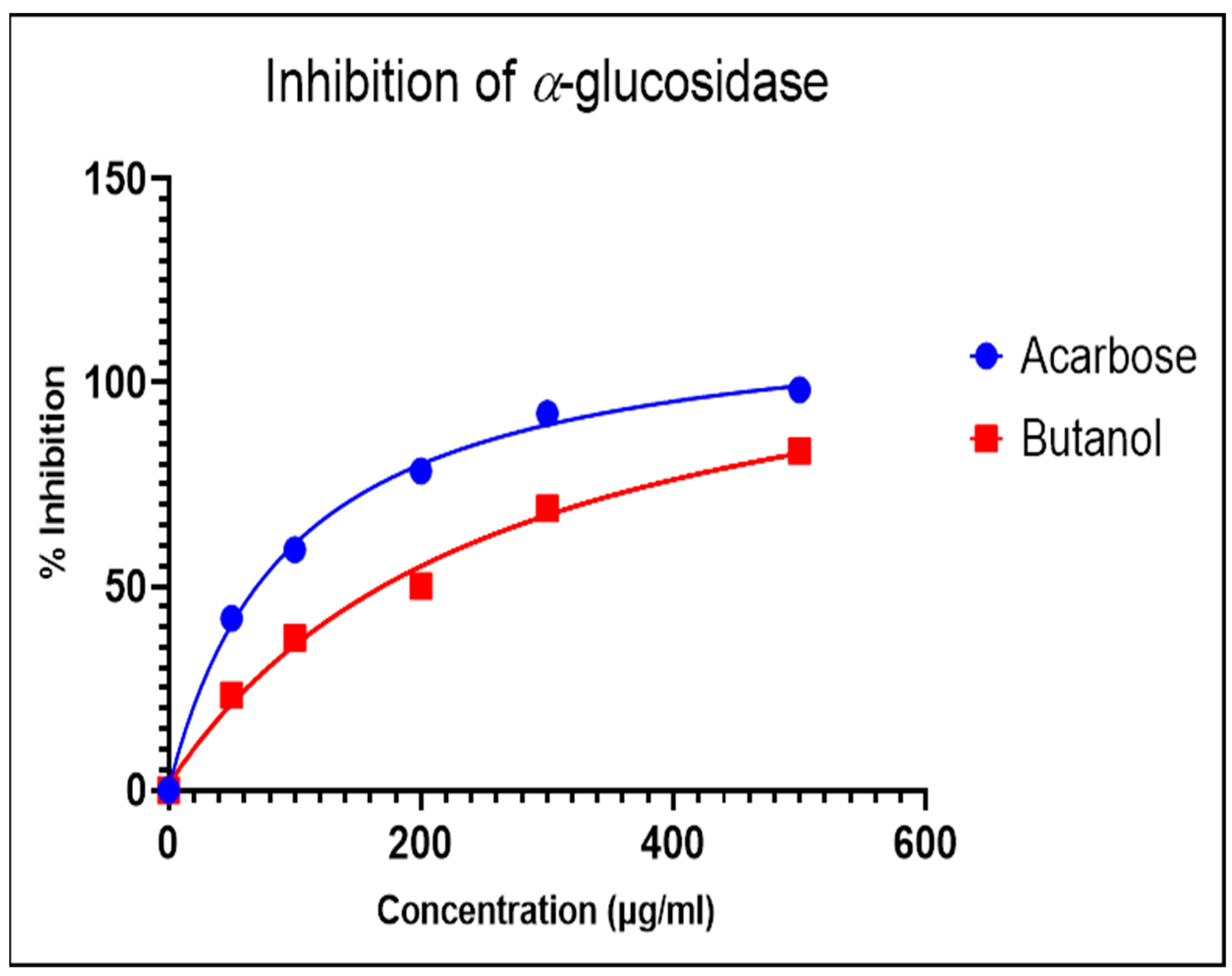
The α-glucosidase enzyme is one of the essential enzymes of our digestive system, located in the small intestine. It plays an important role in the processing and degradation of complex carbohydrates into small, simple, and absorbable ones. Its inhibition is an effective way to delay absorption and also prevent high postprandial blood glucose levels, which may suppress diabetes progression. The n-butanol fraction showed promising activity in comparison to the known α-glucosidase inhibitor, Acarbose. The α-glucosidase inhibitory activity of the n-butanol fraction of the methanol extract of P. javanica is shown in Table 4, Figure 3 and Figure 4.

Table 4.
Inhibition of α-glucosidase by n-butanol fraction and known inhibitor, Acarbose.
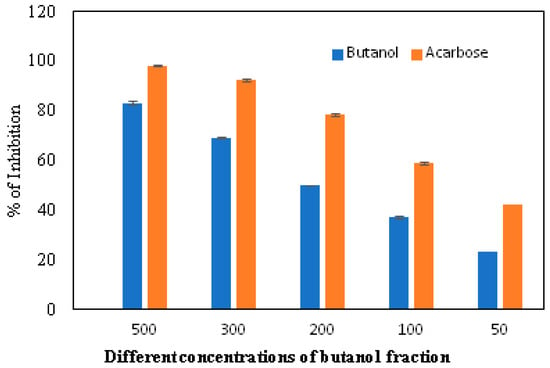
Figure 3.
In vitro inhibition of the n-butanol fraction in comparison to the standard inhibitor, Acarbose.
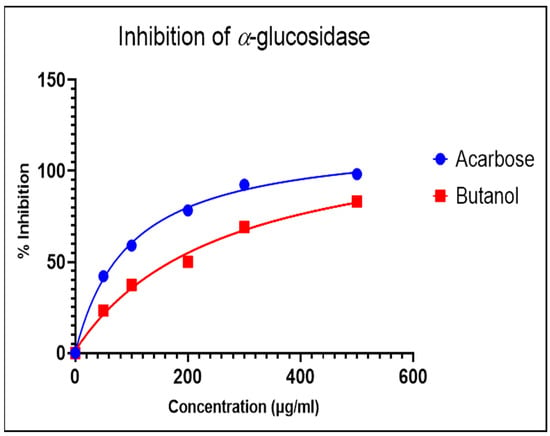
Figure 4.
Dose-dependent response curve of the n-butanol fraction and Acarbose.
2.3.2. In Vivo Analysis in Swiss Albino Mice
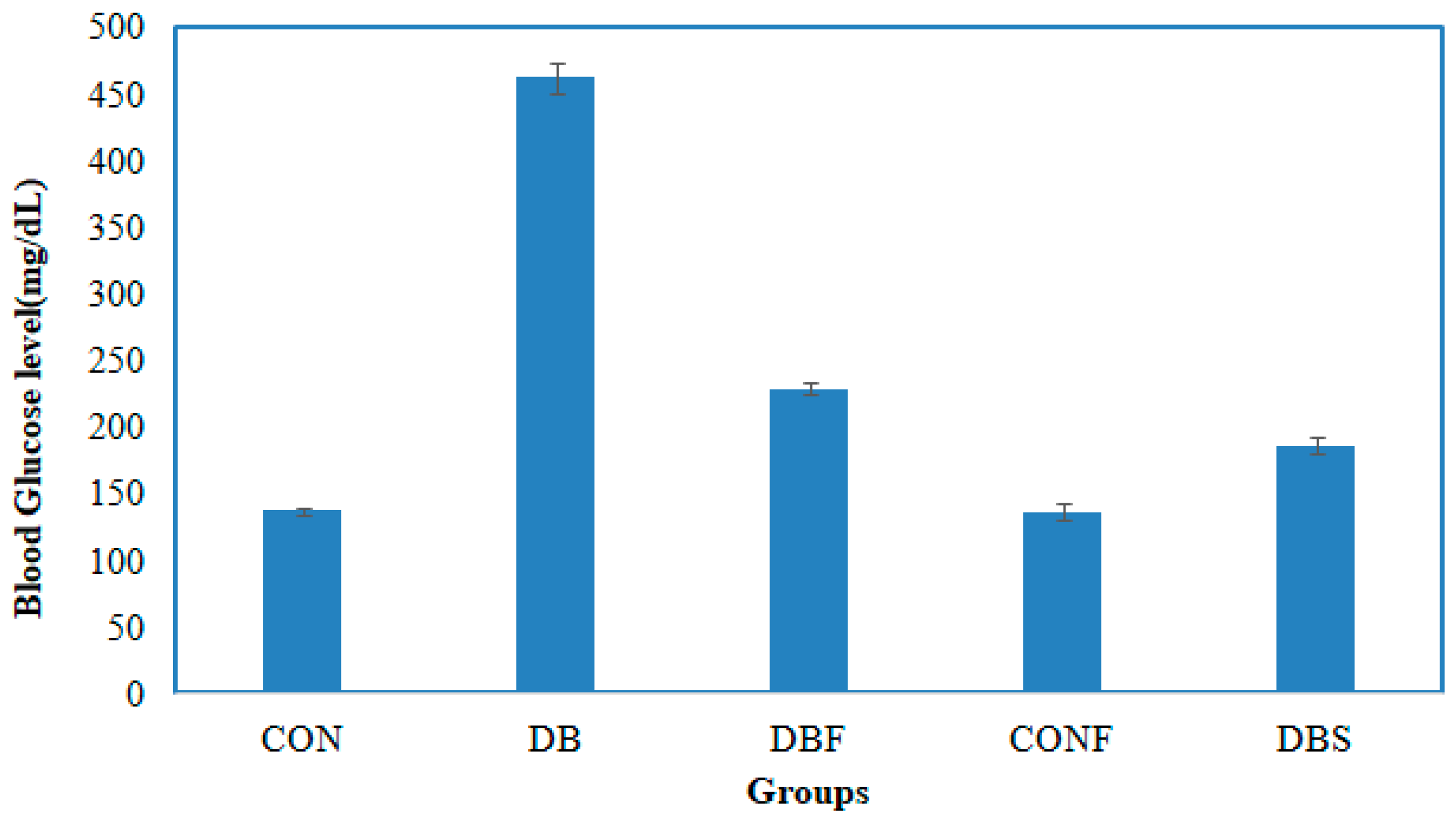
A significant (p < 0.01) increased level of blood glucose was noted in the diabetes group of mice in comparison with the control group of mice. In contrast, treatment with the n-butanol fraction of the methanol extract of pods in diabetic mice significantly (p < 0.01) decreased the blood glucose levels compared to the diabetes group of mice. The blood glucose levels of control mice (CON), diabetic mice (DB), diabetic mice treated with pod extract (DBF), control mice treated with pod extract (CONF), and diabetes mice treated with the standard drug acarbose (DBS) were 137.33, 462.66, 228.66, 136.66, and 186.33 µg/mL, respectively. The changes in blood glucose levels are shown in Figure 5.
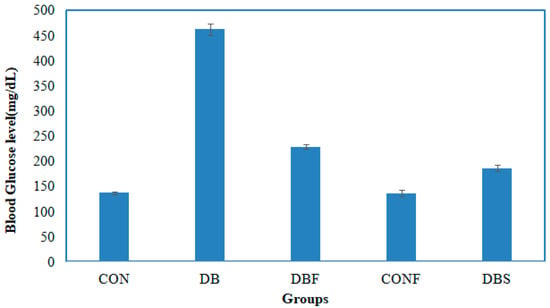
Figure 5.
Effect of the n-butanol fraction on the blood glucose level of diabetic mice (CON—control; DB—diabetic mice; DBF—diabetic mice treated with pod extract; CONF—control treated with pod extract; and DBS—diabetes mice treated with standard drug acarbose).
2.3.3. Oxidative Stress Analysis
Catalase Activity Assay
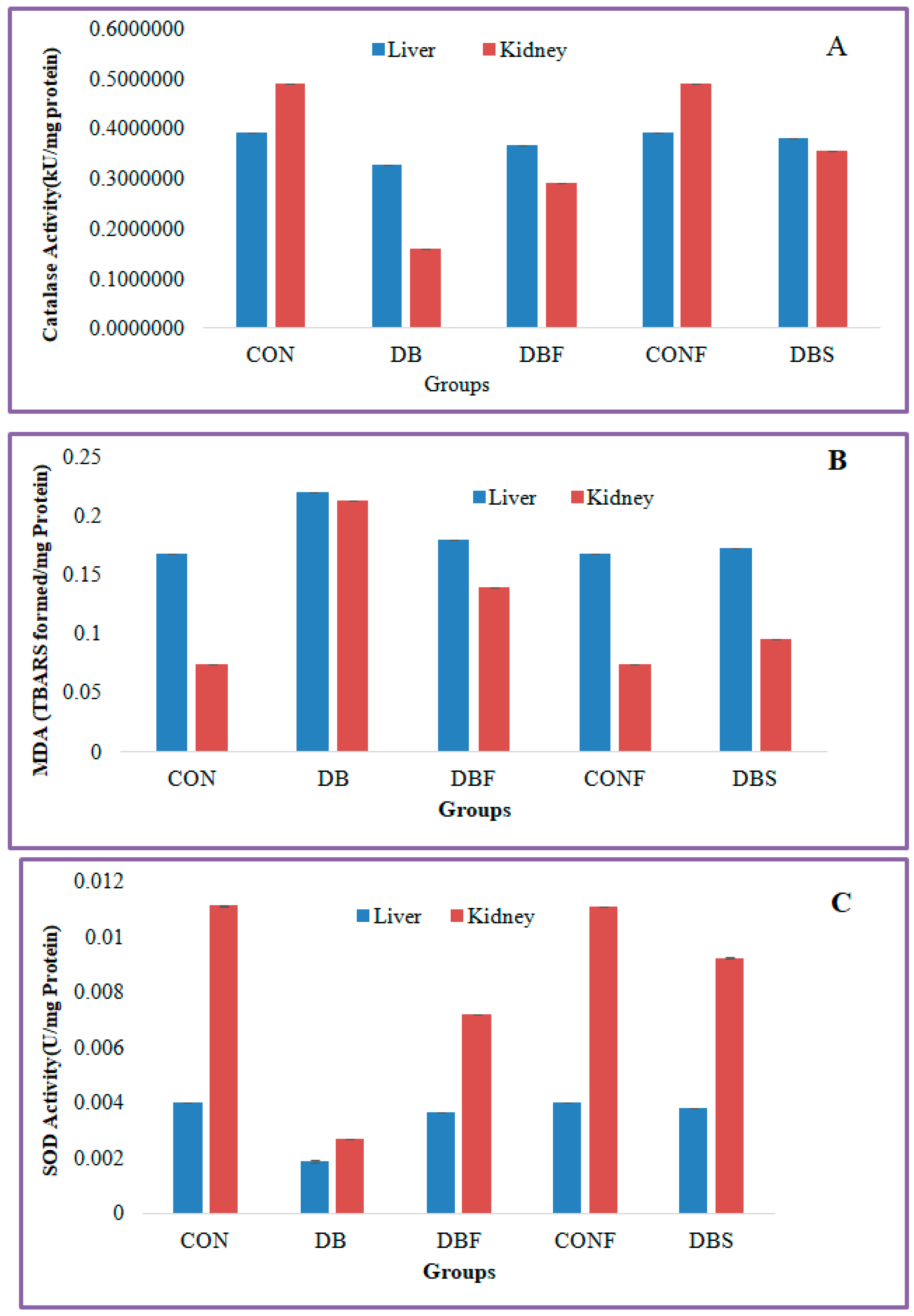
The diabetes group of mice showed significantly (p < 0.01) decreased levels of catalase activity in the STZ-induced diabetic mice liver and kidney tissues in comparison with the control group of mice, whereas significantly (p < 0.01) increased levels of catalase activity were seen in the liver and kidney tissues in extract-treated diabetic mice in comparison to the diabetes group of mice (Figure 6A).
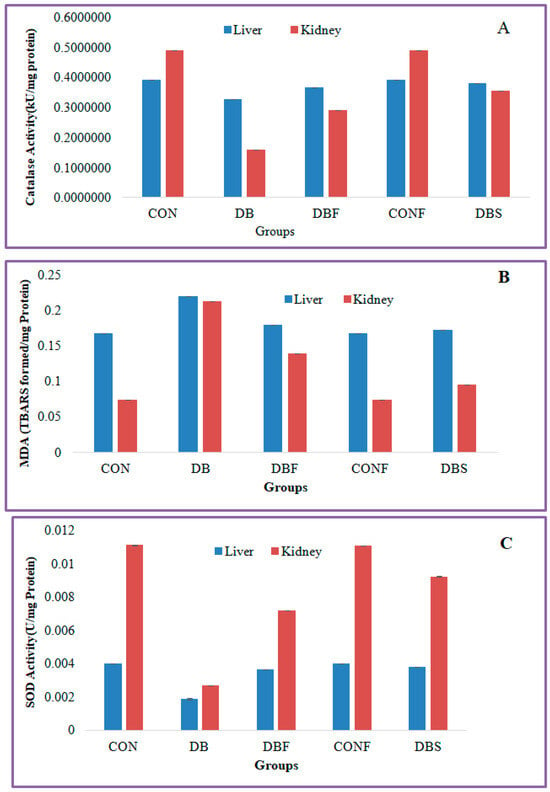
Figure 6.
Effect of n-butanol fraction on oxidative stress in the liver and kidney of diabetic mice ((A): catalase activity, (B): superoxide dismutase, and (C): malondialdehyde). CON—control; CONF—control treated with plant fraction; DB—diabetic mice; DBS—diabetic mice treated with standard drug; and DBF—fraction-treated diabetic mice).
MDA Assay
MDA levels were significantly (p < 0.01) increased in the liver and kidney tissues of the diabetes group of mice in comparison with the control group of mice, whereas extract-treated diabetic mice showed significantly (p < 0.01) decreased levels of MDA in liver and kidney tissues in comparison with the diabetes group of mice (Figure 6B).
SOD Activity Assay
A significant (p < 0.01) decreased level of SOD activity was found in the liver and kidney tissues of the STZ-induced diabetes group of mice compared to the control group of mice. Treatment with a fraction of the extract in diabetic mice significantly (p < 0.01) increased the SOD activity in liver and kidney tissues in comparison with the diabetes group of mice (Figure 6C).
3. Discussion
UHPLC-QTOF-MS/MS, renowned for its exceptional selectivity, sensitivity, and accuracy, has been validated as a potent and efficient instrument for conducting metabolomics analysis [43,44,45,46]. Table 1 and Table 2 display detailed information about the retention time, highly accurate precursor ions, molecular formula, and characteristic m/z of each in both positive and negative ion modes for n-butanol fractions of P. javanica methanol extract. A total of 79 chemical components were tentatively identified in the sample compared with authentic samples, data from the available literature, and the obtained MS data. Glycosides, organic carboxylic and sulphonic acids, amides, phenolics, and their derivatives, ester, coumarins, flavonoids, alkaloids, quinine, and a few other types of chemicals were classified based on their structures. The compounds PJ_02, PJ_03, PJ_06, PJ_15, PJ_21, PJ_22, PJ_25, PJ_29, PJ_36, PJ_37, PJ_39, and PJ_55 showed characteristic [M+Na]+ ion peaks in the positive ionization mode. The compounds PJ_04, PJ_11, PJ_17, PJ_19, PJ_20, PJ_24, PJ_26, PJ_27, PJ_42, PJ_47, and PJ_51 displayed distinctive [M+NH4]+ ion peaks in the positive ionization mode. All other compounds showed distinctive [M+H]+ ion peaks ranging from PJ_01 to PJ_56. The compounds PJ_58 and PJ_77 were identified in negative ionization mode and showed [M+CH3COO]− ion peaks. Under negative ionization mode, distinctive ion peaks corresponding to [M−H]− were observed for compounds from PJ_57 to PJ_79. The compound PJ_61 exhibited an ion peak of [2M+Cl]−.
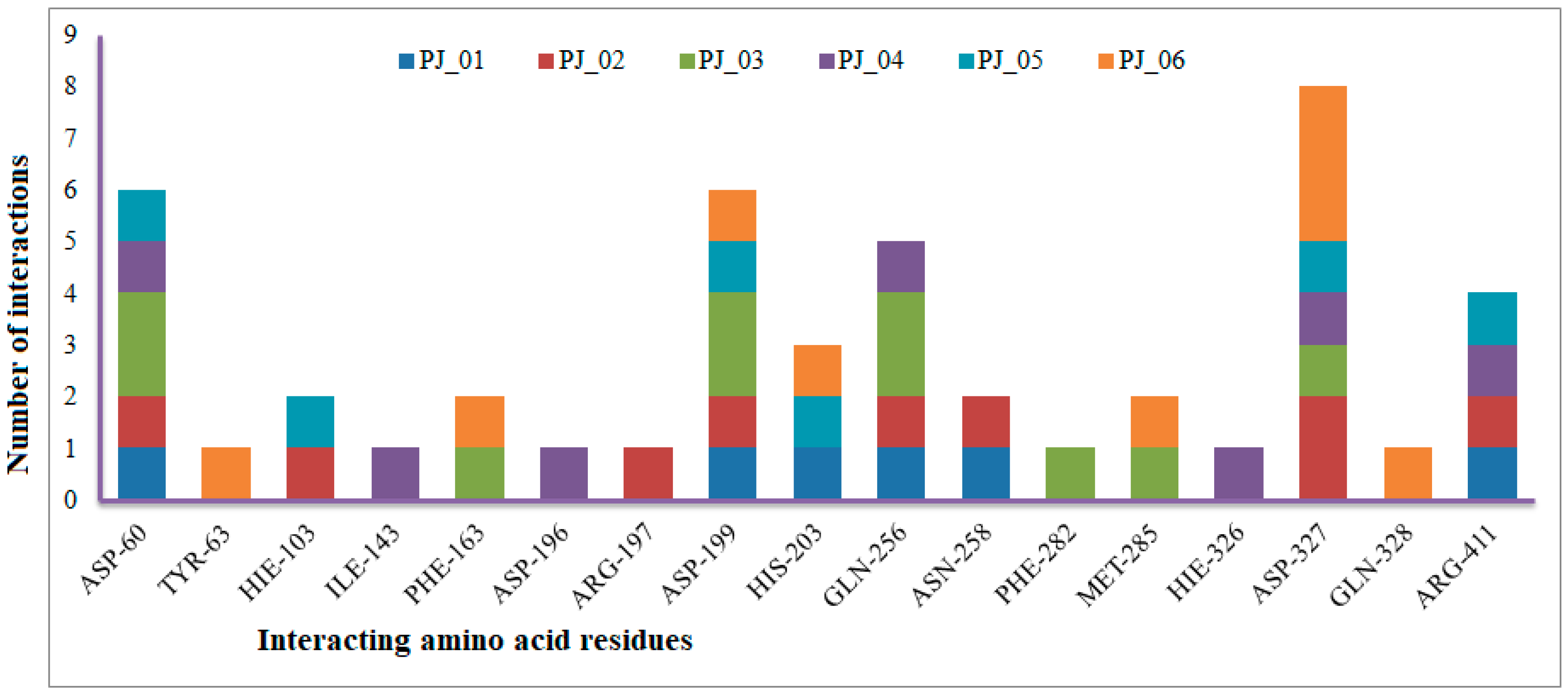
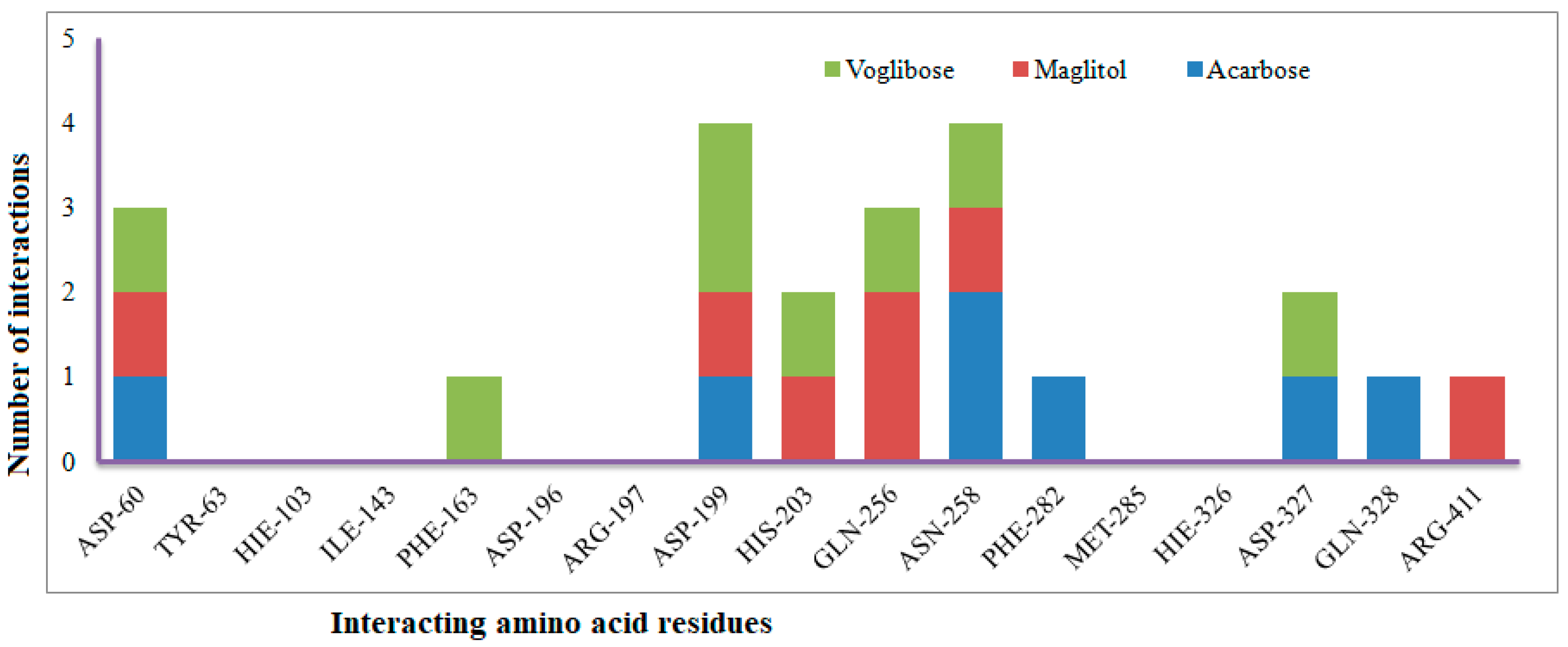
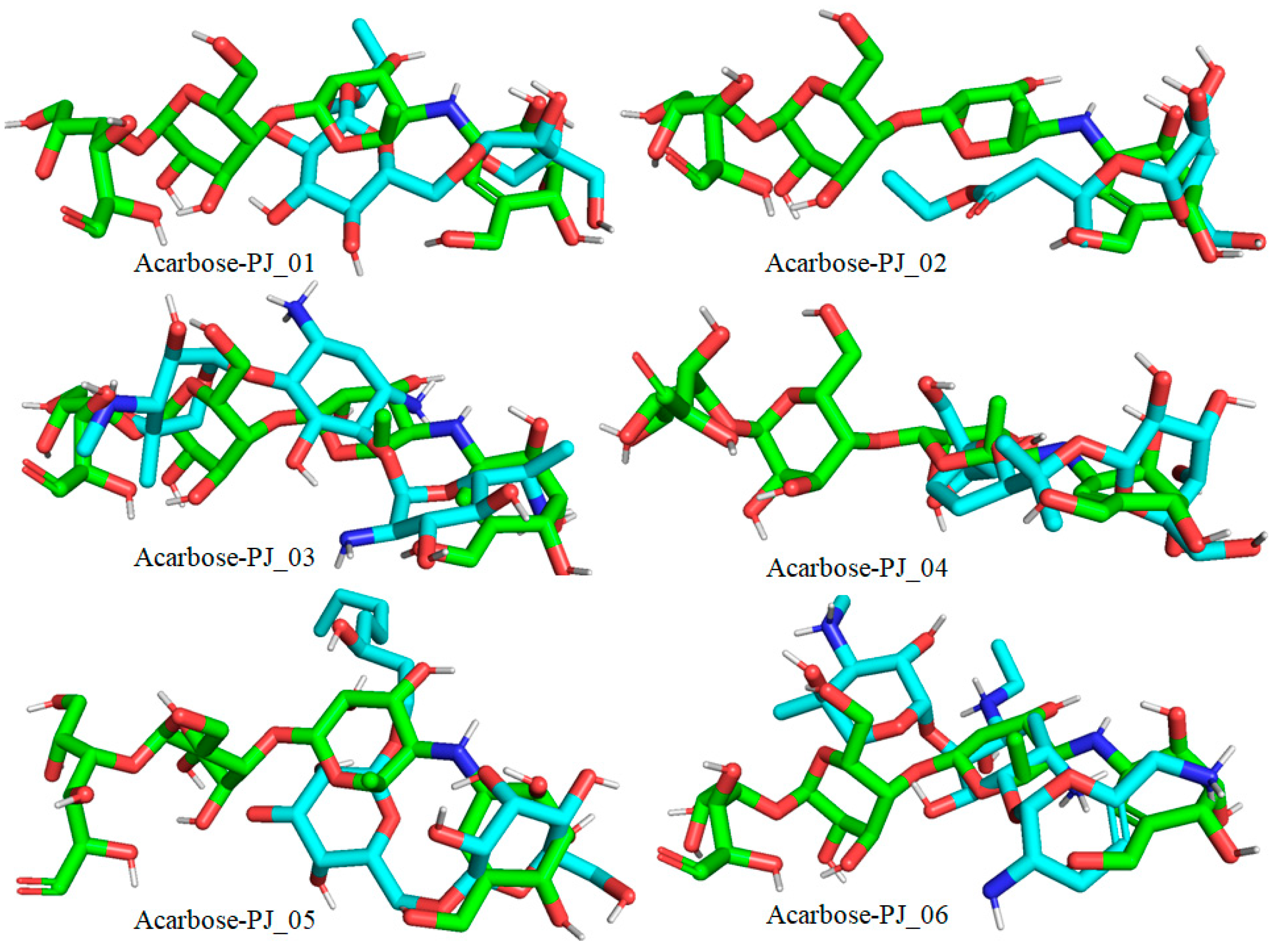
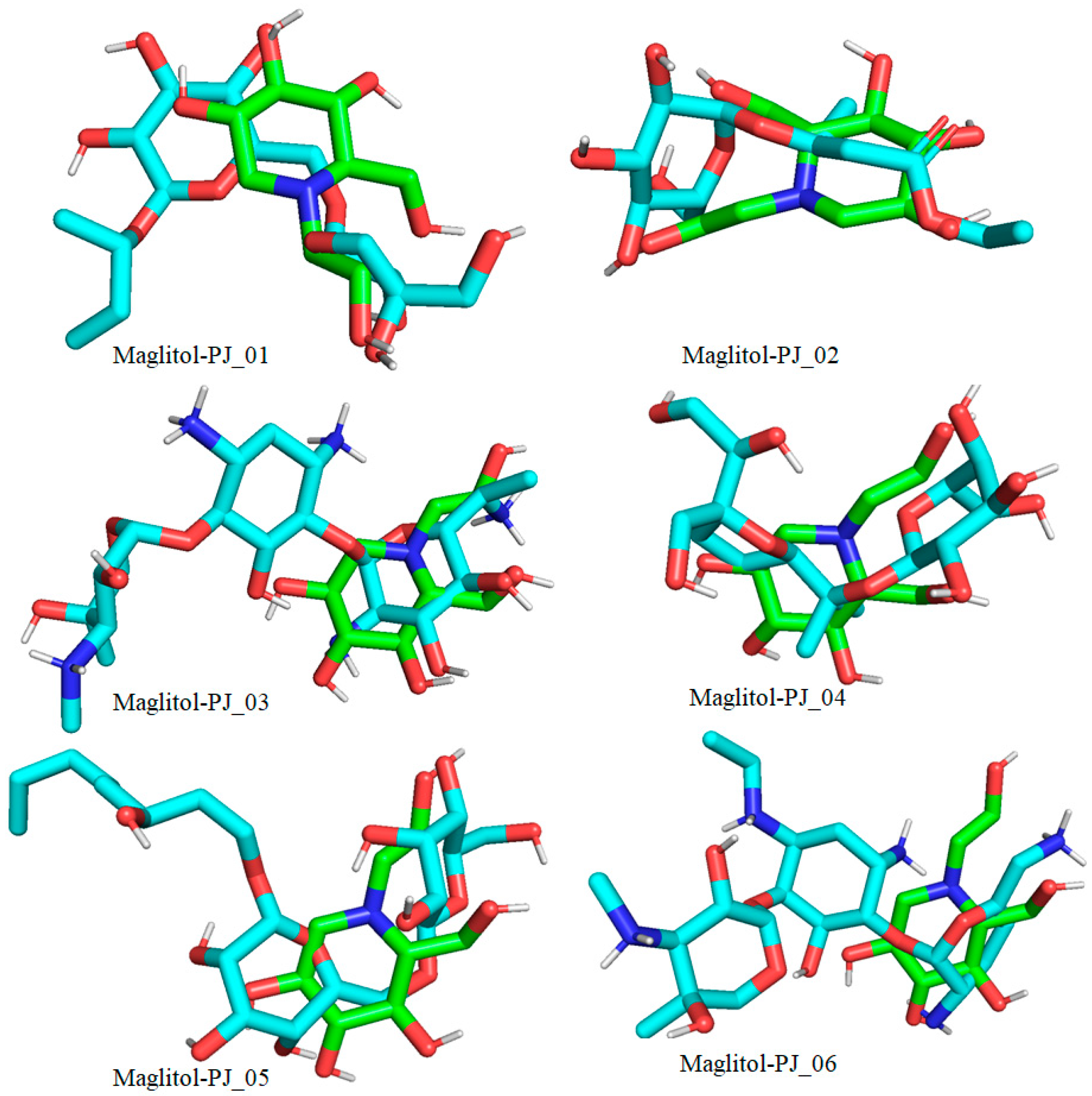
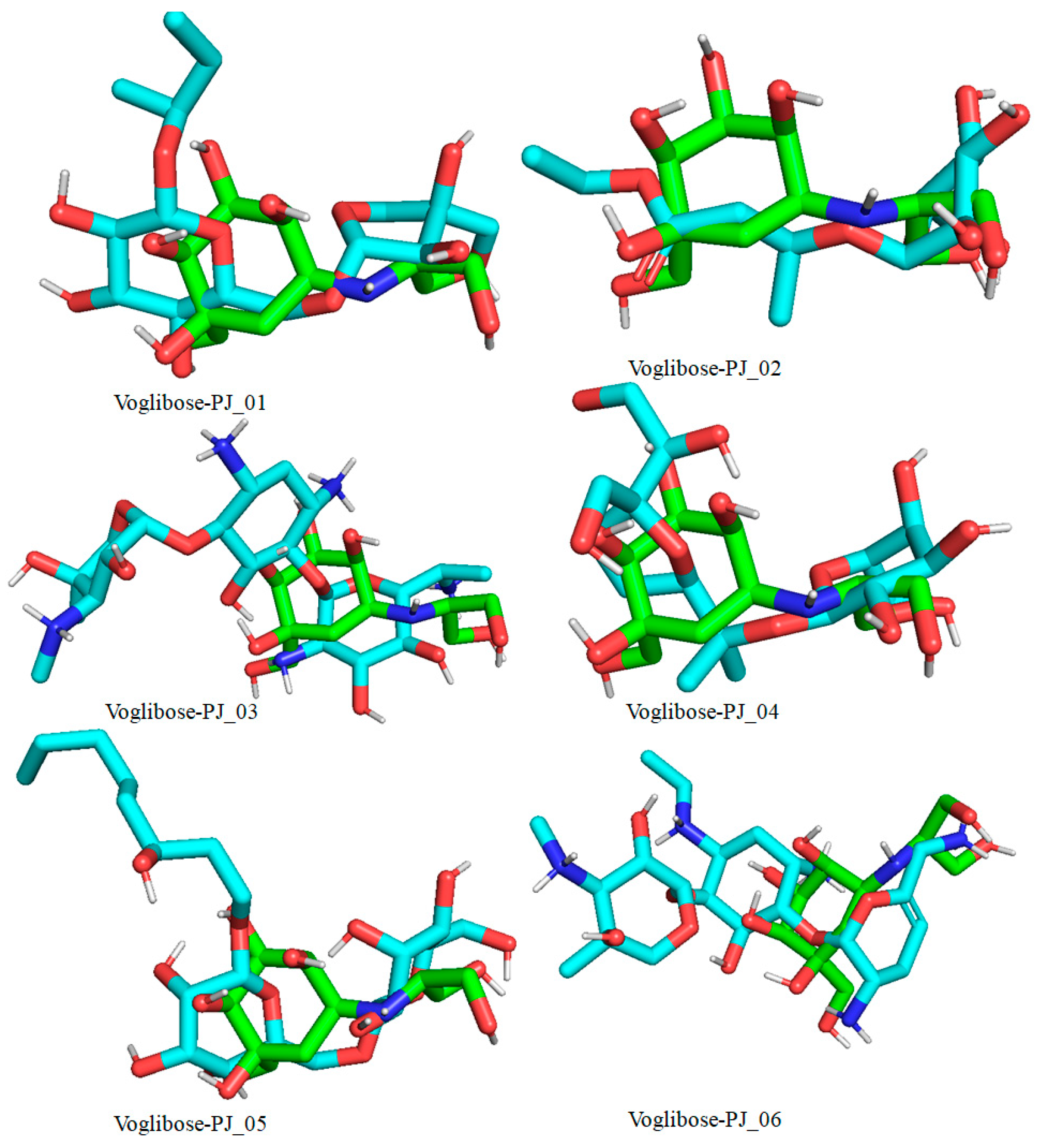
The XP Glide scores of Miglitol and Voglibose were −7.327 and −8.662, respectively. The XP Glide scores of hits PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 were −9.092, −8.579, −13.632, −10.506, −11.013 and −13.818, respectively. The negative sign indicates the energy released after inhibitors bind to α-glucosidase. The XP Glide score of PJ_03 and PJ_05 was also greater than the XP Glide score of the best-known binding inhibitor, Acarbose. All three known inhibitors interact with ASP-60. Similarly, PJ_01, PJ_02, PJ_03, PJ_04, and PJ_05 interact with ASP-60. All known inhibitors exhibit interactions with residues ASP-199 and ASN-258, and PJ_01, PJ_02, PJ_03, PJ_05, and PJ_06 interact with ASP-199, as do PJ_01 and PJ_02 interact with ASN-258. Acarbose and Voglibose showed interactions with ASP-327, as did all other inhibitors except PJ_01. Therefore, most of the selected inhibitors showed similar interactions with known inhibitors. The interactions between known inhibitors and the selected five compounds are shown in Figure 7 and Figure 8. The 3D binding poses of selected hits superimposed on the binding poses of known inhibitors in the active site are shown in Figure 9, Figure 10 and Figure 11. The binding poses of PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 are similar to all known inhibitors, although it depends on the size of the known inhibitors as well as the size of selected hits. The binding poses of PJ_03 were very close to Acarbose. Similarly, the binding poses of PJ_01, PJ_02, and PJ_04 were very similar to the binding poses of Miglitol and Voglibose.
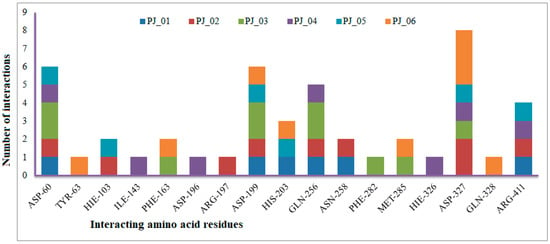
Figure 7.
Number of interactions exhibited by selected top six inhibitors (PJ_01, PJ_02, PJ_03, PJ_04, and PJ_06) with active site amino acid residues of α-glucosidase.
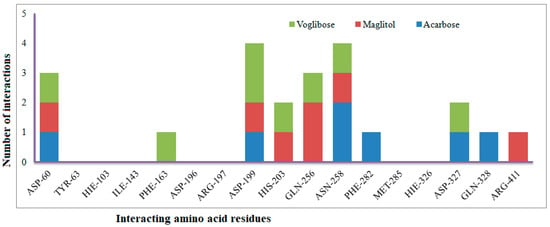
Figure 8.
Number of interactions exhibited by selected known α-glucosidase inhibitors Voglibose, Miglitol, and Acarbose with active site amino acid residues.
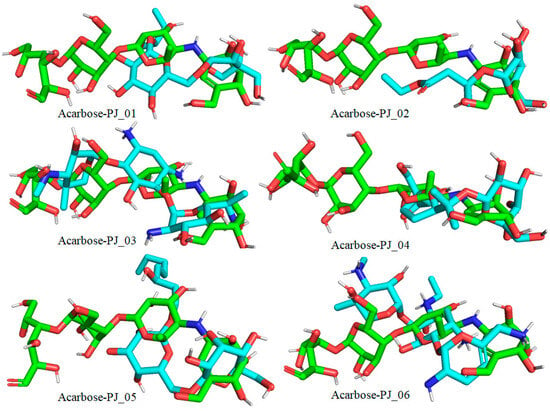
Figure 9.
Docked pose of compounds PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 (cyan color) superimposed on the docked pose of known inhibitor Acarbose (green color) in the active site of α-glucosidase.
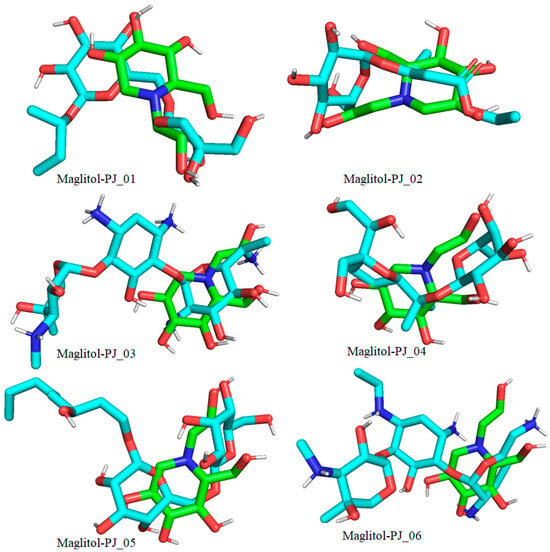
Figure 10.
Docked pose of compounds PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 (cyan color) superimposed on the docked pose of known inhibitor Miglitol (green color) in the active site of α-glucosidase.
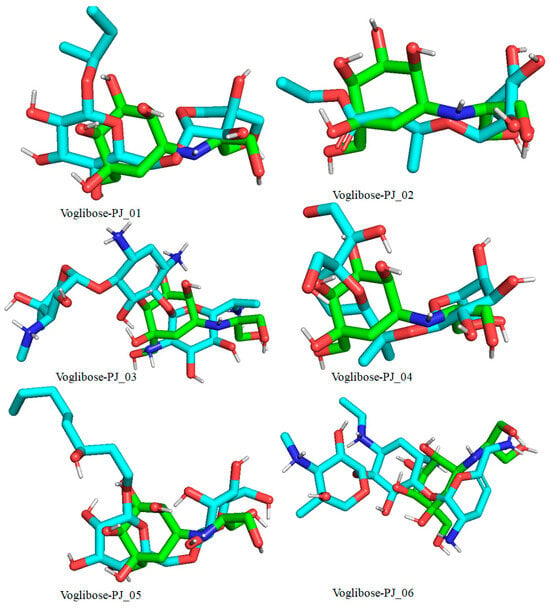
Figure 11.
Docked pose of compounds PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 (cyan color) superimposed on the docked pose of known inhibitor Voglibose (green color) in the active site of α-glucosidase.
The compounds PJ_01, PJ_02, PJ_04, and PJ_05 were glycosides in nature, and compounds PJ_03 and PJ_06 were aminoglycosides. The hydrophilic properties of glycoside molecules make them more water-soluble in general [47,48]. All the compounds are highly polar and, therefore, soluble in water. α-Glucosidase inhibition is an effective measure for the management of diabetes mellitus type 2. α-Glucosidase inhibitors reduce the absorbable form of carbohydrates in the small intestine, thus proving effective control against postprandial hyperglycemia. In the present study, the n-butanol fraction of the methanol extract of P. javanica pod showed good in vitro α-glucosidase inhibitory activity compared to pure compound Acarbose, a commonly used drug for the medication of type-2 diabetes. The % of α-glucosidase inhibition for n-butanol fraction and acarbose (500 µg/mL) were 83.21 and 98.31, respectively (Table 4). The IC50 value of the n-butanol fraction of the extract against α-glucosidase was 261.9 µg/mL, which was 2.7-fold less effective than pure Acarbose. The extract contains a mixture of approx. 79 compounds, of which only 6 exhibited potential in silico binding affinity towards α-glucosidase. Earlier findings also showed that the extract from Hyoscyamus albus L. exhibited an inhibitory effect on α-glucosidase, with an IC50 of 270.43 µg/mL [49], as the extracts contain a number of compounds. Apart from this, P. javanica can be easily found and used as a good food supplement, which could help to prevent T2D management among rural populations. The findings of the present study showed that the blood glucose level of DB mice was 462.66 µg/mL, but the blood glucose level of pod extract-treated mice was 228.66 µg/mL. The blood glucose level of acarbose-treated mice was 186.33 µg/mL, which indicates that the blood glucose level of pod extract-treated mice is comparable with the standard drug acarbose.
The n-butanol fraction not only reduces the blood glucose level in STZ-induced diabetic mice but also reduces oxidative stress in the liver and kidney. STZ-induced type 2 diabetes mellitus is associated with a decrease in catalase expression and a significant reduction in superoxide dismutase activity. However, it elevates polyunsaturated fatty acid oxidation as a result of a substantial increase in MDA in the liver and kidney. The present in vivo investigation revealed that the n-butanol fraction of P. javanica pod significantly increased catalase and SOD activities in the liver and kidney tissues in extract-treated diabetic mice; however, MDA levels in the kidney and liver tissue reduced significantly in the extracted-treated diabetic mice group. These findings provide insight into the fact that ingesting n-butanol fractions is relatively safe for developing anti-diabetes formulations from P. javanica compounds or fractions of extract. Treatment of diabetic mice with 80 mg/Kg/day for 15 consecutive days showed no behavioral changes among the mice.
In vivo oxidative stress analysis showed a significant decrease in MDA levels and an increase in antioxidant enzyme (catalase and SOD) activities in liver and kidney tissue in Parkia javanica pod extract-treated diabetic Swiss albino mice. A recent study showed that oral administration of Parkia speciosa extract significantly increased the levels of SOD and catalase enzymes and decreased MDA levels in the hepatic and renal tissues of diabetic mice [50]. Recent research also showed that hydrogen peroxide acts as a messenger in the signaling mechanism of insulin secretion, although its higher concentration is toxic. Blood catalase is the principal regulator of H2O2 metabolism. Generally, blood catalase activity in type 2 diabetes is highly reduced [51]. The oral administration of SOD decreases the blood glucose level in diabetic rats [52]. Poor metabolic control increases the level of MDA, and it acts as a biomarker in type 2 diabetic patients. Our findings showed excellent results by increasing catalase and SOD activity in the liver and kidney, which may be the cause of the glucose level reduction in experimental mice.
It was reported that consumption of one Parkia roxburghii pod almost every day has no adverse effect [53]. Although many anti-diabetic drugs are available on the market, the use of natural compounds as hypoglycemic agents is increasing, considering that plant-derived drugs are less toxic. Our results corroborate the previous studies on P. roxburghii, P. speciosa, and P. biglobosa [54,55]. Epigallocatechin gallate and hyperin isolated from P. roxburghii and P. speciosa pods can potentially inhibit α-glucosidase activity. Aqueous and methanolic extracts of P. biglobosa fermented seeds showed hypoglycemic activity in alloxan-induced diabetic rats. Our findings strongly support the findings of in silico binding affinity results in which 79 compounds were identified from the n-butanol fraction of the extract using the UPLC–HRMS technique. Six compounds exhibited excellent binding affinity with the α-glucosidase enzyme compared to known α-glucosidase inhibitors. Another interesting reported fact is that these pods also have anti-cancer, antioxidant, anti-angiogenic, and antiproliferative potential; therefore, these edible pods may be used to treat diabetes as well as cancer after detailed studies of diabetic patients with cancer. The compounds showed poor oral absorption rates in humans, indicating that their potential use as anti-diabetes agents is favorable. The majority of the potential compounds will be excreted from the body after inhibition without being absorbed.
4. Materials and Methods
4.1. Collection of Plant Material
Edible pods of P. javanica were procured from the Lake Chowmuhani Market in Agartala, Tripura, India, in March 2023 (Figure 12). The obtained pods have been taken to the Department of Forestry and Biodiversity, Tripura University. The pods were ground into powder, dried without exposure to sunlight, and preserved for further analysis. Primary identification was carried out by Dr. Kausik Majumder, Dept. of Botany, Tripura University. A specimen voucher of the plant (No. BD-01/06) is available in the Central National Herbarium, Government of India, Howrah, Shibpur. The dry powdered pods (0.50 kg) of P. javanica were extracted at room temperature using 1 × 5 L MeOH. A sticky mass containing concentrated extract was stored in vacuum desiccators for three days. About 2.0 g of gummy material has been diluted with 50 mL of distilled water and partitioned sequentially with 300 mL of petroleum ether, CHCl3, and n-butanol each. The yield of the dried n-butanol fraction was 0.50 g. The known α-glucosidase inhibitors, Acarbose, Miglitol, and Voglibose, are polar in nature. Generally, polar compounds and glycosides like acarbose are available in the n-butanol fraction of plant extracts. The n-butanol-soluble fraction underwent a bioactivity assay and UHPLC-QTOF-MS/MS analysis.

Figure 12.
Photographic representation of the tree (A) and pods (B) of P. javanica.
4.2. UHPLC-QTOF-MS/MS
The samples were analyzed using the TOF/Q-TOF Mass Spectrometer, Component Name MS Q-TOF, Component Model G6546A, Ion Source Dual AJS ESI, Min Range (m/z): 100, Max Range (m/z): 1500, Scan Rate (Spectra/Sec): 1.0. The Q-TOF MS system is conjunct with the UPLC (Agilent Technologies, Santa Clara, CA, USA). A unique collection of tools, such as the G7129B auto sampler, the G7116B column compartment, the G7104A Quat. pumps, the G7117A DAD, and the G6546A MS-Q-TOF, were used to accomplish the LC–MS analysis. A reversed-phase ZORBAX RRHD Eclipse Plus C18 analytical column measuring 100 mm by 2.1 mm by 1.8 µm and running at 40 °C was utilized for the compound separation process. Mobile phase A, which included H2O and formic acid (0.1%), and mobile phase B, which included 100% acetonitrile, were used in the elution gradient. The details of solvent composition and timetable with flow rate are given in Table 5 and Table 6, respectively. The solvents (HPLC-grade) methanol, acetonitrile, and formic acid were supplied by Thermo Fisher Scientific (Waltham, MA, USA).

Table 5.
Solvent (water and acetonitrile) composition during HPLC.

Table 6.
Timetable, solvent composition, and flow rate during HPLC.
4.3. Molecular Docking
All 79 structures of the compound were drawn using ChemDraw Professional 15 and saved in .sdf format. The single .sdf file was imported into Maestro-14 [56], and the molecules were prepared using the LigPrep [57] module, Schrodinger-2022-4. Ligprep retains the original chiralities and ionization states of each input structure while generating a single, low-energy, three-dimensional structure. The energy of the molecules was minimized using the OPLS_2005 force field.
The X-ray crystal structure of PDB IDs of α-glucosidase [58,59] PDB ID: 5ZCC was retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) data bank, imported into Maestro, and prepared using “Protein Preparation Wizard” [60]. During protein preparation, hydrogens were added to the protein and co-ligand to fill in the missing side chains and missing loops using prime. The energy of the complexes was then minimized using the OPLS_2005 force field. The co-ligand of the active site was selected to generate the receptor grid box of the 15.0 Å cube.
4.4. Virtual Screening
The method of structure-based virtual screening was employed to determine the binding affinity of compounds of P. javanica to α-glucosidase using Glide [61,62,63,64], Schrodinger-2022-4, LLC, New York, NY, USA. The already prepared molecules are imported into the Virtual Screening Workflow, and the prepared receptor grid is selected. In the VS workflow, 40% of the total molecules from Glide HTVS are forwarded to Glide SP. Then, 40% of the molecules from SP were forwarded to Glide XP, followed by post-processing with prime MM-GBSA. All the computational works were carried out using Dell Inc., System Model: Precision 5820 Tower, Processor: Intel(R) Xeon(R) W-2245 CPU @ 3.90GHz (16 CPUs), ~3.9 GHz, OS: Ubuntu 22.04.1 LTS, 64-bit.
4.5. In Vitro α-Glucosidase Enzyme Inhibitory Assays
The assay analysis is performed by following the procedure of Pistia–Brueggeman and Hollingsworth [65]. Moreover, 20 μL of α-glucosidase (5U/mL) were mixed with 100 μL of n-butanol extract at varying concentrations in 250 μL of 0.1 M phosphate buffer (pH 6.8) and incubated at 37 °C for 15 min. Then, 40 μL of 1.0 M pPNG(4-Nitrophenyl-α-D-glucopyranoside) substrate was added and incubated for 45 min at 37 °C for the reaction. For the termination of the reaction, 100 μL of 0.10 N Na2CO3 was added. The OD value was measured at 405 nm using a spectrophotometer. Acarbose was used as the positive control. The inhibitory activity was calculated using the following formula:
% inhibition of α-glucosidase = (Abscontrol − AbsSample)/(Abscontrol) × 100
4.6. In Vivo Anti-Diabetic Activity
All the experiments with animals and their maintenance were conducted in accordance with institutional practice, within the framework of CCSEA (Committee for Control and Supervision of Experiments on Animals), and as per the Act of the Government of India (2007) for animal welfare. The present study was conducted on Swiss albino mice. The healthy mice of 25.0 g were selected from a mouse colony maintained in the standard laboratory conditions of light (12 h light and 12 h dark), temperature (25.0 ± 2 °C), and humidity (55.0 ± 5%). The experimental mice were kept five per group in polycarbonated cages (43 cm × 27 cm × 14 cm) and fed with mice feed and water ad libitum [66,67].
Mice were intraperitoneally injected with multiple low doses of streptozotocin (STZ) at 50.0 mg/kg body weight for five consecutive days. Freshly prepared 0.10 M citrate buffer (pH 4.5) was used to dissolve the STZ. Experimental mice were kept for 20 h of fasting before STZ administration, and 10% sucrose water was provided during the STZ treatment. The blood was collected from the tail veins of mice to determine the blood glucose level using an Accu-Chek® active (Roche) blood glucose monitoring system at two-day intervals. Experimental mice with blood glucose levels greater than 250 mg/dL were considered to have diabetes [66].
Female mice were divided into control (CON), diabetes (DB), and diabetic mice treated with pod extract (DBF), diabetes mice treated with standard drug acarbose (DBS), and control mice treated with pod extract (CONF) groups (with five mice in each group). The pod extract and acarbose were diluted 100 times in distilled water to prepare the working solution (10 mg of pod extract was dissolved in 1.0 mL of distilled water). Experimental diabetic mice were orally given pod extract (200 µL/mice/day) for 15 consecutive days.
Experimental mice were sacrificed on the 16th day, and blood was collected for blood glucose analysis. The liver and kidney were dissected and kept at −20 °C for oxidative stress analysis.
4.7. Blood Glucose Level Analysis
Blood glucose was determined with the help of the ACCU-CHEK® active (Roche) blood glucose monitoring system [66].
4.8. Oxidative Stress Analysis
Catalase activity assay: Catalase (CAT; EC 1.11.1.6) activity was determined by following the method [68,69].
SOD activity assay: Superoxide dismutase (SOD; EC 1.15.1.1) activity was assayed following the method [70].
MDA assay: Lipid peroxidation was measured based on the reaction between malondialdehyde and thiobarbituric acid (TBA) following the method [71].
4.9. Prediction of Physiochemical Parameters Using SwissADME
SwissADME is a free online tool to assess the pharmacokinetics, drug-likeness, and medicinal chemistry compatibility of small organic molecules [72]. The important physiochemical parameters that were predicted are molecular weight, the numbers of H-bond acceptors and H-bond donors, rotatable bonds, molar refractivity, topological polar surface area, solubility class, gastrointestinal absorption, blood–brain barrier penetration, violation of Lipnskis rule of five, violation of Ghose rule, violation of Veber rule, and bioavailability score [73,74,75,76,77,78].
4.10. Statistical Analysis
The statistical analysis of the data was performed with a one-way analysis of variance (ANOVA). The differences were considered significant at p < 0.01. Statistical Package for the Social Sciences (SPSS-17) and Microsoft Excel programs were used for calculation and graph preparation.
5. Conclusions
The n-butanol fraction of the methanol extract of P. javanica pods comprises seventy-nine compounds identified by the UHPLC-QTOF-MS/MS-based chemical profiling technique. Molecular docking investigations of the identified compounds revealed that PJ_01–PJ_06 showed significant binding affinity toward the target protein of diabetes, α-glucosidase. This study also provided evidence that the n-butanol fraction of the pods of P. javanica extract has potential in vitro and in vivo anti-diabetic activity. The mice treated with acarbose, mice treated with pod extract, and diabetic mice had blood glucose levels of 186.33 µg/mL, 228.66 µg/mL, and 462.66 µg/mL, respectively. The outcomes of acarbose-treated and pod extract-treated mice are comparable. Our findings suggested that the phytochemicals found in the n-butanol fraction of P. javanica have the ability to alleviate hyperglycemia. P. javanica is easily available and can serve as a good food supplement, which could help to prevent T2DM among people. In future studies, bio-assay-guided isolation will be conducted to obtain pure active compounds.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph17070968/s1. Figure S1: UHPLC-QTOF-MS/MS-based spectra of compounds identified by positive ionization mode; Figure S2: UHPLC-QTOF-MS/MS-based spectra of compounds identified by negative ionization mode; Figure S3: Structure of compounds identified by UPLC–HRMS in both positive and negative ionization mode; Table S1: Physiochemical parameters of PJ_01, PJ_02, PJ_03, PJ_04, PJ_05, and PJ_06 inhibitors predicted by SwissADME.
Author Contributions
A.S.: methodology, formal analysis, validation, and the original draft. A.C.: methodology, formal analysis, and validation. S.B.: project administration, and the original draft. B.D.: supervision, review, and editing. S.S.S.: investigation, resources, and writing. R.G.: software and visualization. M.E.A.Z. and S.A.A.-H.: funding acquisition and editing. S.D.: conceptualization, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by DBT, Govt. of India, New Delhi, grant number HRD-11011/32/2021-HRD-DBT. This work was also supported and APC was funded by the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU).
Institutional Review Board Statement
The in vivo experiment was carried out in accordance with the ethical guidelines approved by the Institutional Human Ethics Committee, Tripura University. Approval No. TU/IAEC/2023/I/228.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Acknowledgments
The author, S.B., wishes to thank DBT, Govt. of India, New Delhi, for the Institutional Star College Programme [HRD-11011/32/2021-HRD-DBT]. The authors are also thankful to the Drug Metabolomics Centre, Tripura University, for providing UPLC-MS facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed]
- Sathyanath, S.; Kundapur, R.; Deepthi, R.; Poojary, S.N.; Rai, S.; Modi, B.; Saxena, D. An economic evaluation of diabetes mellitus in India: A systematic review. Diabetes Metab. Syndr. 2022, 16, 102641. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Pervin, R. Chapter 34—Current Antidiabetic Drugs: Review of Their Efficacy and Safety. In Nutritional and Therapeutic Interventions for Diabetes and Metabolic Syndrome, 2nd ed.; Bagchi, D., Nair, S., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 455–473. [Google Scholar]
- William, J.; John, P.; Mumtaz, M.W.; Ch, A.R.; Adnan, A.; Mukhtar, H.; Sharif, S.; Raza, S.A.; Akhtar, M.T. Antioxidant activity, α-glucosidase inhibition and phytochemical profiling of Hyophorbe lagenicaulis leaf extracts. Peer J. 2019, 7, e7022. [Google Scholar] [CrossRef] [PubMed]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2022, 21, 1049–1079. [Google Scholar] [CrossRef] [PubMed]
- Hasimun, P.; Adnyana, I.K. Zingiberaceae family effects on alpha-glucosidase activity: Implication for diabetes. In Bioactive Food as Dietary Interventions for Diabetes; Academic Press: Cambridge, MA, USA, 2019; pp. 387–393. [Google Scholar] [CrossRef]
- Shikov, A.N.; Narkevich, I.A.; Akamova, A.V.; Nemyatykh, O.D.; Flisyuk, E.V.; Luzhanin, V.G.; Povydysh, M.N.; Mikhailova, I.V.; Pozharitskaya, O.N. Medical species used in Russia for the management of diabetes and related disorders. Front. Pharmacol. 2021, 12, 697411. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, K.; Wang, H.; Kaur, J.; Nalbant, G.; Almaqhawi, A.; Kundakci, B.; Panniyammakal, J.; Heinrich, M.; Lewis, S.A.; Greenfield, S.M.; et al. Effectiveness and safety of ayurvedic medicines in type 2 diabetes mellitus management: A systematic review and meta-analysis. Front. Pharmacol. 2022, 13, 821810. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Li, C.; Cui, W.; Guo, Q.; Dong, C.; Zou, H.; Liu, S.; Dong, W.; Miao, L. Review of herbal traditional chinese medicine for the treatment of diabetic nephropathy. J. Diabetes Res. 2016, 2016, 5749857. [Google Scholar] [CrossRef]
- Yuzo, S.; Tomoko, U.; Khookhor, O.; Tatsuro, K.; Tatsuo, T.; Qin, B. Role of herbal medicine (Kampo formulations) on the prevention and treatment of diabetes and diabetic complications. J. Tradit. Med. 2006, 23, 185–195. [Google Scholar]
- Kanwal, S.; Ahmad, S.; Yasmin Begum, M.; Siddiqua, A.; Rao, H.; Ghalloo, B.A.; Shahzad, M.N.; Ahmad, I.; Khan, K.U. Chemical Profiling, in-vitro biological evaluation and molecular docking studies of Ruellia tweediana: An unexplored plant. Saudi Pharm. J. 2024, 32, 101939. [Google Scholar] [CrossRef]
- Nahar, N.; Nazmul Hasan, Z.M.; Biswas, P.; Morsaline, B.M.; Bibi, S.; Albekairi, N.A.; Alshammari, A.; Nazmul Hasan, M. Profiling of secondary metabolite and evaluation of anti-diabetic potency of Crotalaria quinquefolia (L): In-vitro, in-vivo, and in-silico approaches. Saudi Pharm. J. 2024, 32, 101887. [Google Scholar] [CrossRef]
- Ayoola, M.D.; Ogundeko, Y.B.; Obanleowo, T.D.; Omole, D.O.; Chukwu, B.N.; Faloye, K.O. Evaluation of the antidiabetic activities of the fruit of Parquetina nigrescens (Afzel.) bullock and in silico identification of its antidiabetic agent. Bioinform. Biol. Insights 2024, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Akinyede, K.A.; Oyewusi, H.A.; Hughes, G.D.; Ekpo, O.E.; Oguntibeju, O.O. In vitro evaluation of the anti-diabetic potential of aqueous acetone Helichrysum petiolare extract (AAHPE) with molecular docking relevance in diabetes mellitus. Molecules 2022, 27, 155. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Lin, Y.S.; Bartolome, A.P.; Chen, Y.C.; Chiu, S.C.; Yang, W.C. Herbal therapies for type 2 diabetes mellitus: Chemistry, biology, and potential application of selected plants and compounds. Evid. Based Complement Alternat Med. 2013, 2013, 378657. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Ata, A.; Nanjangud, V.A.K.; Sharopov, F.; Ramírez-Alarcón, K.; Ruiz-Ortega, A.; Ayatollahi, S.A.; Fokou, P.V.T.; Kobarfard, F.; Zakaria, Z.A.; et al. Antidiabetic potential of medicinal plants and their active components. Biomolecules 2019, 9, 551. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.D.; Deb, L.; Somkuwar, B.G.; Rana, V.S. Relevance of indian traditional tisanes in the management of type 2 diabetes mellitus: A review. Saudi Pharm. J. 2023, 31, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, D.R.; Parajuli, P.; Subba, G.C. Anti-diabetic plants used by Sikkim and Darjeeling Himalayan tribes, India. J. Ethnopharmacol. 2005, 99, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Blahova, J.; Martiniakova, M.; Babikova, M.; Kovacova, V.; Mondockova, V.; Omelka, R. Pharmaceutical drugs and natural therapeutic products for the treatment of type 2 diabetes mellitus. Pharmaceuticals 2021, 14, 806. [Google Scholar] [CrossRef]
- Daou, M.; Elnaker, N.A.; Ochsenkühn, M.A.; Amin, S.A.; Yousef, A.F.; Yousef, L.F. In vitro α-glucosidase inhibitory activity of Tamarix nilotica shoot extracts and fractions. PLoS ONE 2022, 17, e0264969. [Google Scholar] [CrossRef] [PubMed]
- Mechchate, H.; Es-Safi, I.; Louba, A.; Alqahtani, A.S.; Nasr, F.A.; Noman, O.M.; Farooq, M.; Alharbi, M.S.; Alqahtani, A.; Bari, A.; et al. In vitro alpha-amylase and alpha-glucosidase inhibitory activity and in vivo antidiabetic activity of Withania frutescens L. foliar extract. Molecules 2021, 26, 293. [Google Scholar] [CrossRef]
- Macalalad, M.A.B.; Gonzales, A.A. 3rd. In silico screening and identification of anti-diabetic inhibitors sourced from phytochemicals of Philippine plants against four protein targets of diabetes (PTP1B, DPP-4, SGLT-2, and FBPase). Molecules 2023, 28, 5301. [Google Scholar] [CrossRef]
- Rao, M.M.V.; Hariprasad, T.P.N. In silico analysis of a potential anti-diabetic phytochemical erythrin against therapeutic targets of diabetes. In Silico Pharmacol. 2021, 9, 5. [Google Scholar] [CrossRef]
- Din, A.U.; Khan, M.; Shah, M.Z.; Rauf, A.; Rashid, U.; Khalil, A.A.; Zaman, K.; Al-Awthan, Y.S.; Al-Duais, M.A.; Bahattab, O.; et al. Antidiabetic activity of ficusonolide, a triterpene lactone from Ficus foveolata (Wall. ex Miq.): In vitro, in vivo, and in silico approaches. ACS Omega 2021, 6, 27351–27357. [Google Scholar] [CrossRef] [PubMed]
- Swilam, N.; Nawwar, M.A.M.; Radwan, R.A.; Mostafa, E.S. Antidiabetic activity and in silico molecular docking of polyphenols from Ammannia baccifera L. subsp. Aegyptiaca (willd.) koehne waste: Structure elucidation of undescribed acylated flavonol diglucoside. Plants 2022, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Lei1, M.; Wang, L.; Olatunde, O.O.; Singh, S.; Ovatlarnporn, C.; Basit, A.; Olatunji, O.J. UPLC–ESI–QTOF–MS profiling, antioxidant, anti-diabetic, antibacterial, anti-inflammatory, antiproliferative activities and in silico molecular docking analysis of Barleria strigosa. Chem. Biol. Technol. Agric. 2023, 10, 73. [Google Scholar] [CrossRef]
- Abomughaid, M.M.; El-Shibani, F.A.A.; Abdulkarim, A.K.; Abouzied, A.S.; Sulaiman, G.M.; Abomughayedh, A.M.; Abdulsayid, M.M.F.; Albukhaty, S.; Elrmali, N.; Al-Saffar, A.Z.; et al. Phytochemicals profiling, in vitro and in vivo anti-diabetic activity, and in silico studies on Ajuga iva (L.) Schreb.: A comprehensive approach. Open Chem. 2024, 22, 20230191. [Google Scholar] [CrossRef]
- Majumdar, K.; Datta, B.K. Traditional wild edible fruits for the forest dwellers of Tripura, India. Pleione 2009, 3, 167–178. [Google Scholar]
- Sutradhar, B.; Deb, D.; Majumdar, K.; Datta, B.K. Traditional dye yielding plants of Tripura, Northeast India. Biodiversitas 2015, 16, 121–127. [Google Scholar] [CrossRef]
- Ralte, L.; Khiangte, L.; Thangjam, N.M.; Kumar, A.; Singh, Y.T. GC-MS and molecular docking analyses of phytochemicals from the underutilized plant, Parkia timoriana revealed candidate anti-cancerous and anti-inflammatory agents. Sci. Rep. 2022, 12, 3395. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Das, M.C.; Sandhu, P.; Das, N.; Tribedi, P.; De, U.C.; Yusuf, A.; Bhattacharjee, S. Antibiofilm activity of Parkia javanica against Pseudomonas aeruginosa: A study with fruit extract. RSC Adv. 2017, 7, 5497–5513. [Google Scholar] [CrossRef]
- Khangembam, V.C.; Srivastava, S.K.; Leishangthem, G.D.; Kataria, M.; Thakuria, D. Evaluation of apoptosis inducing ability of Parkia javanica seed extract in cancer cells. Indian J. Pharm. Sci. 2018, 80, 1069–1077. [Google Scholar] [CrossRef]
- Saha, P.; Saha, S.; Sil, S.K. Anti-colon cancer activity of Parkia javanica (Lamk.) Merr. bark extract: An in-vitro study. Int. J. Pharm. Sci. Drug Res. 2021, 13, 536–542. [Google Scholar] [CrossRef]
- Chanu, K.V.; Devi, L.G.; Srivastava, S.K.; Thakuria, D.; Kataria, M.; Telang, A.G. Phytochemical analysis and evaluation of anti-cancer activity of Parkia javanica seeds. Pharma Innov. J. 2018, 7, 305–311. [Google Scholar]
- Chanu, K.V.; Ali, M.A.; Kataria, M. Antioxidant activities of two medicinal vegetables: Parkia javanica and Phlogacanthus thyrsiflorus. Int. J. Pharm. Pharm. Sci. 2012, 4, 102–106. [Google Scholar]
- Saha, R.; Mookerjee, B.J.; Roy, S.; Dinda, B.; Sil, S.K. In vitro activity of Parkia javanica extract against Leishmania donovani parasite. J. Appl. Biosci. 2010, 36, 85–89. [Google Scholar]
- Saleh, M.S.M.; Jalil, J.; Mustafa, N.H.; Ramli, F.F.; Asmadi, A.Y.; Kamisah, Y. UPLC-MS-Based Metabolomics Profiling for α-Glucosidase Inhibiting Property of Parkia speciosa Pods. Life 2021, 11, 78. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, R.; Kalladka, K.; Singha, A.; Pandyanda, N.D.; Ravindra, J.; Vittal, R.; Sil, S.K.; Chakraborty, A.; Chakraborty, G. Investigation of antiproliferative and anti-angiogenic properties of Parkia javanica bark and fruit extracts in zebrafish. PLoS ONE 2023, 18, e0289117. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Sharma, D.; Dash, S.; Dey, S.K.; Sil, S.K. Identification of 2, 4-di-tert-butylphenol (2,4-DTBP) as the major contributor of anti-colon cancer activity of active chromatographic fraction of Parkia javanica (Lamk.) Merr. bark extract. Biomed. Pharmacol. J. 2023, 16, 275–288. [Google Scholar] [CrossRef]
- Dinda, B.; Mohanta, B.C.; Debnath, S.; Ghosh, B.; Arima, S.; Sato, N.; Harigaya, Y. Iridoid glucosides from leaves and stem barks of Parkia javanica. J. Asian Nat. Prod. Res. 2009, 11, 229–235. [Google Scholar] [CrossRef]
- Patra, K.; Jana, S.; Sarkar, A.; Karmakar, S.; Jana, J.; Gupta, M.; Mukherjee, G.; De, U.C.; Mandal, D.P.; Bhattacharjee, S. Parkia javanica extract induces apoptosis in S-180 cells via the intrinsic pathway of apoptosis. Nutr. Cancer 2016, 68, 689–707. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Li, X.; Wang, P.; Tong, Y.; Liu, J.; Shu, G. UHPLC-Q-exactive orbitrap MS/MS-based untargeted metabolomics and molecular networking reveal the differential chemical constituents of the bulbs and flowers of Fritillaria thunbergii. Molecules 2022, 27, 6944. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.U.; Mumtaz, M.W.; Mukhtar, H.; Rashid, U.; Akhtar, M.T.; Raza, S.A.; Nadeem, M. UHPLC-QTOF-MS/MS based phytochemical characterization and anti-hyperglycemic prospective of hydro-ethanolic leaf extract of Butea monosperma. Sci. Rep. 2020, 10, 3530. [Google Scholar] [CrossRef]
- Basit, A.; Ahmad, S.; Khan, K.U.R.; Naeem, A.; Usman, M.; Ahmed, I.; Shahzad, M.N. Chemical profiling of Justicia vahlii Roth. (Acanthaceae) using UPLC-QTOF-MS and GC-MS analysis and evaluation of acute oral toxicity, antineuropathic and antioxidant activities. J. Ethnopharmacol. 2022, 287, 114942. [Google Scholar] [CrossRef] [PubMed]
- Azizah, M.; Pripdeevech, P.; Thongkongkaew, T.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. UHPLC-ESI-QTOF-MS/MS-based molecular networking guided isolation and dereplication of antibacterial and antifungal constituents of Ventilago denticulata. Antibiotics 2020, 9, 606. [Google Scholar] [CrossRef] [PubMed]
- Kren, V.; Martinkova, L. Glycosides in medicine: The role of glycosidic residue in biological activity. Curr. Med. Chem. 2001, 8, 1303–1328. [Google Scholar] [CrossRef]
- Schaub, J.; Zielesny, A.; Steinbeck, C.; Sorokina, M. Description and analysis of glycosidic residues in the largest open natural products database. Biomolecules 2021, 11, 486. [Google Scholar] [CrossRef] [PubMed]
- Lekmine, S.; Benslama, O.; Kadi, K.; Martín-García, A.I.; Yilmaz, M.A.; Akkal, S.; Boumegoura, A.; Alhomida, A.S.; Ola, M.S.; Ali, A. LC/MS-MS analysis of phenolic compounds in Hyoscyamus albus L. extract: In vitro antidiabetic activity, in silico molecular docking, and in vivo investigation against STZ-induced diabetic mice. Pharmaceuticals 2023, 16, 1015. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, W.; Yang, L.; Fan, H.; Olatunji, O.J. Stink bean (Parkia speciosa) empty pod: A potent natural antidiabetic agent for preventing pancreatic and hepatorenal dysfunction in high-fat diet/streptozotocin-induced type 2 diabetes in rats. Arch. Physiol. Biochem. 2023, 129, 261–267. [Google Scholar] [CrossRef]
- László, G. Catalase deficiency and type 2 diabetes. Diabetes Care 2008, 31, e93. [Google Scholar] [CrossRef]
- Guo, J.; Liu, H.; Zhao, D. Glucose-lowering effects of orally administered superoxide dismutase in type 2 diabetic model rats. Npj Sci. Food 2022, 6, 36. [Google Scholar] [CrossRef]
- Sheikh, Y.; Maibam, B.C.; Talukdar, N.C.; Deka, D.C.; Borah, J.C. In vitro and in vivo anti-diabetic and hepatoprotective effects of edible pods of Parkia roxburghii and quantification of the active constituent by HPLC-PDA. J. Ethnopharmacol. 2016, 191, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.S.M.; Jalil, J.; Zainalabidin, S.; Asmadi, A.Y.; Mustafa, N.H.; Kamisah, Y. Genus Parkia: Phytochemical, medicinal uses, and pharmacological properties. Int. J. Mol. Sci. 2021, 22, 618. [Google Scholar] [CrossRef]
- Odetola, A.A.; Akinloye, O.; Egunjobi, C.; Adekunle, W.A.; Ayoola, A.O. Possible anti-diabetic and antihyperlipidaemic effect of fermented Parkia biglobosa (JACQ) extract in alloxan-induced diabetic rats. Clin. Exp. Pharmacol. Physiol. 2006, 33, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Maestro; Schrödinger, LLC: New York, NY, USA, 2022.
- LigPrep; Schrödinger, LLC: New York, NY, USA, 2022.
- Auiewiriyanukul, W.; Saburi, W.; Kato, K.; Yao, M.; Mori, H. Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity. FEBS Lett. 2018, 592, 2268–2281. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Saburi, W.; Yao, M. Crystal structure of Alpha-glucosidase in complex with maltose. Deposited: 2018-02-16 Released: 2018-12-26. [CrossRef]
- Protein Preparation Wizard; Epik, Schrödinger, LLC: New York, NY, USA; Impact, Schrödinger, LLC: New York, NY, USA; Prime, Schrödinger, LLC: New York, NY, USA, 2022.
- Glide; Schrödinger, LLC: New York, NY, USA, 2022.
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Shaw, D.E.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Pistia-Brueggeman, G.; Hollingsworth, R.I. A preparation and screening strategy for glycosidase inhibitors. Tetrahedron 2001, 57, 8773–8778. [Google Scholar] [CrossRef]
- Sutradhar, S.; Deb, A.; Singh, S.S. Melatonin attenuates diabetes-induced oxidative stress in spleen and suppression of splenocyte proliferation in laboratory mice. Arch. Physiol. Biochem. 2022, 128, 1401–1412. [Google Scholar] [CrossRef]
- Poovitha, S.; Parani, M. In vitro and in vivo α-amylase and α-glucosidase inhibiting activities of the protein extracts from two varieties of bitter gourd (Momordica charantia L.). BMC Complement. Altern. Med. 2016, 16, 185. [Google Scholar] [CrossRef]
- Sinha, A.K. Colorimetric assay of catalase. Anal. Biochem. 1972, 47, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Hadwan, M.H. New method for assessment of serum catalase activity. Indian J. Sci. Technol. 2016, 9, 1–5. [Google Scholar] [CrossRef]
- Das, K.; Samanta, L.; Chainy, G.B.N. A modified spectrophotometric assay of superoxide dismutase using nitrite formation by superoxide radicals. Indian J. Biochem. Biophys. 2000, 37, 201–204. [Google Scholar]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. Swiss ADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V.A. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. Chem. Med. Chem. 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).