Transmission Dynamics and Novel Treatments of High Risk Carbapenem-Resistant Klebsiella pneumoniae: The Lens of One Health
Abstract
:1. Introduction
2. The Epidemiology of Klebsiella pneumoniae between Environment, Animals, and Humans
3. Antibiotic Resistance among Klebsiella pneumoniae
4. Evolution of KL 64 Carbapenem-Resistant Klebsiella pneumoniae in China
5. Evolution of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae
6. The Treatment of Klebsiella pneumoniae
6.1. Aztreonam
6.2. Polymyxins
6.3. Tigecycline
6.4. Ceftazidime-Avibactam
6.5. Cefiderocol
6.6. Nanoparticles
6.7. Phage Therapy
6.8. Antimicrobial Peptides
7. Vaccine
8. Infection Control
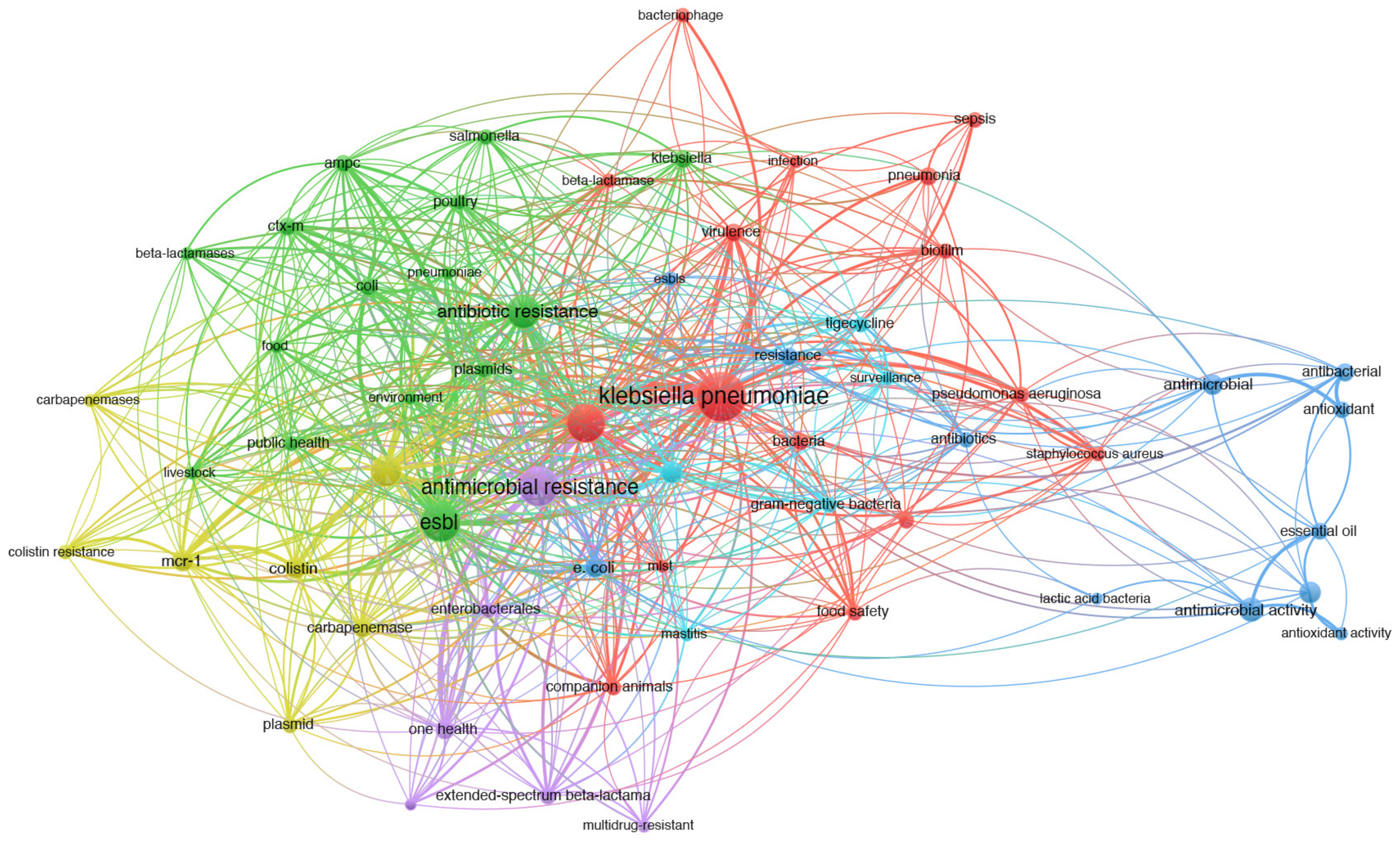
9. Co-Occurrence Analysis
- Colistin resistance. Given the indispensability of polymyxins in human therapy, the WHO accords them the utmost priority among antibiotics for curbing animal usage. The implementation of a colistin withdrawal strategy and subsequent decline in its agricultural application have significantly contributed to mitigating colistin resistance in both humans and animals. Despite limited knowledge on this matter, the persistence of mcr-1 and colistin resistance suggests that factors beyond animal colistin usage may influence CRKP epidemiology. Therefore, continuous monitoring of colistin is imperative to serve as an alert system for promoting prudent use in hospitals. Furthermore, the rational design of nano-antibiotics and phages emerges as a promising approach to combat the crisis posed by colistin resistance.
- One health. The concept of One Health is exemplified by the global impact of antibiotic usage in animals, highlighting the imperative for enhanced collaboration across environmental, animal (domestic and wildlife), and human health sectors. However, similar power imbalances and inefficiencies that persist in global health governance also influence the governance of One Health. To facilitate informed decision-making ensuring both food security and public health protection, integrated approaches are required to elucidate the relationship between antibiotic use in animals and the burden of antibiotic resistance in humans. We advocate for novel infection prevention and control programs, founded on the pillars of One Health, to reduce Gram-positive hospital-associated pathogen transmission.
- Microbiome. Technological advancements such as next-generation sequencing [50,61], RNA sequencing [60], bacterial droplet-based single-cell RNA-seq [169], and deep learning approaches [81] have enhanced our proficiency in identifying and investigating antibiotic resistance. These emerging technologies and methodologies complement established culture-based methods, enabling rapid and accurate determination of resistance in both cultivable and non-cultivable bacteria for clinical and surveillance applications. Enhanced in vitro sequencing methodologies and computational algorithms for genomic data organization and antibiotic resistance prediction are being employed to address the specific challenges associated with comprehending and evaluating genetic determinants of resistance using sequencing data. However, ensuring the accuracy of reference databases is imperative for all subsequent analyses. Therefore, the systematic annotation of recently identified genes and the avoidance of misinterpretations will greatly benefit both fundamental science and public health.
10. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riley, L.W. Extraintestinal Foodborne Pathogens. Annu. Rev. Food Sci. Technol. 2020, 11, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Drigo, B.; Brunetti, G.; Aleer, S.C.; Bell, J.M.; Short, M.D.; Vasileiadis, S.; Turnidge, J.; Monis, P.; Cunliffe, D.; Donner, E. Inactivation, removal, and regrowth potential of opportunistic pathogens and antimicrobial resistance genes in recycled water systems. Water Res. 2021, 201, 117324. [Google Scholar] [CrossRef] [PubMed]
- Chirabhundhu, N.; Luk-In, S.; Phuadraksa, T.; Wichit, S.; Chatsuwan, T.; Wannigama, D.L.; Yainoy, S. Occurrence and mechanisms of tigecycline resistance in carbapenem- and colistin-resistant Klebsiella pneumoniae in Thailand. Sci. Rep. 2024, 14, 5215. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives T H 1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef]
- Wang, M.; Earley, M.; Chen, L.; Hanson, B.M.; Yu, Y.; Liu, Z.; Salcedo, S.; Cober, E.; Li, L.; Kanj, S.S.; et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): A prospective, multicentre, cohort study. Lancet Infect. Dis. 2022, 22, 401–412. [Google Scholar] [CrossRef]
- Sabota, J.M.; Hoppes, W.L.; Ziegler, J.R.; DuPont, H.; Mathewson, J.; Rutecki, G.W. A new variant of food poisoning: Enteroinvasive Klebsiella pneumoniae and Escherichia coli sepsis from a contaminated hamburger. Am. J. Gastroenterol. 1998, 93, 118–119. [Google Scholar] [CrossRef]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Weaver, B.; Aziz, M.; Gauld, L.; Grande, H.; Bigler, R.; Horwinski, J.; Porter, S.; et al. Intermingled Klebsiella pneumoniae Populations Between Retail Meats and Human Urinary Tract Infections. Clin. Infect. Dis. 2015, 61, 892–899. [Google Scholar] [CrossRef]
- Calbo, E.; Freixas, N.; Xercavins, M.; Riera, M.; Nicolas, C.; Monistrol, O.; Sole, M.d.m.; Sala, M.R.; Vila, J.; Garau, J. Foodborne Nosocomial Outbreak of SHV1 and CTX-M-15-producing Klebsiella pneumoniae: Epidemiology and Control. Clin. Infect. Dis. 2011, 52, 743–749. [Google Scholar] [CrossRef]
- Rennie, R.P.; Anderson, C.M.; Wensley, B.G.; Albritton, W.L.; Mahony, D.E. Klebsiella pneumoniae gastroenteritis masked by Clostridium perfringens. J. Clin. Microbiol. 1990, 28, 216–219. [Google Scholar] [CrossRef]
- Houri, H.; Aghdaei, H.A.; Firuzabadi, S.; Khorsand, B.; Soltanpoor, F.; Rafieepoor, M.; Tanhaei, M.; Soleymani, G.; Azimirad, M.; Sadeghi, A.; et al. High Prevalence Rate of Microbial Contamination in Patient-Ready Gastrointestinal Endoscopes in Tehran, Iran: An Alarming Sign for the Occurrence of Severe Outbreaks. Microbiol. Spectr. 2022, 10, e0189722. [Google Scholar] [CrossRef]
- Thakur, S.; Gray, G.C. The Mandate for a Global “One Health” Approach to Antimicrobial Resistance Surveillance. Am. J. Trop. Med. Hyg. 2019, 100, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Mavingui, P.; Boetsch, G.; Boissier, J.; Darriet, F.; Duboz, P.; Fritsch, C.; Giraudoux, P.; Le Roux, F.; Morand, S.; et al. The One Health Concept: 10 Years Old and a Long Road Ahead. Front. Vet. Sci. 2018, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Okello, A.L.; Bardosh, K.; Smith, J.; Welburn, S.C. One Health: Past successes and future challenges in three African contexts. PLoS Negl. Trop. Dis. 2014, 8, e2884. [Google Scholar] [CrossRef] [PubMed]
- Siu, L.K.; Fung, C.-P.; Chang, F.-Y.; Lee, N.; Yeh, K.-M.; Koh, T.H.; Ip, M. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 2011, 49, 3761–3765. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, Q.; Li, X.; Kang, J.; Song, Y.; Song, J.; Yin, D.; Duan, J. Successful control of the first carbapenem-resistant Klebsiella pneumoniae outbreak in a Chinese hospital 2017–2019. Antimicrob. Resist. Infect. Control 2020, 9, 91. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Zhang, S.; Chen, N.; Sun, Y.; Ma, K.; Hong, D.; Li, L.; Du, Y.; Lu, X.; et al. TPGS-based and S-thanatin functionalized nanorods for overcoming drug resistance in Klebsiella pneumonia. Nat. Commun. 2022, 13, 3731. [Google Scholar] [CrossRef]
- Yang, X.; Sun, Q.; Li, J.; Jiang, Y.; Li, Y.; Lin, J.; Chen, K.; Chan, E.W.-C.; Zhang, R.; Chen, S. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg. Microbes Infect. 2022, 11, 841–849. [Google Scholar] [CrossRef]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef]
- Yang, J.; Liu, S.; Lu, J.; Sun, T.; Wang, P.; Zhang, X. An area under the concentration–time curve threshold as a predictor of efficacy and nephrotoxicity for individualizing polymyxin B dosing in patients with carbapenem-resistant gram-negative bacteria. Crit. Care 2022, 26, 320. [Google Scholar] [CrossRef]
- Ludden, C.; Moradigaravand, D.; Jamrozy, D.; Gouliouris, T.; Blane, B.; Naydenova, P.; Hernandez-Garcia, J.; Wood, P.; Hadjirin, N.; Radakovic, M.; et al. A One Health Study of the Genetic Relatedness of Klebsiella pneumoniae and Their Mobile Elements in the East of England. Clin. Infect. Dis. 2020, 70, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-T.; Zhang, X.-Y.; Wan, S.-W.; Hao, J.-J.; Jiang, R.-D.; Song, F.-J. Characteristics of Carbapenem-Resistant Enterobacteriaceae in Ready-to-Eat Vegetables in China. Front. Microbiol. 2018, 9, 1147. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Runcharoen, C.; Moradigaravand, D.; Blane, B.; Paksanont, S.; Thammachote, J.; Anun, S.; Parkhill, J.; Chantratita, N.; Peacock, S.J. Whole genome sequencing reveals high-resolution epidemiological links between clinical and environmental Klebsiella pneumoniae. Genome Med. 2017, 9, 6. [Google Scholar] [CrossRef]
- Bai, R.; Wang, X.; Zou, Z.; Zhou, W.; Tan, C.; Cao, Y.; Fu, B.; Zhai, W.; Hu, F.; Wang, Y.; et al. Limited transmission of carbapenem-resistant Klebsiella pneumoniae between animals and humans: A study in Qingdao. Emerg. Microbes Infect. 2024, 13, 2387446. [Google Scholar] [CrossRef]
- Richter, L.; du Plessis, E.M.; Duvenage, S.; Allam, M.; Ismail, A.; Korsten, L. Whole Genome Sequencing of Extended-Spectrum- and AmpC- β-Lactamase-Positive Enterobacterales Isolated From Spinach Production in Gauteng Province, South Africa. Front. Microbiol. 2021, 12, 734649. [Google Scholar] [CrossRef]
- Rodrigues, C.; Hauser, K.; Cahill, N.; Ligowska-Marzęta, M.; Centorotola, G.; Cornacchia, A.; Garcia Fierro, R.; Haenni, M.; Nielsen, E.M.; Piveteau, P.; et al. High Prevalence of Klebsiella pneumoniae in European Food Products: A Multicentric Study Comparing Culture and Molecular Detection Methods. Microbiol. Spectr. 2022, 10, e0237621. [Google Scholar] [CrossRef]
- Theocharidi, N.A.; Balta, I.; Houhoula, D.; Tsantes, A.G.; Lalliotis, G.P.; Polydera, A.C.; Stamatis, H.; Halvatsiotis, P. High Prevalence of Klebsiella pneumoniae in Greek Meat Products: Detection of Virulence and Antimicrobial Resistance Genes by Molecular Techniques. Foods 2022, 11, 708. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Wei, L.; Wu, L.; Wen, H.; Feng, Y.; Zhu, S.; Liu, Y.; Tang, L.; Doughty, E.; van Schaik, W.; McNally, A.; et al. Spread of Carbapenem-Resistant Klebsiella pneumoniae in an Intensive Care Unit: A Whole-Genome Sequence-Based Prospective Observational Study. Microbiol. Spectr. 2021, 9, e0005821. [Google Scholar] [CrossRef]
- Zenati, K.; Sahli, F.; Garcia, V.; Bakour, S.; Belhadi, D.; Rolain, J.M.; Touati, A. Occurrence and clonal diversity of multidrug-resistant Klebsiella pneumoniae recovered from inanimate surfaces in Algerian hospital environment: First report of armA, qnrB and aac(6′)-Ib-cr genes. J. Glob. Antimicrob. Resist. 2017, 10, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Jian, Z.; Liu, W.; Li, J.; Pei, N. One Health Analysis of mcr-Carrying Plasmids and Emergence of mcr-10.1 in Three Species of Klebsiella Recovered from Humans in China. Microbiol. Spectr. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Guo, M.; Yang, H.; Wen, Y.; Zhu, Z.; Wang, T.; Zhu, J.; Chen, L.; Du, H. blaKPC-24-Harboring Aeromonas veronii from the Hospital Sewage Samples in China. Microbiol. Spectr. 2022, 10, e0055522. [Google Scholar] [CrossRef]
- Ota, Y.; Prah, I.; Nukui, Y.; Koike, R.; Saito, R. blaKPC-2-Encoding IncP-6 Plasmids in Citrobacter freundii and Klebsiella variicola Strains from Hospital Sewage in Japan. Appl. Environ. Microbiol. 2022, 88, e0001922. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, R.; Wang, S.; Zhang, A.; Duan, Q.; Sun, S.; Jin, L.; Wang, X.; Zhang, Y.; Wang, C.; et al. Expansion and transmission dynamics of high risk carbapenem-resistant Klebsiella pneumoniae subclones in China: An epidemiological, spatial, genomic analysis. Drug Resist. Updat. 2024, 74, 101083. [Google Scholar] [CrossRef]
- Naas, T.; Nordmann, P.; Vedel, G.; Poyart, C. Plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 2005, 49, 4423–4424. [Google Scholar] [CrossRef]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef]
- Moellering, R.C. NDM-1—A Cause for Worldwide Concern. N. Engl. J. Med. 2010, 363, 2377–2379. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Karlowsky, J.A.; Lob, S.H.; Kazmierczak, K.M.; Badal, R.E.; Young, K.; Motyl, M.R.; Sahm, D.F. In Vitro Activity of Imipenem against Carbapenemase-Positive Enterobacteriaceae Isolates Collected by the SMART Global Surveillance Program from 2008 to 2014. J. Clin. Microbiol. 2017, 55, 1638–1649. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Fu, B.; Shi, X.; Sun, C.; Liu, Z.; Wang, S.; Shen, Z.; Walsh, T.R.; Cai, C.; Wang, Y.; et al. Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ. Int. 2020, 139, 105715. [Google Scholar] [CrossRef] [PubMed]
- Teban-Man, A.; Szekeres, E.; Fang, P.; Klümper, U.; Hegedus, A.; Baricz, A.; Berendonk, T.U.; Pârvu, M.; Coman, C. Municipal Wastewaters Carry Important Carbapenemase Genes Independent of Hospital Input and Can Mirror Clinical Resistance Patterns. Microbiol. Spectr. 2022, 10, e0271121. [Google Scholar] [CrossRef] [PubMed]
- Hooban, B.; Fitzhenry, K.; O’Connor, L.; Miliotis, G.; Joyce, A.; Chueiri, A.; Farrell, M.L.; DeLappe, N.; Tuohy, A.; Cormican, M.; et al. A Longitudinal Survey of Antibiotic-Resistant Enterobacterales in the Irish Environment, 2019–2020. Sci. Total Environ. 2022, 828, 154488. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.-A.; Matsumura, Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019, 33, e00102-19. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 2012, 56, 559–562. [Google Scholar] [CrossRef]
- Zhu, J.; Ju, Y.; Zhou, X.; Chen, T.; Zhuge, X.; Dai, J. Epidemiological characteristics of SHV, cmlv, and FosA6-producing carbapenem-resistant Klebsiella pneumoniae based on whole genome sequences in Jiangsu, China. Front. Microbiol. 2023, 14, 1219733. [Google Scholar] [CrossRef]
- Walker, M.K.; Diao, G.; Warner, S.; Babiker, A.; Neupane, M.; Strich, J.R.; Yek, C.; Kadri, S.S. Carbapenem use in extended-spectrum cephalosporin-resistant Enterobacterales infections in US hospitals and influence of IDSA guidance: A retrospective cohort study. Lancet Infect. Dis. 2024, 24, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Follador, R.; Heinz, E.; Wyres, K.L.; Ellington, M.J.; Kowarik, M.; Holt, K.E.; Thomson, N.R. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2016, 2, e000073. [Google Scholar] [CrossRef] [PubMed]
- Diago-Navarro, E.; Motley, M.P.; Ruiz-Peréz, G.; Yu, W.; Austin, J.; Seco, B.M.S.; Xiao, G.; Chikhalya, A.; Seeberger, P.H.; Fries, B.C. Novel, Broadly Reactive Anticapsular Antibodies against Carbapenem-Resistant Klebsiella pneumoniae Protect from Infection. MBio 2018, 9, e00091-18. [Google Scholar] [CrossRef] [PubMed]
- Diago-Navarro, E.; Calatayud-Baselga, I.; Sun, D.; Khairallah, C.; Mann, I.; Ulacia-Hernando, A.; Sheridan, B.; Shi, M.; Fries, B.C. Antibody-Based Immunotherapy To Treat and Prevent Infection with Hypervirulent Klebsiella pneumoniae. Clin. Vaccine Immunol. 2017, 24, e00456-16. [Google Scholar] [CrossRef] [PubMed]
- Malachowa, N.; Kobayashi, S.D.; Porter, A.R.; Freedman, B.; Hanley, P.W.; Lovaglio, J.; Saturday, G.A.; Gardner, D.J.; Scott, D.P.; Griffin, A.; et al. Vaccine Protection against Multidrug-Resistant Klebsiella pneumoniae in a Nonhuman Primate Model of Severe Lower Respiratory Tract Infection. MBio 2019, 10, e02994-19. [Google Scholar] [CrossRef]
- Hu, F.; Pan, Y.; Li, H.; Han, R.; Liu, X.; Ma, R.; Wu, Y.; Lun, H.; Qin, X.; Li, J.; et al. Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: A longitudinal, multi-centre study. Nat. Microbiol. 2024, 9, 814–829. [Google Scholar] [CrossRef]
- Wang, J.; Feng, Y.; Zong, Z. The Origins of ST11 KL64 Klebsiella pneumoniae: A Genome-Based Study. Microbiol. Spectr. 2023, 11, e0416522. [Google Scholar] [CrossRef]
- Chen, T.; Wang, Y.; Zhou, Y.; Zhou, W.; Chi, X.; Shen, P.; Zheng, B.; Xiao, Y. Recombination Drives Evolution of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 KL47 to KL64 in China. Microbiol. Spectr. 2023, 11, e0110722. [Google Scholar] [CrossRef]
- Guo, M.-Q.; Wang, Y.-T.; Wang, S.-S.; Chen, L.-K.; Xu, Y.-H.; Li, G. Genomic epidemiology of hypervirulent carbapenem-resistant Klebsiella pneumoniae at Jinshan local hospital, Shanghai, during 2014-2018. J. Microbiol. Immunol. Infect. 2024, 57, 128–137. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, A.; Sun, S.; Yin, G.; Wu, X.; Ding, Q.; Wang, Q.; Chen, F.; Wang, S.; van Dorp, L.; et al. Increase in antioxidant capacity associated with the successful subclone of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11-KL64. Nat. Commun. 2024, 15, 67. [Google Scholar] [CrossRef]
- Zhou, K.; Xue, C.X.; Xu, T.; Shen, P.; Wei, S.; Wyres, K.L.; Lam, M.M.C.; Liu, J.; Lin, H.; Chen, Y.; et al. A point mutation in recC associated with subclonal replacement of carbapenem-resistant Klebsiella pneumoniae ST11 in China. Nat. Commun. 2023, 14, 2464. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Dong, N.; Zheng, Z.; Lin, D.; Huang, M.; Wang, L.; Chan, E.W.-C.; Shu, L.; Yu, J.; Zhang, R.; et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 2018, 18, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Hu, D. The making of hypervirulent Klebsiella pneumoniae. J. Clin. Lab. Anal. 2022, 36, e24743. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Yang, X.; Chan, E.W.-C.; Zhang, R.; Chen, S. Klebsiella species: Taxonomy, hypervirulence and multidrug resistance. EBioMedicine 2022, 79, 103998. [Google Scholar] [CrossRef]
- Pu, D.; Zhao, J.; Chang, K.; Zhuo, X.; Cao, B. “Superbugs” with hypervirulence and carbapenem resistance in Klebsiella pneumoniae: The rise of such emerging nosocomial pathogens in China. Sci. Bull. 2023, 68, 2658–2670. [Google Scholar] [CrossRef]
- Wang, Y.; Dagan, T. The evolution of antibiotic resistance islands occurs within the framework of plasmid lineages. Nat. Commun. 2024, 15, 4555. [Google Scholar] [CrossRef]
- Beamud, B.; Benz, F.; Bikard, D. Going viral: The role of mobile genetic elements in bacterial immunity. Cell Host Microbe 2024, 32, 804–819. [Google Scholar] [CrossRef]
- Grodner, B.; Shi, H.; Farchione, O.; Vill, A.C.; Ntekas, I.; Diebold, P.J.; Wu, D.T.; Chen, C.-Y.; Kim, D.M.; Zipfel, W.R.; et al. Spatial mapping of mobile genetic elements and their bacterial hosts in complex microbiomes. Nat. Microbiol. 2024, 9, 2262–2277. [Google Scholar] [CrossRef]
- Li, R.; Cheng, J.; Dong, H.; Li, L.; Liu, W.; Zhang, C.; Feng, X.; Qin, S. Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents 2020, 55, 105952. [Google Scholar] [CrossRef]
- Yang, X.; Wai-Chi Chan, E.; Zhang, R.; Chen, S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat. Microbiol. 2019, 4, 2039–2043. [Google Scholar] [CrossRef]
- Liao, W.; Liu, Y.; Zhang, W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: A review over the last 10 years. J. Glob. Antimicrob. Resist. 2020, 23, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wyres, K.L.; Duchêne, S.; Wick, R.R.; Judd, L.M.; Gan, Y.-H.; Hoh, C.-H.; Archuleta, S.; Molton, J.S.; Kalimuddin, S.; et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 2018, 9, 2703. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Kong, X.; Hao, J.; Liu, J. Epidemiological Characteristics and Formation Mechanisms of Multidrug-Resistant Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2020, 11, 581543. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-H.; Huang, T.-W.; Juan, C.-H.; Chou, S.-H.; Tseng, Y.-Y.; Chen, T.-W.; Yang, T.-C.; Lin, Y.-T. Tigecycline-non-susceptible hypervirulent Klebsiella pneumoniae strains in Taiwan. J. Antimicrob. Chemother. 2019, 75, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Kochan, T.J.; Nozick, S.H.; Valdes, A.; Mitra, S.D.; Cheung, B.H.; Lebrun-Corbin, M.; Medernach, R.L.; Vessely, M.B.; Mills, J.O.; Axline, C.M.R.; et al. Klebsiella pneumoniae clinical isolates with features of both multidrug-resistance and hypervirulence have unexpectedly low virulence. Nat. Commun. 2023, 14, 7962. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Lu, M.-C.; Tang, H.-L.; Liu, H.-C.; Chen, C.-H.; Liu, K.-S.; Lin, C.; Chiou, C.-S.; Chiang, M.-K.; Chen, C.-M.; et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: Comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. 2011, 11, 50. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, L.; Ouyang, P.; Wang, Q.; Wang, R.; Wang, J.; Gao, H.; Wang, X.; Wang, H. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: A multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 2020, 75, 327–336. [Google Scholar] [CrossRef]
- Russo, T.A.; Olson, R.; Fang, C.-T.; Stoesser, N.; Miller, M.; MacDonald, U.; Hutson, A.; Barker, J.H.; La Hoz, R.M.; Johnson, J.R. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 2018, 56, e00776-18. [Google Scholar] [CrossRef]
- Kaur, J.N.; Singh, N.; Smith, N.M.; Klem, J.F.; Cha, R.; Lang, Y.; Chen, L.; Kreiswirth, B.; Holden, P.N.; Bulitta, J.B.; et al. Next generation antibiotic combinations to combat pan-drug resistant Klebsiella pneumoniae. Sci. Rep. 2024, 14, 3148. [Google Scholar] [CrossRef]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [PubMed]
- Shaw, E.; Rombauts, A.; Tubau, F.; Padullés, A.; Càmara, J.; Lozano, T.; Cobo-Sacristán, S.; Sabe, N.; Grau, I.; Rigo-Bonnin, R.; et al. Clinical outcomes after combination treatment with ceftazidime/avibactam and aztreonam for NDM-1/OXA-48/CTX-M-15-producing Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 2018, 73, 1104–1106. [Google Scholar] [CrossRef] [PubMed]
- Karlowsky, J.A.; Kazmierczak, K.M.; de Jonge, B.L.M.; Hackel, M.A.; Sahm, D.F.; Bradford, P.A. In Vitro Activity of Aztreonam-Avibactam against Enterobacteriaceae and Pseudomonas aeruginosa Isolated by Clinical Laboratories in 40 Countries from 2012 to 2015. Antimicrob. Agents Chemother. 2017, 61, e00472-17. [Google Scholar] [CrossRef] [PubMed]
- Doijad, S.P.; Gisch, N.; Frantz, R.; Kumbhar, B.V.; Falgenhauer, J.; Imirzalioglu, C.; Falgenhauer, L.; Mischnik, A.; Rupp, J.; Behnke, M.; et al. Resolving colistin resistance and heteroresistance in Enterobacter species. Nat. Commun. 2023, 14, 140. [Google Scholar] [CrossRef]
- Liu, X.; Chen, Y.; Yang, H.; Li, J.; Yu, J.; Yu, Z.; Cao, G.; Wu, X.; Wang, Y.; Wu, H.; et al. Acute toxicity is a dose-limiting factor for intravenous polymyxin B: A safety and pharmacokinetic study in healthy Chinese subjects. J. Infect. 2021, 82, 207–215. [Google Scholar] [CrossRef]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updat. 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.-Y.; Zhang, Y.; Liu, X.; Cui, Z.-H.; Ma, X.-Y.; Feng, Y.; Fang, L.-X.; Lian, X.-L.; et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, C.; Cheng, H.; Yang, X.; Huang, Z.; Chen, X.; Ju, Z.; Zhang, H.; Wang, H. Widespread Dissemination of Plasmid-Mediated Tigecycline Resistance Gene tet(X4) in Enterobacterales of Porcine Origin. Microbiol. Spectr. 2022, 10, e0161522. [Google Scholar] [CrossRef]
- Dai, S.; Liu, D.; Han, Z.; Wang, Y.; Lu, X.; Yang, M.; Zhang, Y. Mobile tigecycline resistance gene tet(X4) persists with different animal manure composting treatments and fertilizer receiving soils. Chemosphere 2022, 307, 135866. [Google Scholar] [CrossRef]
- Peng, Z.; Hu, Z.; Li, Z.; Zhang, X.; Jia, C.; Li, T.; Dai, M.; Tan, C.; Xu, Z.; Wu, B.; et al. Antimicrobial resistance and population genomics of multidrug-resistant Escherichia coli in pig farms in mainland China. Nat. Commun. 2022, 13, 1116. [Google Scholar] [CrossRef]
- Tilahun, M.; Kassa, Y.; Gedefie, A.; Ashagire, M. Emerging Carbapenem-Resistant Enterobacteriaceae Infection, Its Epidemiology and Novel Treatment Options: A Review. Infect. Drug Resist. 2021, 14, 4363–4374. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. A resurgence of β-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int. J. Antimicrob. Agents 2015, 46, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Brennan-Krohn, T.; Manetsch, R.; O’Doherty, G.A.; Kirby, J.E. New strategies and structural considerations in development of therapeutics for carbapenem-resistant Enterobacteriaceae. Transl. Res. 2020, 220, 14–32. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Lok, J.J.; Earley, M.; Cober, E.; Richter, S.S.; Perez, F.; Salata, R.A.; Kalayjian, R.C.; Watkins, R.R.; Doi, Y.; et al. Colistin Versus Ceftazidime-Avibactam in the Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae. Clin. Infect. Dis. 2018, 66, 163–171. [Google Scholar] [CrossRef]
- Tumbarello, M.; Raffaelli, F.; Giannella, M.; Mantengoli, E.; Mularoni, A.; Venditti, M.; De Rosa, F.G.; Sarmati, L.; Bassetti, M.; Brindicci, G.; et al. Ceftazidime-Avibactam Use for Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae Infections: A Retrospective Observational Multicenter Study. Clin. Infect. Dis. 2021, 73, 1664–1676. [Google Scholar] [CrossRef]
- Wong, J.L.C.; Romano, M.; Kerry, L.E.; Kwong, H.S.; Low, W.W.; Brett, S.J.; Clements, A.; Beis, K.; Frankel, G. OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat. Commun. 2019, 10, 3957. [Google Scholar] [CrossRef]
- Di Pilato, V.; Principe, L.; Andriani, L.; Aiezza, N.; Coppi, M.; Ricci, S.; Giani, T.; Luzzaro, F.; Rossolini, G.M. Deciphering variable resistance to novel carbapenem-based β-lactamase inhibitor combinations in a multi-clonal outbreak caused by Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam. Clin. Microbiol. Infect. 2023, 29, 537.e1–537.e8. [Google Scholar] [CrossRef]
- Shields, R.K.; Chen, L.; Cheng, S.; Chavda, K.D.; Press, E.G.; Snyder, A.; Pandey, R.; Doi, Y.; Kreiswirth, B.N.; Nguyen, M.H.; et al. Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne blaKPC-3 Mutations during Treatment of Carbapenem-Resistant Klebsiella pneumoniae Infections. Antimicrob. Agents Chemother. 2017, 61, e02097-16. [Google Scholar] [CrossRef]
- Nicola, F.; Cejas, D.; González-Espinosa, F.; Relloso, S.; Herrera, F.; Bonvehí, P.; Smayevsky, J.; Figueroa-Espinosa, R.; Gutkind, G.; Radice, M. Outbreak of Klebsiella pneumoniae ST11 Resistant To Ceftazidime-Avibactam Producing KPC-31 and the Novel Variant KPC-115 during COVID-19 Pandemic in Argentina. Microbiol. Spectr. 2022, 10, e0373322. [Google Scholar] [CrossRef]
- Bassetti, M.; Echols, R.; Matsunaga, Y.; Ariyasu, M.; Doi, Y.; Ferrer, R.; Lodise, T.P.; Naas, T.; Niki, Y.; Paterson, D.L.; et al. Efficacy and safety of cefiderocol or best available therapy for the treatment of serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): A randomised, open-label, multicentre, pathogen-focused, descriptive, phase 3 trial. Lancet Infect. Dis. 2021, 21, 226–240. [Google Scholar] [CrossRef]
- Liu, C.; Yi, J.; Lu, M.; Yang, P.; Du, C.; Jiang, F.; Du, P.; Shen, N. Dynamic within-host cefiderocol heteroresistance caused by blaSHV-12 amplification in pandrug-resistant and hypervirulent Klebsiella pneumoniae sequence type 11. Drug Resist. Updat. 2024, 73, 101038. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Mesini, A.; Mariani, M.; Tavella, E.; Sette, C.; Ugolotti, E.; Bartalucci, C.; Palmero, C.; Bandettini, R.; Castagnola, E. Reduce susceptibility to cefiderocol in gram negative bacteria in children: Is hope already lost before it’s even arrived? J. Infect. Public Health 2024, 17, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Martín, T.; Alonso-García, I.; González-Pinto, L.; Outeda-García, M.; Guijarro-Sánchez, P.; López-Hernández, I.; Pérez-Vázquez, M.; Aracil, B.; López-Cerero, L.; Fraile-Ribot, P.; et al. Activity of cefiderocol and innovative β-lactam/β-lactamase inhibitor combinations against isogenic strains of Escherichia coli expressing single and double β-lactamases under high and low permeability conditions. Int. J. Antimicrob. Agents 2024, 63, 107150. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Gao, M.; Yun, Y.; Malmsten, M.; Rotello, V.M.; Zboril, R.; Akhavan, O.; Kraskouski, A.; Amalraj, J.; Cai, X.; et al. Antibacterial Nanomaterials: Mechanisms, Impacts on Antimicrobial Resistance and Design Principles. Angew. Chem. Int. Ed. 2023, 62, e202217345. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, W.; Wei, G.; Liu, Q.; Shao, G.; Gu, X.; Cui, X.; Zhou, Z.; Wang, Y.; Zhao, S.; et al. Bioinspired Nanozymes as Nanodecoys for Urinary Tract Infection Treatment. ACS Nano 2024, 18, 9019–9030. [Google Scholar] [CrossRef]
- He, X.; Lv, Y.; Lin, Y.; Yu, H.; Zhang, Y.; Tong, Y.; Zhang, C. Platinum Nanoparticles Regulated V2C MXene Nanoplatforms with NIR-II Enhanced Nanozyme Effect for Photothermal and Chemodynamic Anti-Infective Therapy. Adv. Mater. 2024, 36, e2400366. [Google Scholar] [CrossRef]
- Jiang, P.; Zhang, L.; Liu, X.; Ye, C.; Zhu, P.; Tan, T.; Wang, D.; Wang, Y. Tuning oxidant and antioxidant activities of ceria by anchoring copper single-site for antibacterial application. Nat. Commun. 2024, 15, 1010. [Google Scholar] [CrossRef]
- Pan, Y.; Zheng, H.; Li, G.; Li, Y.; Jiang, J.; Chen, J.; Xie, Q.; Wu, D.; Ma, R.; Liu, X.; et al. Antibiotic-Like Activity of Atomic Layer Boron Nitride for Combating Resistant Bacteria. ACS Nano 2022, 16, 7674–7688. [Google Scholar] [CrossRef]
- Alizadeh, N.; Salimi, A. Multienzymes activity of metals and metal oxide nanomaterials: Applications from biotechnology to medicine and environmental engineering. J. Nanobiotechnol. 2021, 19, 26. [Google Scholar] [CrossRef]
- Stabryla, L.M.; Johnston, K.A.; Diemler, N.A.; Cooper, V.S.; Millstone, J.E.; Haig, S.-J.; Gilbertson, L.M. Role of bacterial motility in differential resistance mechanisms of silver nanoparticles and silver ions. Nat. Nanotechnol. 2021, 16, 996–1003. [Google Scholar] [CrossRef]
- Radzig, M.A.; Nadtochenko, V.A.; Koksharova, O.A.; Kiwi, J.; Lipasova, V.A.; Khmel, I.A. Antibacterial effects of silver nanoparticles on gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action. Colloids Surf. B Biointerfaces 2013, 102, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Chen, F.; Yue, L.; Luo, Y.; Wang, Z.; Xing, B. CeO(2) Nanoparticles Regulate the Propagation of Antibiotic Resistance Genes by Altering Cellular Contact and Plasmid Transfer. Environ. Sci. Technol. 2020, 54, 10012–10021. [Google Scholar] [CrossRef] [PubMed]
- Zha, Y.; Li, Z.; Zhong, Z.; Ruan, Y.; Sun, L.; Zuo, F.; Li, L.; Hou, S. Size-dependent enhancement on conjugative transfer of antibiotic resistance genes by micro/nanoplastics. J. Hazard. Mater. 2022, 431, 128561. [Google Scholar] [CrossRef] [PubMed]
- Panáček, A.; Kvítek, L.; Smékalová, M.; Večeřová, R.; Kolář, M.; Röderová, M.; Dyčka, F.; Šebela, M.; Prucek, R.; Tomanec, O.; et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 2018, 13, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Xia, X.; Xia, N.; Zhang, S.; Guo, X. Modification of Fatty acids in membranes of bacteria: Implication for an adaptive mechanism to the toxicity of carbon nanotubes. Environ. Sci. Technol. 2014, 48, 4086–4095. [Google Scholar] [CrossRef]
- Natan, M.; Banin, E. From Nano to Micro: Using nanotechnology to combat microorganisms and their multidrug resistance. FEMS Microbiol. Rev. 2017, 41, 302–322. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Maffei, E.; Woischnig, A.K.; Burkolter, M.R.; Heyer, Y.; Humolli, D.; Thürkauf, N.; Bock, T.; Schmidt, A.; Manfredi, P.; Egli, A.; et al. Phage Paride can kill dormant, antibiotic-tolerant cells of Pseudomonas aeruginosa by direct lytic replication. Nat. Commun. 2024, 15, 175. [Google Scholar] [CrossRef]
- Reardon, S. Phage therapy gets revitalized. Nature 2014, 510, 15–16. [Google Scholar] [CrossRef]
- Onsea, J.; Uyttebroek, S.; Chen, B.; Wagemans, J.; Lood, C.; Van Gerven, L.; Spriet, I.; Devolder, D.; Debaveye, Y.; Depypere, M.; et al. Bacteriophage Therapy for Difficult-to-Treat Infections: The Implementation of a Multidisciplinary Phage Task Force (The PHAGEFORCE Study Protocol). Viruses 2021, 13, 1543. [Google Scholar] [CrossRef]
- Jędrusiak, A.; Fortuna, W.; Majewska, J.; Górski, A.; Jończyk-Matysiak, E. Phage Interactions with the Nervous System in Health and Disease. Cells 2023, 12, 1720. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Young, R.; Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Caflisch, K.M.; Suh, G.A.; Patel, R. Biological challenges of phage therapy and proposed solutions: A literature review. Expert Rev. Anti-Infect. Ther. 2019, 17, 1011–1041. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Loh, B.; Gordillo Altamirano, F.; Yu, Y.; Hua, X.; Leptihn, S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 2021, 10, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.Y.K.; Nang, S.C.; Chan, H.-K.; Li, J. Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv. Drug Deliv. Rev. 2022, 187, 114378. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Advances in Bacteriophage Therapy against Relevant MultiDrug-Resistant Pathogens. Antibiotics 2021, 10, 672. [Google Scholar] [CrossRef]
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, A.; et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022, 185, 2879–2898.e24. [Google Scholar] [CrossRef]
- Hesse, S.; Malachowa, N.; Porter, A.R.; Freedman, B.; Kobayashi, S.D.; Gardner, D.J.; Scott, D.P.; Adhya, S.; DeLeo, F.R. Bacteriophage Treatment Rescues Mice Infected with Multidrug-Resistant Klebsiella pneumoniae ST258. MBio 2021, 12, e00034-21. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, J.; Guo, C.; Ge, C.; Wang, X.; Wei, D.; Li, X.; Si, H.; Hu, C. Identification and complete genome of lytic “Kp34likevirus” phage vB_KpnP_Bp5 and therapeutic potency in the treatment of lethal Klebsiella pneumoniae infections in mice. Virus Res. 2021, 297, 198348. [Google Scholar] [CrossRef]
- Corbellino, M.; Kieffer, N.; Kutateladze, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Tsertsvadze, G.; Rimoldi, S.G.; Nizharadze, D.; Hoyle, N.; et al. Eradication of a Multidrug-Resistant, Carbapenemase-Producing Klebsiella pneumoniae Isolate Following Oral and Intra-rectal Therapy With a Custom Made, Lytic Bacteriophage Preparation. Clin. Infect. Dis. 2019, 70, 1998–2001. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Wu, N.; Bao, J.; Shi, X.; Ou, H.; Ye, S.; Zhao, W.; Wei, Z.; Cai, J.; Li, L.; et al. Heterogeneous Klebsiella pneumoniae Co-infections Complicate Personalized Bacteriophage Therapy. Front. Cell. Infect. Microbiol. 2021, 10, 608402. [Google Scholar] [CrossRef] [PubMed]
- Yacoby, I.; Shamis, M.; Bar, H.; Shabat, D.; Benhar, I. Targeting antibacterial agents by using drug-carrying filamentous bacteriophages. Antimicrob. Agents Chemother. 2006, 50, 2087–2097. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Tzipilevich, E.; Habusha, M.; Ben-Yehuda, S. Acquisition of Phage Sensitivity by Bacteria through Exchange of Phage Receptors. Cell 2017, 168, 186–199.e12. [Google Scholar] [CrossRef]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef]
- Schooley, R.T.; Strathdee, S. Treat phage like living antibiotics. Nat. Microbiol. 2020, 5, 391–392. [Google Scholar] [CrossRef]
- Adler, B.A.; Hessler, T.; Cress, B.F.; Lahiri, A.; Mutalik, V.K.; Barrangou, R.; Banfield, J.; Doudna, J.A. Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing. Nat. Microbiol. 2022, 7, 1967–1979. [Google Scholar] [CrossRef]
- Nick, J.A.; Dedrick, R.M.; Gray, A.L.; Vladar, E.K.; Smith, B.E.; Freeman, K.G.; Malcolm, K.C.; Epperson, L.E.; Hasan, N.A.; Hendrix, J.; et al. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection. Cell 2022, 185, 1860–1874.e12. [Google Scholar] [CrossRef]
- Guan, J.; Oromí-Bosch, A.; Mendoza, S.D.; Karambelkar, S.; Berry, J.D.; Bondy-Denomy, J. Bacteriophage genome engineering with CRISPR–Cas13a. Nat. Microbiol. 2022, 7, 1956–1966. [Google Scholar] [CrossRef]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef] [PubMed]
- Hohn, B.; Murray, K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc. Natl. Acad. Sci. USA 1977, 74, 3259–3263. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.; Diamond, S.; Cress, B.F.; Crits-Christoph, A.; Lou, Y.C.; Borges, A.L.; Shivram, H.; He, C.; Xu, M.; Zhou, Z.; et al. Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 2022, 7, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-free gene expression: An expanded repertoire of applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef]
- Würstle, S.; Lee, A.; Kortright, K.E.; Winzig, F.; An, W.; Stanley, G.L.; Rajagopalan, G.; Harris, Z.; Sun, Y.; Hu, B.; et al. Optimized preparation pipeline for emergency phage therapy against Pseudomonas aeruginosa at Yale University. Sci. Rep. 2024, 14, 2657. [Google Scholar] [CrossRef]
- Wang, G. Database-Guided Discovery of Potent Peptides to Combat HIV-1 or Superbugs. Pharmaceuticals 2013, 6, 728–758. [Google Scholar] [CrossRef]
- Abbas, M.; Ovais, M.; Atiq, A.; Ansari, T.M.; Xing, R.; Spruijt, E.; Yan, X. Tailoring supramolecular short peptide nanomaterials for antibacterial applications. Coord. Chem. Rev. 2022, 460, 214481. [Google Scholar] [CrossRef]
- Kolano, L.; Knappe, D.; Volke, D.; Sträter, N.; Hoffmann, R. Ribosomal Target-Binding Sites of Antimicrobial Peptides Api137 and Onc112 Are Conserved among Pathogens Indicating New Lead Structures To Develop Novel Broad-Spectrum Antibiotics. Chembiochem 2020, 21, 2628–2634. [Google Scholar] [CrossRef]
- van der Weide, H.; Vermeulen-de Jongh, D.M.C.; van der Meijden, A.; Boers, S.A.; Kreft, D.; ten Kate, M.T.; Falciani, C.; Pini, A.; Strandh, M.; Bakker-Woudenberg, I.A.J.M.; et al. Antimicrobial activity of two novel antimicrobial peptides AA139 and SET-M33 against clinically and genotypically diverse Klebsiella pneumoniae isolates with differing antibiotic resistance profiles. Int. J. Antimicrob. Agents 2019, 54, 159–166. [Google Scholar] [CrossRef]
- Sugrue, I.; Ross, R.P.; Hill, C. Bacteriocin diversity, function, discovery and application as antimicrobials. Nat. Rev. Microbiol. 2024, 22, 556–571. [Google Scholar] [CrossRef] [PubMed]
- Santos-Júnior, C.D.; Torres, M.D.T.; Duan, Y.; Rodríguez del Río, Á.; Schmidt, T.S.B.; Chong, H.; Fullam, A.; Kuhn, M.; Zhu, C.; Houseman, A.; et al. Discovery of antimicrobial peptides in the global microbiome with machine learning. Cell 2024, 187, 3761–3778.e16. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.M.; da Silva, Á.P.; Júnior, N.G.O.; Martínez, O.F.; Franco, O.L. Peptides as a therapeutic strategy against Klebsiella pneumoniae. Trends Pharmacol. Sci. 2022, 43, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Sun, Q.; Xiao, D.; Zhang, M.; Gao, S.; Guo, B.; Lin, Y. Tetrahedral framework nucleic acids/hyaluronic acid-methacrylic anhydride hybrid hydrogel with antimicrobial and anti-inflammatory properties for infected wound healing. Int. J. Oral Sci. 2024, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Cong, Z.; Yan, Z.; Xiao, X.; Liu, L.; Luo, Z.; Zou, J.; Chen, M.; Wu, Y.; Zhou, M.; Liu, R. Peptide-mimicking poly(2-oxazoline) displaying potent antibacterial and antibiofilm activities against multidrug-resistant Gram-positive pathogenic bacteria. J. Mater. Sci. Technol. 2024, 214, 233–244. [Google Scholar] [CrossRef]
- Kidd, T.J.; Mills, G.; Sá-Pessoa, J.; Dumigan, A.; Frank, C.G.; Insua, J.L.; Ingram, R.; Hobley, L.; Bengoechea, J.A. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 2017, 9, 430–447. [Google Scholar] [CrossRef]
- Yu, W.; Guo, X.; Li, Q.; Li, X.; Wei, Y.; Shao, C.; Zhang, L.; Wang, J.; Shan, A. Revolutionizing Antimicrobial Biomaterials: Integrating an Enzyme Degradation-Resistant Sequence into Self-Assembled Nanosystems to Overcome Stability Limitations of Peptide-Based Drugs. Adv. Fiber Mater. 2024, 6, 1188–1211. [Google Scholar] [CrossRef]
- Wantuch, P.L.; Knoot, C.J.; Robinson, L.S.; Vinogradov, E.; Scott, N.E.; Harding, C.M.; Rosen, D.A. Heptavalent O-Antigen Bioconjugate Vaccine Exhibiting Differential Functional Antibody Responses Against Diverse Klebsiella pneumoniae Isolates. J. Infect. Dis. 2024, jiae097. [Google Scholar] [CrossRef]
- Gorden, P.J.; Kleinhenz, M.D.; Ydstie, J.A.; Brick, T.A.; Slinden, L.M.; Peterson, M.P.; Straub, D.E.; Burkhardt, D.T. Efficacy of vaccination with a Klebsiella pneumoniae siderophore receptor protein vaccine for reduction of Klebsiella mastitis in lactating cattle. J. Dairy Sci. 2018, 101, 10398–10408. [Google Scholar] [CrossRef]
- Li, W.; Hu, Y.; Zhang, Q.; Hua, L.; Yang, Z.; Ren, Z.; Zheng, X.; Huang, W.; Ma, Y. Development of Drug-Resistant Klebsiella pneumoniae Vaccine via Novel Vesicle Production Technology. ACS Appl. Mater. Interfaces 2021, 13, 32703–32715. [Google Scholar] [CrossRef]
- Serrano, E.; Demanez, J.P.; Morgon, A.; Chastang, C.; Van Cauwenberge, P. Effectiveness of ribosomal fractions of Klebsiella pneumoniae, Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae and the membrane fraction ofKp (Ribomunyl) in the prevention of clinical recurrences of infectious rhinitis. Eur. Arch. Oto-Rhino-Laryngol. 1997, 254, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Naveed, M.; Jabeen, K.; Aziz, T.; Mughual, M.S.; Ul-Hassan, J.; Sheraz, M.; Rehman, H.M.; Alharbi, M.; Albekairi, T.H.; Alasmari, A.F. Whole proteome analysis of MDR Klebsiella pneumoniae to identify mRNA and multiple epitope based vaccine targets against emerging nosocomial and lungs associated infections. J. Biomol. Struct. Dyn. 2023, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Zhang, X.; Zeng, X.; Zhao, Z.; Sun, T.; Xia, Z.; Jing, H.; Yuan, Y.; Chen, Z.; Gou, Q.; et al. A rational designed multi-epitope vaccine elicited robust protective efficacy against Klebsiella pneumoniae lung infection. Biomed. Pharmacother. 2024, 174, 116611. [Google Scholar] [CrossRef] [PubMed]
- Douradinha, B. Exploring the journey: A comprehensive review of vaccine development against Klebsiella pneumoniae. Microbiol. Res. 2024, 287, 127837. [Google Scholar] [CrossRef]
- Assoni, L.; Couto, A.J.M.; Vieira, B.; Milani, B.; Lima, A.S.; Converso, T.R.; Darrieux, M. Animal models of Klebsiella pneumoniae mucosal infections. Front. Microbiol. 2024, 15, 1367422. [Google Scholar] [CrossRef]
- Chang, D.; Sharma, L.; Dela Cruz, C.S.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef]
- Chen, T.; Zhu, J.; Wang, G.; Sun, J.; Ma, X.; Tian, L.; Zhang, M.; Wang, F.; Yu, Z. The global state of research in stem cells therapy for spinal cord injury (2003–2022): A visualized analysis. Front. Neurosci. 2024, 18, 1323383. [Google Scholar] [CrossRef]
- Chen, T.; Zhu, J.; Zhao, Y.; Li, H.; Li, P.; Fan, J.; Wei, X. The global state of research in pain management of osteoarthritis (2000-2019): A 20-year visualized analysis. Medicine 2021, 100, E23944. [Google Scholar] [CrossRef]
- Ma, P.; Amemiya, H.M.; He, L.L.; Gandhi, S.J.; Nicol, R.; Bhattacharyya, R.P.; Smillie, C.S.; Hung, D.T. Bacterial droplet-based single-cell RNA-seq reveals antibiotic-associated heterogeneous cellular states. Cell 2023, 186, 877–891.e14. [Google Scholar] [CrossRef]


| Agent | Class A | Class B | Class D | Mechanisms |
|---|---|---|---|---|
| Aztreonam | − | + | + | Inhibits bacterial cell wall synthesis |
| Fosfomcymin | + | + | + | Inhibits bacterial cell wall synthesis |
| Colistin, Polymyxin B | +/− | +/− | +/− | Destruction of bacterial cell membrane |
| Tigecycline | +/− | +/− | +/− | Inhibits protein synthesis |
| Ceftazidime- avibactam | + | - | + | β-lactam-β-lactamase inhibitor combinations |
| Meropenem- vaborbactam | + | - | - | β-lactam-β-lactamase inhibitor combinations |
| Imipenem- relebactam | + | - | - | Non-β-lactam bicyclic DBO β-lactamase inhibitor |
| Cefiderocol | + | + | + | “Trojan horse” straegy |
| Plazomicin | + | + | + | Inhibits protein synthesis |
| Nacubactam | + | +/− | + | β-lactamase inhibitor |
| Zidebactam | + | +/− | + | β-lactamase inhibitor |
| Taniborbactam | + | + | + | β-lactamase inhibitor |
| Phage therapy | + | + | + | Use of host machinery for their replication |
| Nanoparticles | + | + | + | Destruction of bacterial cell membrane; disruption electron transport chain; catalytic killing; ionic killing; cell division arrest |
| Antimicrobial peptide | + | + | + | Interacts with LPS and/or phospholipids /DNA/capsule/protein |
| Vaccine | + | + | + | Humoral response |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Chen, T.; Ju, Y.; Dai, J.; Zhuge, X. Transmission Dynamics and Novel Treatments of High Risk Carbapenem-Resistant Klebsiella pneumoniae: The Lens of One Health. Pharmaceuticals 2024, 17, 1206. https://doi.org/10.3390/ph17091206
Zhu J, Chen T, Ju Y, Dai J, Zhuge X. Transmission Dynamics and Novel Treatments of High Risk Carbapenem-Resistant Klebsiella pneumoniae: The Lens of One Health. Pharmaceuticals. 2024; 17(9):1206. https://doi.org/10.3390/ph17091206
Chicago/Turabian StyleZhu, Jiaying, Taoyu Chen, Yanmin Ju, Jianjun Dai, and Xiangkai Zhuge. 2024. "Transmission Dynamics and Novel Treatments of High Risk Carbapenem-Resistant Klebsiella pneumoniae: The Lens of One Health" Pharmaceuticals 17, no. 9: 1206. https://doi.org/10.3390/ph17091206







